Abstract
Background: It is recognized that Candida dubliniensis commonly colonizes oral and subgingival sites in immunocompetent subjects with periodontal disease.
Objective: Since there are few data available on genetic characterization of C. dubliniensis in periodontal pockets and other oral sites, the aim of this study was to characterize subgingival and mucosal C. dubliniensis isolates recovered from immunocompetent subjects and to assay the genetic similarity of such isolates from both niches in the same patient by random amplified polymorphic DNA (RAPD).
Design: C. dubliniensis recovered from subgingival plaque and from buccal cavity samples were studied in 240 immunocompetent non-smoking individuals. Arbitrary amplification was carried out by RAPD-polymerase chain reaction (PCR).
Results: RAPD analysis showed identical genotypes of C. dubliniensis in different sampling sites (buccal cavity and subgingival areas) in eight of 10 patients except for those derived from two participants who presented presumably unrelated isolates.
Conclusions: On the basis of the findings presented, the origin of the colonization of C. dubliniensis in subgingival biofilm seems to be the buccal cavity in a single patient. Consequently, it may be assumed that most of C. dubliniensis in these sites arise from the endogenous commensal strains.
Yeasts of the Candida genus are opportunistic commensals in the human mouth, representing potential sources of oropharyngeal candidiasis, as well as other more serious forms of the disease, such as esophageal and systemic diseases Citation1Citation2. Candida species have also been recovered from periodontal pockets in a large number (7.1–19.6%) of patients with periodontal disease Citation3Citation4Citation5Citation6Citation7Citation8.
Although the yeast most frequently associated to this type of infection is Candida albicans, other less prevalent emerging species have been isolated Citation4Citation6Citation7Citation9Citation10Citation11Citation12.
Mucosal surfaces are the primary body reservoirs for these microorganisms Citation13. This reflects the ability of the yeast to colonize different oral surfaces and the variety of factors which predispose the host to Candida colonization and subsequent infection. Colonization of the oral cavity appears to be facilitated by several specific adherence interactions between Candida species and oral surfaces which enable the yeast to resist host clearance mechanisms. Mucosal and subgingival Candida dubliniensis colonization has been reported in immunocompetent subjects with periodontal disease Citation6Citation7Citation14 and both oral sites are appropriate niches for multiplication of this species.
It would seem relevant to know about epidemiological aspects of subgingival C. dubliniensis, given its capacity to adhere to bacteria in the oral microbiota, and to coaggregate, that enables it to colonize the depth of subgingival biofilm Citation4Citation11Citation15Citation16.
In recent years, several molecular typing methods have been used to characterize Candida species isolates and to delineate strain relatedness, although polymerase chain reaction (PCR) based methods remain the most widely used. Among these, the random amplified polymorphic DNA (RAPD) method of DNA fingerprinting has become quite popular for all infectious fungi and has been successfully applied to assess the genetic relatedness of Candida species Citation17Citation18Citation19Citation20Citation21Citation22Citation23Citation24. These methods have greatly enhanced knowledge about the epidemiology of oral and subgingival Candida species, and they can provide valuable information by their capacity to distinguish distinct isolates of the same species. Some studies have demonstrated that commensal yeasts dominate in oral candidiasis, whereas controversial evidence shows that genetically homogeneous, hypervirulent strains of C. albicans are involved in the disease Citation25Citation26Citation27Citation28.
Since there are few data available on genetic characterization of C. dubliniensis in periodontal pockets and other oral sites, the aim of this study was to characterize subgingival and mucosal C. dubliniensis isolates recovered from immunocompetent subjects and to assay the genetic similarity of C. dubliniensis isolates from both niches in the same patient by RAPD.
Material and methods
Sources of isolates
A total of 21 isolates of C. dubliniensis, 10 from the buccal cavity and 11 from subgingival biofilm, were collected from 240 non-smoking individuals who attended the dental clinic of the University of Buenos Aires, Argentina. Periodontal evaluations included clinical examination and radiographs with clinical measurements: clinical attachment level (CAL), pocket depth (PD), plaque index Citation29, gingival index Citation30, and bleeding on probing (BOP). Location of the gingival margin was determined and tooth mobility was assessed. Measurements were made at four sites per tooth (mesial, buccal, distal, and lingual positions) at 15 teeth excluding the third molar.
Participation in our survey was voluntary and all the patients provided a written informed consent.
Sampling, culture conditions, and identification of isolates
All the volunteers were requested to thoroughly rinse their mouths with sterile distilled water. The dental professional then isolated the area to be sampled by means of cotton rolls and a high-speed suction device. Following removal of the supragingival plaque, in order to avoid contamination, four subgingival plaque samples were taken from each patient: the upper right and lower central incisor, the first upper right molar and the first lower left molar, to mimic the more common periodontal pockets infected in periodontitis, by using a 7/8 sterile Gracey curette. Samples were pooled and cultured on a differential chromogenic medium (CHROMagar Candida, Paris, France). Buccal cavity samples, including palatal, buccal mucosa, and tongue, were collected by sterile cotton-tipped swabs and were streaked directly onto the chromogenic medium. Isolated yeasts were identified by conventional mycological methods: colony color on the chromogenic medium, micromorphology in agar milk with 1% Tween 80 Citation31, and carbohydrate assimilation tests using the commercially available kit API ID 32C (®BioMérieux, France). Further studies were conducted for characterization of C. dubliniensis, including chlamydospore formation on Staib agar after incubation for 72 h at 28°C Citation32 and specific PCR with primers from actin gene (ACT)-associated intron sequences of C. dubliniensis, DUBF (GTATTTGTCCTTCCCCTTTTC) and DUBR (GTGTTGTGTGCACTAACGTC) Citation7Citation33.
Random amplified polymorphic DNA (RAPD) analysis
Yeast DNA was isolated according to a previously described technique Citation7Citation34Citation35. The DNA was quantified and its purity was evaluated at 260 nm (SmartSpecTM 3000 Spectrophotometer BIO-RAD).
Five different primers were included in the typing assays. Primer sequences were as follows: OPA 02 (TGCCGAGCTG), OPA 09 (GGGTAACGCC), M13F (CGACGTTGTAAAACGACGCCCAGT), M13R (CAGGAAACAGCTATGAC), and OCP 5 (GATGACCGCC). All were used in RAPD-PCR following the method of Williams et al. Citation36.
Arbitrary amplification was performed in a total volume of 50 µl containing: 1×buffer 2.5 mM MgCl2, 0.2 mM each of the dNTP, 0.5 µM of the primer, 1.25 U Taq DNA polymerase (Invitrogen), and 75 ng of template DNA. The cycling program was made up of 4 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 25°C, 2 min at 72°C followed by a final extension of 5 min at 72°C. These steps were carried out in a Minicycler DNA thermal cycler (TM MJ Research Inc., NY, USA). Products were separated by electrophoresis in 2% agarose gel and stained with ethidium bromide. They were visualized under UV light and digitalized by the image analyzer software (EPI-Chemi Darkroom. UVP Laboratory Products, California, USA). Band profiles were analyzed and compared visually. Each band was scored as positive or negative for all isolates, and for each isolate, the presence or absence of each band was registered. The resulting matrix was interpreted using the Treecon program, where isolates were grouped according to their pattern resemblance. Based on matrix of similarity coefficients (SC), a dendrogram was generated by the unweighted pair group method using arithmetic averages (UPGMA). The criterion used for genotyping was as follows: arbitrary threshold at an SC of 90% for high relatedness isolates Citation20.
Statistical analysis
Statistical analysis was performed by using Statistix 7.0 and SPSS 11.0 versions. Confidence intervals (CI) were calculated at 95% employing the Epi-Info 6.04 program (Atlanta University, USA).
Results
Clinical features
The 240 subjects included in the study ranged in age from 18 to 75 years (mean age 37), 55% were females (132/240) and had not received any antibacterial or antifungal agents before sampling. shows clinical periodontal parameters (PD and CAL Mean±SD) of subjects at the time of sampling. Patients were classified into groups according to the periodontal health status as shown in : healthy (n=53), gingivitis (n=58), and chronic periodontitis (n=129). Gingivitis and chronic periodontitis patients were BOP positive.
Table 1. Clinical periodontal parameters (Mean±SD and 95% Confidence interval) of subjects at the time of sampling according to periodontal health status*
As expected, periodontitis sites showed significantly more signs of disease, including higher mean PD (p<0.001) and mean CAL (p<0.001) than healthy patients.
The carriage of C. dubliniensis
summarizes species distribution of yeast isolates in the buccal cavity and the subgingival biofilm according to the periodontal health status in 240 immunocompetent individuals. The yeast microflora had similar species in both studied sites ().
Table 2. Species distribution of yeast isolates in buccal cavity (BC) and subgingival biofilm (SB) according to periodontal health status* in 240 immunocompetent individuals
Out of the 203 recovered yeasts, C. albicans was the most frequent species corresponding to 26.7% (64/240) in the buccal cavity and 21.7% (52/240) in the subgingival biofilm, as shown in .
C. dubliniensis was isolated in 4.2% (10/240) and 4.6% (11/240) of patients in both niches, respectively. Distribution of C. dubliniensis isolates from the subingival biofilm according to periodontal health status (), was as follows: 2.5% from subjects with chronic periodontitis, 1.7% with gingivitis, and 0.4% from healthy individuals.
RAPD-PCR assay
We selected five RAPD primers, based on their reproducibility, after pre-screening to analyze 21 isolates of C. dubliniensis. The number of bands ranged from three splitters (M13r) to 12 (M13f). Two of the five primers were the most informative (M13f and OPC5) and they generated the highest number of band patterns (between 10 and 12).
shows the dendrogram of RAPD fingerprints of C. dubliniensis isolates, the SC ranged from 50 to 100%. Eight genetic clusters and five main genotypes were obtained at an SC of 90%, genotypes I, II, III, IV, and V, as shown in .
Figure 1. Dendrogram generated by UPGMA clustering method, using the coefficient of similarity between RAPD-PCR of C. dubliniensis in oral cavity (a) and subgingival biofilm (b) in immunocompetent individuals.
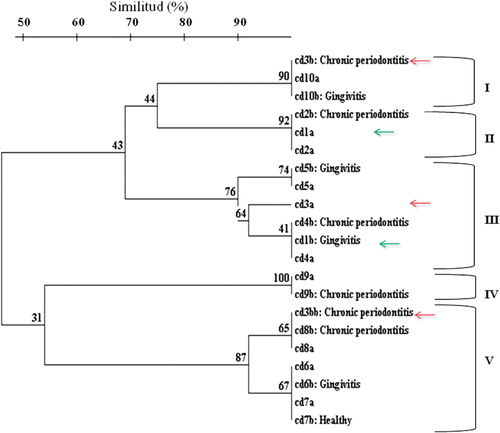
All isolates unclustered or belonging to different clusters by RAPD analysis were observed to differ by three or more bands.
RAPD analysis showed identical genotypes of C. dubliniensis in different sampling sites in eight of the 10 patients (buccal cavity and subgingival areas), except for those observed in two participants. One case was patient 1, who presented presumably unrelated isolates (, green arrows). The other patient, number 3 presented two strains in subgingival biofilm (Cd3b and Cd3bb), which represented genotype I and IV, respectively (). Moreover, participant 3 presented a mouth C. dubliniensis strain, Cd3a, which corresponded with genotype III (, red arrows). Those yeasts were presumably unrelated isolates Citation20.
Discussion
C. dubliniensis is a yeast species that is characterized by its in vitro resistance to the antifungal azole group in HIV patients, for their capacity to adhere in vitro to human buccal epithelial cells and other microorganisms in the oral microbiota, as well as the high proteinase activity Citation9Citation15Citation37Citation38. Originally, this species was isolated from oropharyngeal candidiasis in AIDS patients. Later, numerous papers have been published indicating its presence in all types of clinical samples as well as in healthy individuals Citation7Citation10Citation14Citation33Citation39Citation40Citation41Citation42Citation43Citation44Citation45Citation46Citation47Citation48Citation49Citation50.
In this study, C. dubliniensis showed a prevalence of 4.2% in the buccal mucosa from 240 patients. These results do not match those found by other authors, who found low prevalence of this species in healthy HIV-negative patients Citation14Citation44Citation51Citation52Citation53. This disagreement could be due to the different study populations evaluated (healthy subjects and periodontitis patients).
In this study, the prevalence of C. dubliniensis was 4.6% (95% CI: 2.4–8.3) in subgingival biofilm in patients from Buenos Aires, Argentina. This species was isolated both in healthy patients and in patients with periodontal disease. shows an increased prevalence of this species in individuals with chronic periodontitis, compared to healthy individuals and individuals with gingivitis, which is not a statistical significant difference. Based on the findings in this study, it is confirmed that C. dubliniensis can colonize subgingival biofilm in immunocompetent individuals with periodontal disease and healthy ones Citation6Citation7. Similar findings have been reported by Song et al., since these authors found that C. dubliniensis dominated among Candida species other than C. albicans, being most prominent in periodontal pockets Citation54.
Others authors have not found C. dubliniensis in the gingival sulcus in healthy individuals Citation4Citation10Citation11.
The RAPD-based typing was used to assess the diversity of Candida species isolates, as it has been described as a simple, rapid, and reliable discriminatory method for clinical epidemiological studies of oral Candida infections Citation17Citation23Citation24Citation27Citation35. In spite of this, and considering the reproducibility pitfalls of the RAPD method, it was considered adequate to compare the reliability of the RAPD-based clustering with that obtained with pulsed field gel electrophoresis analysis, referred to as a more robust method Citation20Citation23Citation55.
In the present study, the RAPD analysis showed similar genotypes of C. dubliniensis in different sampling sites from the same patient (buccal cavity and subgingival areas), except for those observed in two patients ().
RAPD profiles of C. dubliniensis isolates from the participants were generally distinct; therefore almost every individual harbored his/her own specific isolate.
Such genetic heterogeneity within isolates was reported from other oral and non-oral sources in C. albicans Citation25Citation26Citation56.
These results led us to the conclusion that, the origin of the colonization of C. dubliniensis in subgingival biofilm seems to be the buccal cavity, consequently, it may be assumed that most of C. dubliniensis in these sites arise from endogenous commensal strains. As the only mode of reproduction known for C. dubliniensis is asexual, these results suggest a common clonal origin of isolates in both niches Citation57.
In our series, C. dubliniensis genotypes had some differences between oral and subgingival isolates in two patients. Participants 1 and 3, harbored subgingival strains genetically presumably unrelated, to the isolated ones from their buccal cavity. Pizzo et al. Citation25 noted the presence of a different C. albicans DNA type in subgingival sites. This observation suggests that the presence of C. albicans or C. dubliniensis could also occur due to the colonization with subgingivally adapted strains, possibly as a result of genetic variations such as gene conversion and/or chromosomal translations Citation19Citation25Citation26Citation27.
Similar C. dubliniensis genotypes may be distributed among healthy and periodontal disease individuals, as shown in , genotypes I, II, III, and V included isolates from individuals that were healthy or had periodontal disease. Therefore, a hypervirulent strain of C. dubliniensis that is involved in disease patients should be excluded. Other authors also observed that genetically identical yeasts appeared in both healthy and diseased subjects when they investigated C. albicans isolates Citation22Citation25Citation27.
To date, no studies have been aimed at genetic characterization of subgingival C. dubliniensis isolates. Therefore, our yeast isolates were subjected to RAPD-PCR analysis, which has proved to be a rapid, simple, cost-effective and discriminatory technique for molecular typing of C. dubliniensis.
This is the first survey in Argentina to study the molecular characterization of C. dubliniensis by RAPD-PCR clinical isolates in different ecological niches of the oral cavity. Such findings may be useful as baseline information on subgingival C. dubliniensis colonization in our country.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors.
Acknowledgements
This study was supported by grants UBACYT M426 and UBACYT O016 from the University of Buenos Aires.
References
- Almirante B, Rodriguez D, Park BJ, Cuenca-Estrella M, Planes AM, Almela M, et al.. Epidemiology and predictors of mortality in cases of Candida bloodstream infection: results from population-based surveillance, Barcelona, Spain, from 2002 to 2003. J Clin Microbiol. 2005; 43: 1829–35.
- Ascioglu S, Rex JH, de Pauw B, Bennett JE, Bille J, Crokaert F, et al.. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin Infect Dis. 2002; 34: 7–14.
- Patel M, Coogan M, Galpin JS. Periodontal pathogens in subgingival plaque of HIV-positive subjects with chronic periodontitis. Oral Microbiol Immunol. 2003; 18: 199–201.
- Portela MB, Souza IP, Costa EM, Hagler AN, Soares RM, Santos AL. Differential recovery of Candida species from subgingival sites in human immunodeficiency virus-positive and healthy children from Rio de Janeiro, Brazil. J Clin Microbiol. 2004; 42: 5925–7.
- Dahlen G, Wikstrom M. Occurrence of enteric rods, Staphylococci and Candida in subgingival samples. Oral Microbiol Immunol. 1995; 10: 42–6.
- Jewtuchowicz VM, Brusca MI, Mujica MT, Gliosca LA, Finquelievich JL, Lovannitti CA, et al.. Subgingival distribution of yeast and their antifungal susceptibility in immunocompetent subjects with and without dental devices. Acta Odontol Latinoam. 2007; 20: 17–22.
- Jewtuchowicz VM, Mujica MT, Brusca MI, Sordelli N, Malzone MC, Pola SJ, et al.. Phenotypic and genotypic identification of Candida dubliniensis from subgingival sites in immunocompetent subjects in Argentina. Oral Microbiol Immunol. 2008; 23: 505–9.
- Reynaud AH, Nygaard-Ostby B, Boygard GK, Eribe ER, Olsen I, Gjermo P. Yeasts in periodontal pockets. J Clin Periodontol. 2001; 28: 860–4.
- Martinez M, Lopez-Ribot JL, Kirkpatrick WR, Coco BJ, Bachmann SP, Patterson TF. Replacement of Candida albicans with C. dubliniensis in human immunodeficiency virus-infected patients with oropharyngeal candidiasis treated with fluconazole. J Clin Microbiol. 2002; 40: 3135–9.
- Urzua B, Hermosilla G, Gamonal J, Morales-Bozo I, Canals M, Barahona S, et al.. Yeast diversity in the oral microbiota of subjects with periodontitis: Candida albicans and Candida dubliniensis colonize the periodontal pockets. Med Mycol. 2008; 46: 783–93.
- Jabra-Rizk MA, Ferreira SM, Sabet M, Falkler WA, Merz WG, Meiller TF. Recovery of Candida dubliniensis and other yeasts from human immunodeficiency virus-associated periodontal lesions. J Clin Microbiol. 2001; 39: 4520–2.
- Waltimo TM, Orstavik D, Meurman JH, Samaranayake LP, Haapasalo MP. In vitro susceptibility of Candida albicans isolates from apical and marginal periodontitis to common antifungal agents. Oral Microbiol Immunol. 2000; 15: 245–8.
- Cannon RD, Holmes AR, Mason AB, Monk BC. Oral Candida: clearance, colonization, or candidiasis?. J Dent Res. 1995; 74: 1152–61.
- Luque AG, Biasoli MS, Tosello ME, Binolfi A, Lupo S, Magaro HM. Oral yeast carriage in HIV-infected and non-infected populations in Rosario, Argentina. Mycoses. 2009; 52: 53–9.
- Jabra-Rizk MA, Falkler WAJr, Merz WG, Baqui AA, Kelley JI, Meiller TF. Cell surface hydrophobicity-associated adherence of Candida dubliniensis to human buccal epithelial cells. Rev Iberoam Micol. 2001; 18: 17–22.
- Jabra-Rizk MA, Falkler WAJr, Merz WG, Kelley JI, Baqui AA, Meiller TF. Coaggregation of Candida dubliniensis with Fusobacterium nucleatum. J Clin Microbiol. 1999; 37: 1464–8.
- Jain P, Khan ZK, Bhattacharya E, Ranade SA. Variation in random amplified polymorphic DNA (RAPD) profiles specific to fluconazole-resistant and -sensitive strains of Candida albicans. Diagn Microbiol Infect Dis. 2001; 41: 113–9.
- Dassanayake RS, Samaranayake LP. Amplification-based nucleic acid scanning techniques to assess genetic polymorphism in Candida. Crit Rev Microbiol. 2003; 29: 1–24.
- Samaranayake YH, Samaranayake LP, Dassanayake RS, Yau JY, Tsang WK, Cheung BP, et al.. Genotypic shuffling’ of sequential clones of Candida albicans in HIV-infected individuals with and without symptomatic oral candidiasis. J Med Microbiol. 2003; 52: 349–59.
- Soll DR. The ins and outs of DNA fingerprinting the infectious fungi. Clin Microbiol Rev. 2000; 13: 332–70.
- Montour L, Tey R, Xu J. Isolation of Candida dubliniensis in an aboriginal community in Ontario, Canada. J Clin Microbiol. 2003; 41: 3423–6.
- Hellstein J, Vawter-Hugart H, Fotos P, Schmid J, Soll DR. Genetic similarity and phenotypic diversity of commensal and pathogenic strains of Candida albicans isolated from the oral cavity. J Clin Microbiol. 1993; 31: 3190–9.
- Costa F, Manaia CM, Figueiral MH, Pinto E. Genotypic analysis of Candida albicans isolates obtained from removable prosthesis wearers. Lett Appl Microbiol. 2008; 46: 445–9.
- Pujol C, Joly S, Lockhart SR, Noel S, Tibayrenc M, Soll DR. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive DNA probe Ca3 for fingerprinting Candida albicans. J Clin Microbiol. 1997; 35: 2348–58.
- Pizzo G, Barchiesi F, Falconi Di Francesco L, Giuliana G, Arzeni D, Milici ME, et al.. Genotyping and antifungal susceptibility of human subgingival Candida albicans isolates. Arch Oral Biol. 2002; 47: 189–96.
- Pizzo G, Giammanco GM, Pecorella S, Campisi G, Mammina C, D'Angelo M. Biotypes and randomly amplified polymorphic DNA (RAPD) profiles of subgingival Candida albicans isolates in HIV infection. New Microbiol. 2005; 28: 75–82.
- Song X, Eribe ER, Sun J, Hansen BF, Olsen I. Genetic relatedness of oral yeasts within and between patients with marginal periodontitis and subjects with oral health. J Periodontal Res. 2005; 40: 446–52.
- Lamster IB, Grbic JT, Mitchell-Lewis DA, Begg MD, Mitchell A. New concepts regarding the pathogenesis of periodontal disease in HIV infection. Ann Periodontol Am Acad Periodontol. 1998; 3: 62–75.
- Silness J, Loe H. Periodontal disease in pregnancy II. Correlation between oral hygiene and periodontal condtion. Acta Odontol Scand. 1964; 22: 121–35.
- Loe H, Silness J. Periodontal disease in pregnancy I. Prevalence and severity. Acta Odontol Scand. 1963; 21: 533–51.
- Jitsurong S, Kiamsiri S, Pattararangrong N. New milk medium for germ tube and chlamydoconidia production by Candida albicans. Mycopathologia. 1993; 123: 95–8.
- Staib P, Morschhauser J. Chlamydospore formation on Staib agar as a species-specific characteristic of Candida dubliniensis. Mycoses. 1999; 42: 521–4.
- Donnelly SM, Sullivan DJ, Shanley DB, Coleman DC. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1 intron and exon sequences. Microbiology (Reading, England). 1999; 145: 1871–82.
- Scherer S, Stevens DA. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987; 25: 675–9.
- Duran EL, Mujica MT, Jewtuchowicz VM, Finquelievich JL, Pinoni MV, Iovannitti CA. Examination of the genetic variability among biofilm-forming Candida albicans clinical isolates. Rev Iberoam Micol. 2007; 24: 268–71.
- Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990; 18: 6531–5.
- Gilfillan GD, Sullivan DJ, Haynes K, Parkinson T, Coleman DC, Gow NA. Candida dubliniensis: phylogeny and putative virulence factors. Microbiology (Reading, England). 1998; 144: 829–38.
- Sullivan DJ, Moran G, Donnelly S, Gee S, Pinjon E, McCartan B, et al.. Candida dubliniensis: an update. Rev Iberoam Micol. 1999; 16: 72–6.
- Binolfi A, Biasoli MS, Luque AG, Tosello ME, Magaro HM. High prevalence of oral colonization by Candida dubliniensis in HIV-positive patients in Argentina. Med Mycol. 2005; 43: 431–7.
- Blignaut E, Pujol C, Joly S, Soll DR. Racial distribution of Candida dubliniensis colonization among South Africans. J Clin Microbiol. 2003; 41: 1838–42.
- Diaz-Guerra TM, Mellado E, Cuenca Estrella M, Laguna F, Rodriguez-Tudela JL. Molecular characterization by PCR-fingerprinting of Candida dubliniensis strains isolated from two HIV-positive patients in Spain. Diagn Microbiol Infect Dis. 1999; 35: 113–9.
- Fotedar R, Al-Hedaithy SS. Candida dubliniensis at a university hospital in Saudi Arabia. J Clin Microbiol. 2003; 41: 1907–11.
- Kamei K, McCullough MJ, Stevens DA. Initial case of Candida dubliniensis infection from Asia: non-mucosal infection. Med Mycol. 2000; 38: 81–3.
- Kirkpatrick WR, Revankar SG, McAtee RK, Lopez-Ribot JL, Fothergill AW, McCarthy DI, et al.. Detection of Candida dubliniensis in oropharyngeal samples from human immunodeficiency virus-infected patients in North America by primary CHROMagar Candida screening and susceptibility testing of isolates. J Clin Microbiol. 1998; 36: 3007–12.
- Polacheck I, Strahilevitz J, Sullivan D, Donnelly S, Salkin IF, Coleman DC. Recovery of Candida dubliniensis from non-human immunodeficiency virus-infected patients in Israel. J Clin Microbiol. 2000; 38: 170–4.
- Silva V, Cabrera M, Diaz MC, Abarca C, Hermosilla G. Prevalence of Candida albican serotypes in blood isolates in Chile, and first report of Candida dubliniensis candidemia. Rev Iberoam Micol. 2003; 20: 46–51.
- Sullivan D, Haynes K, Bille J, Boerlin P, Rodero L, Lloyd S, et al.. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J Clin Microbiol. 1997; 35: 960–4.
- Tamura M, Watanabe K, Mikami Y, Yazawa K, Nishimura K. Molecular characterization of new clinical isolates of Candida albicans and C. dubliniensis in Japan: analysis reveals a new genotype of C. albicans with group I intron. J Clin Microbiol. 2001; 39: 4309–15.
- Tintelnot K, Haase G, Seibold M, Bergmann F, Staemmler M, Franz T, et al.. Evaluation of phenotypic markers for selection and identification of Candida dubliniensis. J Clin Microbiol. 2000; 38: 1599–608.
- Pineda G, Scollo K, Santiso G, Lehmann E, Arechavala A. Isolation of Candida dubliniensis in different clinical samples. Analysis of phenotypical methods to differenciate from Candida albicans. Revista Argentina de Microbiología. 2008; 40: 211–7.
- Giammanco GM, Pizzo G, Pecorella S, Distefano S, Pecoraro V, Milici ME. Identification of Candida dubliniensis among oral yeast isolates from an Italian population of human immunodeficiency virus-infected (HIV + ) subjects. Oral Microbiol Immunol. 2002; 17: 89–94.
- Meiller TF, Jabra-Rizk MA, Baqui A, Kelley JI, Meeks VI, Merz WG, et al.. Oral Candida dubliniensis as a clinically important species in HIV-seropositive patients in the United States. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1999; 88: 573–80.
- Milan EP, de Laet Sant’ Ana P, de Azevedo Melo AS, Sullivan DJ, Coleman DC, Lewi D, et al.. Multicenter prospective surveillance of oral Candida dubliniensis among adult Brazilian human immunodeficiency virus-positive and AIDS patients. Diagn Microbiol Infect Dis. 2001; 41: 29–35.
- Song X, Sun J, Hansen BF, Olsen I. Oral distribution of genera, species and biotypes of yeasts in patients with marginal periodontitis. Microbial Ecol Health Dis. 2003; 15: 114–9.
- Lopez-Ribot JL, McAtee RK, Kirkpatrick WR, Perea S, Patterson TF. Comparison of DNA-based typing methods to assess genetic diversity and relatedness among Candida albicans clinical isolates. Rev Iberoam Micol. 2000; 17: 49–54.
- Waltimo TM, Dassanayake RS, Orstavik D, Haapasalo MP, Samaranayake LP. Phenotypes and randomly amplified polymorphic DNA profiles of Candida albicans isolates from root canal infections in a Finnish population. Oral Microbiol Immunol. 2001; 16: 106–12.
- Badoc C, De Meeus T, Bertout S, Odds FC, Mallie M, Bastide JM. Clonality structure in Candida dubliniensis. FEMS Microbiol Lett. 2002; 209: 249–54.