Abstract
Background: Periodontitis is recognized as a complex polymicrobial disease, however, the impact of the bacterial interactions among the 700–1,000 different species of the oral microbiota remains poorly understood. We conducted an in vitro screen for oral bacteria that mitigate selected virulence phenotypes of the important periodontal pathogen, Porphyromonas gingivalis. Methods: We isolated and identified oral anaerobic bacteria from subgingival plaque of dental patients. When cocultured with P. gingivalis W83, specific isolates reduced the cytopathogenic effects of P. gingivalis on oral epithelial cells. Results: In an initial screen of 103 subgingival isolates, we identified 19 distinct strains from nine species of bacteria (including Actinomyces naeslundii, Streptococcus oralis, Streptococcus mitis, and Veilonella dispar) that protect oral epithelial cells from P. gingivalis-induced cytotoxicity. We found that some of these strains inhibited P. gingivalis growth in plate assays through the production of organic acids, whereas some decreased the gingipain activity of P. gingivalis in coculture or mixing experiments. Conclusion: In summary, we identified 19 strains isolated from human subgingival plaque that interacted with P. gingivalis, resulting in mitigation of its cytotoxicity to oral epithelial cells, inhibition of growth, and/or reduction of gingipain activity. Understanding the mechanisms of interaction between bacteria in the oral microbial community may lead to the development of new probiotic agents and new strategies for interrupting the development of periodontal disease.
Periodontitis is an inflammatory condition characterized by damage to gingival tissue and periodontal ligaments, resorption of alveolar bone, and loss of teeth Citation1. The destruction of tissue seen in periodontitis is thought to be due to a combination of factors produced by oral bacteria and the induction of host inflammatory mediators. Although periodontitis is linked to the presence of certain bacterial species, no single bacterium is either necessary or sufficient to cause human disease Citation2Citation3Citation4. In the periodontal pocket of either healthy or diseased teeth, there are many different bacteria that coexist in close proximity to each other, and it has been postulated that specific interactions can modulate the effects on host tissue destruction. For example, one well-described interaction between different species of bacteria is in the formation of the biofilm that coats the teeth and gingival tissues, where specific, nonpathogenic bacteria may be required to start the initial biofilm that allows pathogenic bacteria to attach and cause disease Citation5Citation6. Antagonistic interactions have also been documented, most notably between Streptococcus mutans and Streptococcus gordonii in which the former interferes with the latter's colonization through production of species-specific proteases Citation7.
Porphyromonas gingivalis is a Gram-negative asaccharolytic anaerobe that is a member of the ‘red-complex’ bacteria that have been shown to be critical species in the development of periodontitis Citation8Citation9. P. gingivalis expresses a variety of virulence factors that include fimbriae, lectin-like adhesins, capsular polysaccharide, lipopolysaccharide, hemagglutinins, hemolysins, outer membrane vesicles, and various proteolytic enzymes that cause chronic inflammation of the gingiva leading to tissue damage and loss of teeth Citation10Citation11Citation12. Although these virulence factors are important for the growth and survival of P. gingivalis, some of these have also been shown to assist in the pathogenesis of other oral microbes. For example, P. gingivalis gingipains cleave, inactivate, and cause loss of activity of complement factors, immunoglobulins, antimicrobial peptides, cytokines (IL-6, IL-12, IL-1b, TNF-alpha, and IFN-gamma), and chemokines (IL-8), protecting other bacteria in close proximity to P. gingivalis from the action of these host defense mechanisms Citation13. Similarly, P. gingivalis proteases liberate amino acids from complex proteins as well as iron from heme-containing compounds (hemoglobin, hemopexin, haptoglobin, and transferrin) that can serve as nutrients for itself or other nearby bacteria Citation14Citation15. Antagonistic activity of oral commensals or pathogens against growth of other periodontal pathogens has been described Citation4Citation16Citation17Citation18Citation19. However, the exact mechanisms of interactions between the members of the dental microbiota that either ameliorate or aggravate periodontitis are not well understood. Here, we show that specific strains of clinically isolated oral bacteria can inhibit growth and/or virulence of P. gingivalis in vitro. These in vitro findings are a first step toward understanding the potential contribution of ‘commensal’ oral bacteria protection against periodontitis.
Methods
Bacterial strains and growth conditions
Porphyromonas gingivalis strain W83 (ATCC BAA-308) was cultured anaerobically in blood agar containing tryptic soy (TS; MP Biomedicals) with yeast extract (1 g/l), hemin (5 µg/ml), Vitamin K1 (1 µg/ml) and 5% sheep blood, or brain heart infusion (BHI; Beckton, Dickson and Company) broth supplemented with yeast extract (5 g/l), hemin (5 µg/ml), Vitamin K1 (1 µg/ml), and l-cysteine according to standard protocols Citation13. P. gingivalis was incubated in BHI broth at 37°C inside anaerobic gas pouches (BD GasPak Easy). Bacterial cells were collected by centrifugation and suspended to OD600=1 in phosphate-buffered saline (PBS).
Other oral bacteria were isolated from subgingival plaque samples collected during routine cleaning from 20 patients who were diagnosed as having periodontitis in the periodontal clinic at Tufts University School of Dental Medicine. The protocol for sample collection was reviewed and approved by the Tufts University Institutional Review Board. Subgingival plaque samples from the deepest part of three periodontal pockets, including the mesial side of two single-rooted teeth, and a molar tooth per patient were obtained. Upon removing the supragingival plaque and isolation of the area, sterile paper points were inserted into the periodontal pockets for 20 sec, placed immediately into BHI broth with glycerol and frozen prior to use. Glycerol stocks were streaked onto blood agar plates. Plates were incubated anaerobically at 37°C, and single colonies of single isolates were selected and re-streaked three times on blood agar plates to ensure purity.
Identification of clinical isolates
The identity of isolates was determined by 16S rRNA sequencing. The 16S rRNA genes of the clinical isolates were amplified using universal primers, primers 8UA (5′-AGAGTTTGATCCTGGCTCAG-3′), and 907B (5′-CCGTCAATTCMTTTRAGTTT-3′) using the following cycling conditions: 3 min at 95°C; 30 sec at 96°C, 30 sec at 45°C, and 1 min at 72°C for 35 cycles; and 10 min at 72°C Citation20. The resulting sequences were compared with microbial genome databases using NCBI (www.ncbi.org) and BLAST2TREE (http://bioinfo.unice.fr/blast). The most closely related species was identified by percent sequence similarity. In cases where less than 97% similarity was obtained, sequence analysis was repeated using a second set of 16S rRNA universal primers, 774A (5′-GTAGTCCACGCTGTAAACGATG-3′) and 1485B (5′-TACGGTTACCTTGTTACGAC-3′) using the same cycling conditions Citation20. The nucleotide sequences have been deposited in GenBank (Accession Numbers JF496718-JF496736).
Host cell culture
The immortalized oral epithelial cell line OKF6/TERT-2 (kind gift of Dr. J. Rheinwald) was cultured in Keratinocyte – serum-free medium (K-sfm, Gibco) supplemented with epidermal growth factor (Gibco) and bovine pituitary extract (Gibco) as described Citation21. Primary human gingival epithelial cells (HGEC) were cultured in Progenitor Cell Targeted Oral Epithelium Medium (CELLnTEC Advanced Cell Systems) according to the manufacturer's instructions.
Bacterial and cellular cocultures
The effect of individual strains of isolated oral bacteria on the cytotoxicity of P. gingivalis was determined by measuring the ability of mixed bacterial cell suspensions to cause morphological changes in oral epithelial cells adapted from the method described by Shah et al. Citation22. P. gingivalis with or without another strain of oral bacteria were added to monolayers of OKF6/TERT-2 or HGEC primary cells at a multiplicity of infection (MOI) of 100. The cocultures were incubated in K-sfm for 16 hrs at 37°C in a humidified atmosphere with 5% CO2. Uninfected host cells or cells infected with P. gingivalis alone were used as the negative and positive controls, respectively. Cells were reviewed by blinded investigators for rounding and detachment using an inverted phase-contrast microscope at 200× magnification (Nikon TMS) and scored on a scale of ‘–’ for no visible inhibition of cytotoxicity (complete cell rounding and detachment), ‘ + ’ weak inhibition (partial cell rounding and detachment), and ++ strong inhibition (minimal cell rounding and detachment). We also tested for wound healing in the presence of oral bacterial isolates and P. gingivalis using the method described by Furuta et al. Citation23. Monolayers of OKF6/TERT-2 or HGEC cells grown in 48-well tissue culture plates were wounded by scratching with a pipet tip and infected as above. The cells were examined by blinded investigators after 16 hrs and scored for growth of cells into the wounded area. Experiments were performed two times in duplicate.
Assay for inhibition of gingipain activity
Determination of the effects of microbial interactions on gingipain production of P. gingivalis was performed using a modification of the strategy described by Bakri & Douglas Citation24. First, we measured the effects of coculturing P. gingivalis with individual isolated strains on the resulting gingipain activity of the bacterial cultures. Briefly, P. gingivalis was grown anaerobically in BHI, while bacterial strains to be tested were anaerobically grown in TS broth for 48 hrs. Each bacterial culture (10 µl) was mixed with P. gingivalis culture (10 µl) and further incubated in BHI broth for 24 hrs. Combined cultures (20 µl) were mixed with the arginine-gingipain substrate benzoyl-arginine naphthylamide (BANA, 440 µg/ml) in 100 mM Tris–HCl buffer (with 2 mm dithiothreitol), incubated at 37°C for 2 hrs, and centrifuged to remove bacterial cells. Fast Blue BB detection solution (350 µg/ml Diazotized 4′-amino-2′,5′-diethoxybenzanilide zinc chloride salt in ethylene glycol, MP Biomedicals) was added as a coupling reagent to bind napthylamine liberated by the action of gingipains, and the optical density at 490 nm was measured after 30 min using a spectrophotometer. To assess the effects of independently cultured bacterial strains on the gingipain activity of P. gingivalis, bacterial cultures (10 µl) grown anaerobically to stationary phase in TS broth were mixed with P. gingivalis culture (10 µl) of BANA substrate, and immediately processed as described above. Assays were performed three times in triplicate.
Assays for inhibition of P. gingivalis growth
Determination of the capacity of isolated strains of oral bacteria to inhibit growth of P. gingivalis was performed by plating P. gingivalis from an overnight BHI broth culture (100 µl) onto a blood agar plate Citation16. Overnight broth cultures of the individual clinical isolates of nonP. gingivalis bacteria (10 µl) were spotted on blood agar plates containing the lawn of P. gingivalis. Inhibitory activity was determined by measuring the zone of inhibition between the tested strains and P. gingivalis lawn after 72 hrs of anaerobic incubation at 37°C. Inhibition experiments were also performed using UV-killed bacteria (resuspended to OD600=1 in PBS and spotted on the P. gingivalis lawn) and bacteria-free supernatants (10 µl aliquots deposited onto round wells cut into the agar plate and allowed to diffuse before inoculation with P. gingivalis). Experiments were performed three times in triplicate.
Assays for acid production
Identification of strains that produce organic acid metabolites was performed by incubating the clinical strains anaerobically in TS broth containing 2.5 g/l of glucose, at 37°C for 48 hrs. The pH of broth cultures was measured using Hydrion pH papers (range of 3.5–8.0, Microlab Essentials). Experiments were performed three times in triplicate.
Assay for production of hydrogen peroxide (H2O2)
Testing for H2O2 production by selected oral bacterial isolates was performed using two different assays: Citation1 alpha hemolysis of red blood cells and Citation2 a hydrogen peroxide colorimetric assay. Hundred microliters of an overnight culture of P. gingivalis were spread onto the surface of blood agar plates. After drying, 10 µl of overnight broth cultures of isolates to be tested were spotted on the plates, which were then incubated anaerobically at 37°C for 72 hrs, and observed for alpha hemolysis. Hemolysis was recorded as either present (+) or absent (−).
For quantitative measurements of H2O2 production, TS broth cultures (15 µl of each isolate) was mixed with Hydrogen Peroxide Substrate Solution (85 µl) containing ferrous iron and xylenol orange (Hydrogen Peroxide Assay Kit, National Diagnostics). The mixture was incubated at room temperature for 30 min. The OD595 was measured for each sample and compared with a standard curve. Standards were prepared from two-fold dilutions of 0.015% H2O2. The limit of detection of the assay was 15 ng/ml of H2O2. Experiments were performed three times in triplicate.
Results
Effects of clinical isolates on P. gingivalis-induced changes in host cells
We isolated a total of 103 strict and facultative anaerobic bacteria representing 26 different bacterial species from 20 subjects with periodontitis (). The majority of the isolates were identified to be Gram-positive species by 16S rRNA sequencing. Although Streptococcus and Actinomyces were the predominant genera identified, the nature of the plaque sample, media type, and incubation conditions are likely to have influenced the species of bacteria isolated. We sought to determine whether any of these bacteria may have an effect on virulence of the pathogenic bacteria, P. gingivalis. We used two in vitro models of P. gingivalis – host cell interaction. First, we used a cellular cytotoxicity assay Citation22Citation25 where P. gingivalis (strain W83) was added to confluent cell cultures resulting in morphological changes in both the oral epithelial cell line OKF6/TERT-2 and in HGEC. These changes included loss of adhesion, cell rounding, and detachment (A). We also used a wound-healing model Citation23 where confluent cell cultures of OKF6/TERT-2 or HGEC were injured by creating a linear streak. Wounded cells that were not exposed to P. gingivalis repaired the gap and fully recovered within 12 hrs. In contrast, cells treated with P. gingivalis showed greatly impaired ability to grow and repair the wound (B).
Fig. 1. Effects of Porphyromonas gingivalis (Pg) on oral epithelial cells. (A) Pg W83, Streptococcus oralis 180B (So180B), or Pg+So180B were added to OKF6/TERT2 cells. Cocultures were allowed to incubate in 5% CO2 at 37°C for 16 hr and then examined by blinded investigators. (B) OKF6/TERT-2 cells were grown to confluence and used in the wound-healing assay. A linear streak was made in the cell monolayer. Pg, So180B, or Pg+So180B were added to the cultures at MOI = 100. After incubation for 2 hrs, the plates were washed to remove bacteria and fresh media was added. The cultures were incubated for 16 hrs and read by blinded investigators.
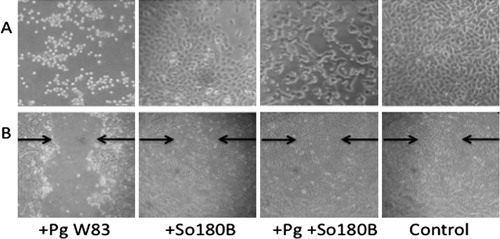
Table 1. Inhibition of Porphyromonas gingivalis (Pg) cytotoxicity by clinical strains
To determine whether the presence of commensal oral bacteria could modify the host cell effects induced by P. gingivalis, individual clinical isolates were added to cocultures of P. gingivalis and oral epithelial cells. Differences in cytotoxicity were observed upon the coinfections of host cells with combinations of P. gingivalis and some clinical isolates (). Results are shown for the cytotoxicity assay. Results for the wound-healing model were in agreement with the cytotoxicity assay (a representative result is shown in ). Nineteen clinical isolates that weakly or strongly reduced the cytotoxic effects of P. gingivalis were identified and selected for further study. Not all strains from a specific species of bacteria showed similar effects on P. gingivalis cytotoxicity. Of note, not all bacteria grew equally well under these conditions; although we added equivalent numbers of bacteria at the beginning of the assay, some bacterial strains grew to a higher density by the end of the assay. P. gingivalis, which is an anaerobic bacterium, survived but did not show growth under these conditions (as determined by plating of the cocultures on blood agar plates after incubation). There was no direct correlation between growth of the commensal bacteria and inhibition of P. gingivalis cytotoxicity, suggesting that inhibition by the 19 isolates cannot be explained solely on the basis of competition for nutrients.
Effects of clinical isolates on gingipain activity of P. gingivalis
We next sought to identify mechanisms by which the subset of 19 clinical isolates could affect the ability of P. gingivalis to induce changes in host cells. Cell detachment, cell adhesion molecule cleavage, and apoptosis have been attributed to gingipains, the trypsin-like cysteine proteases produced by P. gingivalis Citation26Citation27. We cocultured the 19 bacterial strains identified above with P. gingivalis W83 and measured arginine-gingipain (Rgp) activity using the BANA substrate test. We found that 5 of 19 strains showed statistically significant inhibition of gingipain activity by greater than 25% when cocultured simultaneously with P. gingivalis (). Six strains also showed inhibition of gingipain activity when cultured separately from P. gingivalis. When cultures of these bacteria were mixed with cultures of P. gingivalis and analyzed immediately, there was also decreased Rgp activity, suggesting that the inhibition of gingipains by these strains occurs through mechanisms that involve direct inhibition of gingipain activity ().
Table 2. Effects on Porphyromonas gingivalis (Pg) gingipain activity and production of acid and hydrogen peroxide by oral bacterial strain
Effects of clinical isolates on growth of P. gingivalis
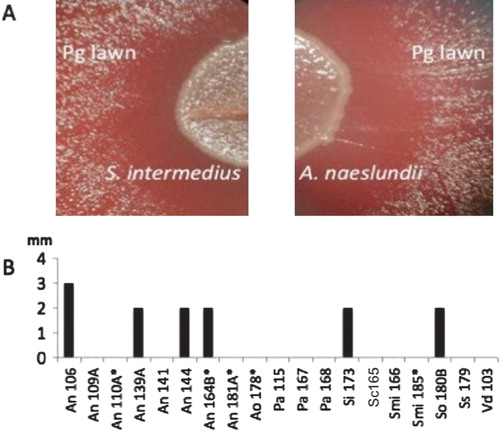
Another potential mechanism by which interactions between clinical isolates may modulate the cytotoxicity caused by P. gingivalis is through affecting bacterial replication. The ability of the 19 isolated strains to affect the growth of P. gingivalis was investigated by agar plate assay Citation16. Four Actinomyces naeslundii isolates, one Streptococcus Intermedius, and one Streptococcus oralis inhibited the growth of P. gingivalis as determined by a zone of inhibition when grown on a lawn of P. gingivalis on blood agar plates (). The inhibitory properties of these strains appeared to require active bacterial growth or the presence of outer membrane components that may be affected by UV exposure because neither cell-free supernatants nor UV-killed cells affected P. gingivalis growth.
Fig. 2. Effect of oral bacterial isolates on Porphyromonas gingivalis (Pg) growth. Broth cultures of 19 clinical strains identified as inhibitory to Pg cytotoxicity were spotted on blood agar inoculated with a lawn of Pg and incubated anaerobically at 37°C for 48 hrs. The growth of Pg was inhibited near the colonies of 6 of 19 strains including (A) Streptococcus intermedius and Actinomyces naeslundii. (B) Growth inhibition was measured as the size of the clear zone (in mm) between the edge of the colony and the lawn of P. gingivalis. Bars represent the average of triplicate measurements confirmed in three independent experiments. Abbreviations used: An, Actinomyces naeslundii; Pa, Propionibacterium acnes; Si, Streptococcus intermedius; Smi, Streptococcus mitis; Sc, Streptococcus constellatus; So, Streptococcus oralis; Streptococcus sanguinis; Vd, Veillonella dispar. *less than 98% 16S rRNA sequence similarity to the closest type strain.
Acid production by clinical isolates
Production of acid is a mechanism used by some oral bacteria to control the growth of other bacteria Citation28. We determined the acid -producing ability of the 19 strains of bacteria identified as inhibitory to P. gingivalis cytotoxicity (). Thirteen strains were shown to be acid producers in broth cultures, including seven strains that reduced the pH of the media to less than 6.0. All 8 A. naeslundii isolates tested positive for acid production. Among the 6 Streptococcus strains studied, S. intermedius 173, Streptococcus mitis 166 and S. oralis 180B lowered the pH of the cultures to less than 6.5. All but one strain that inhibited the growth of P. gingivalis reduced culture pH to less than 6.0 and the one strain that did not was also shown to be a producer of H2O2 (see below). There was a statistically significant relationship between the ability to produce acid and the ability to inhibit the growth of P. gingivalis (P=0.044, Fisher's Exact Test).
H2O2 production by clinical isolates
Production of reactive oxygen species through the generation of H2O2 is another mechanism used by bacteria to control growth of other bacteria Citation29Citation30. Five of the 19 inhibitory isolates were H2O2 producers in broth cultures (). Two strains of A. naeslundii, a S. mitis strain and a S. oralis strain, were identified as H2O2 producers. Only one of these strains was capable of inhibiting the growth of P. gingivalis (S. oralis 180B), whereas none of the other strains, including the highest H2O2-producing strain, showed any inhibition of P. gingivalis growth. No significant association was observed between the ability to produce H2O2 and the inhibition of growth of P. gingivalis. There was a statistically significant relationship between the ability to produce H2O2 and the ability to inhibit Rgp activity of P. gingivalis when cultured separately and mixed and analyzed immediately (P=0.017, Fisher's Exact Test).
Discussion
A complex community of bacteria populates and interacts within the oral cavity, with estimates of between 700–1,000 species in the ‘global’ oral microbiome and 150–350 within a given mouth Citation31. How these interactions may contribute to the development or prevention of disease is just beginning to be understood. In this study, we used P. gingivalis-induced cytotoxicity as a model to study how interspecies interactions influence the virulence potential of a periodontal pathogen in vitro. Of the over 100 bacterial isolates tested, we identified 19 strains from 8 different species of bacteria that were able to weakly or strongly inhibit host cell rounding and detachment mediated by P. gingivalis W83, a virulent strain in animal models of infection Citation32. Previous studies have shown that the cytotoxicity assays we utilized measured a mixture of P. gingivalis virulence components’ effects on cells, including but not limited to, expression/activity of proteases such as gingipains, invasion, end-product metabolite production, the effects of cell surface components such as lipopolysaccharides and fimbriae, and the ability to induce apoptosis Citation33. We choose to start with a cytotoxicity screen that is affected by multiple different interactions because we felt that it was likely that different microbes may involve multiple factors and affect different pathways in coculture conditions.
The ability to inhibit the host-cell effects of P. gingivalis was spread among several bacterial species that belong to the phyla Actinobacteria and Firmicutes, notably of the genera Actinomyces, Streptococcus, and Veillonella. Actinobacteria and Firmicutes are known to precede P. gingivalis in oral biofilm colonization Citation34. Interestingly, even within species of bacteria, not all strains were similarly capable of blocking the effects of P. gingivalis on host cells, suggesting that there is significant strain-to-strain variability in interactions with P. gingivalis. This has implications for future studies of interspecies interactions between bacteria as it cannot be assumed that all members of a certain species of bacteria, observed as ‘inhibitory’ to one type of pathogenic bacteria, are always inhibitory. Until the genes associated with the inhibitory functions of a particular strain are identified, the determination of the inhibitory capacity of each specific strain will need to be tested phenotypically. Furthermore, given the differences in the expression of virulence factors between known laboratory and clinical isolates of P. gingivalis, it is likely that some of the bacteria that inhibit P. gingivalis strain W83, may not inhibit cytotoxicity of other P. gingivalis strains. In limited testing of other strains of P. gingivalis, we have found that some of the inhibitory strains also inhibit cytotoxicity of these strains, whereas others do not (unpublished data, E. Tenorio).
Although the goal of this study was not to provide an in-depth understanding of the mechanisms of inhibition of P. gingivalis toxicity by individual isolates, we did perform tests of effects of these clinical isolates on well-established virulence factors of P. gingivalis. P. gingivalis is known for producing an array of proteolytic enzymes (as many as 46 different proteases) that are important in tissue destruction, activation of host matrix-metalloproteinases, and immunomodulation Citation35. Gingipains (encoded by the genes rgpA, rgpB, and kgp) are the major proteolytic enzymes of P. gingivalis and have been linked to the severity of periodontal disease Citation35. Gingipains are a major class of virulence factors linked to the destruction of periodontal tissues, alveolar bone loss, and modulation of the host immune system Citation36Citation37. In addition, gingipains are involved in the adherence of P. gingivalis to other bacteria and human epithelial cells Citation38. Gingipain-specific inhibitors have been shown to suppress the pathogenicity of P. gingivalis in vitro Citation39. In this study, we identified strains of Actinomyces and Streptococcus genera that affected arginine-gingipain activity. These strains were nonidentical to the strains that inhibited P. gingivalis growth, suggesting that for at least a portion of the strains, the inhibition of gingipain activity was separate from the effects on growth. A subset of the strains studied inhibited gingipain activity when cocultured simultaneously with P. gingivalis, whereas others showed inhibition when cultured separately from P. gingivalis and mixed and analyzed immediately. Interestingly, many were not the same strains that inhibited in coculture, suggesting that they may have different mechanisms of inhibition. For example, those strains that inhibit in coculture may be acting by suppressing the growth of P. gingivalis or gingipain expression by direct interaction between the organisms resulting in decreased production of gingipains. Those that act immediately may be producing an inhibitor of gingipains. Further studies will be required to differentiate these possibilities.
Although gingipains have been identified as major virulence factors for P. gingivalis, only a minority of the strains that mitigated its cytotoxic effects appeared to affect gingipain activity. It is not unexpected that different bacterial species may have evolved different/multiple mechanisms for blocking P. gingivalis-induced toxicity. Another classic mechanism by which bacteria may interact with each other is through affecting the growth of the other organism. This may occur through competition for nutrients, through quorum-sensing mechanisms or production of bacteriostatic or bactericidal agents. We found that among the 19 isolates that mitigated the cytotoxic effects of P. gingivalis, four Actinomyces and two Streptococcus isolates had a direct, negative effect on growth of P. gingivalis in in vitro coculture conditions. The inhibitory effect was observed with live bacteria but not with culture supernatants or UV-killed cells under the conditions tested.
Possible ways that oral bacteria may affect P. gingivalis growth include the production of acid or H2O2. Of the six A. naeslundii and streptococcal strains that inhibited the growth of P. gingivalis, all were capable of producing acid. However, there were some acid-producing strains of bacteria that did not inhibit the growth of P. gingivalis, suggesting that acid production alone does not regulate the growth of P. gingivalis. One hypothesis is that there is production of an enzyme or regulator by the strains that inhibit P. gingivalis growth that requires a low pH for activation. Of note, gingipain expression and activity under anaerobic conditions remains high between pH 6.5–9.5 with Lysine-gingipain showing a maximum activity around 7.5–8.5 but is decreased at lower pHs, so lower pH may affect virulence even without directly affecting growth Citation40.
H2O2 production did not appear to have a clear role in the growth inhibition of P. gingivalis. Many of the strains that produced H2O2 did not inhibit P. gingivalis growth (including the highest producers). Whether H2O2 production by these strains may have a role in vivo is unknown. S. oralis has been previously reported to exert H2O2-mediated probiotic effects in vivo Citation41. Interestingly, we observed that strains that produced H2O2 were more likely to inhibit the Rgp activity of P. gingivalis in independent cultures. Addition of H2O2 has been shown to strongly decrease the Rgp activity of P. gingivalis, while only moderately affecting its growth in complex culture medium Citation42.
The role of the inhibitory strains of bacteria in controlling P. gingivalis growth and colonization in the human mouth is unknown. All of our bacteria were recovered from patients with some degree of periodontitis, and we did not observe a simple correlation between the number of inhibitory strains recovered and the severity of periodontitis (data not shown). Interestingly, we did not recover P. gingivalis from any tooth where an inhibitory strain was also recovered. However, as our study was not designed to assess the presence and effects of inhibitory strains on P. gingivalis growth in vivo and there are numerous alternative explanations for this finding, the true role of these inhibitory strains on the presence of P. gingivalis in vivo and on the development of periodontitis awaits future-controlled trials in healthy and diseased patients.
Conclusions
This study provides in vitro data on how interactions between P. gingivalis and other oral bacteria may influence its pathogenic potential. We have identified a number of oral bacterial strains that are capable of reducing the cytotoxic effects of P. gingivalis in vitro and interfering with its growth and proteolytic activity through multiple different mechanisms. It is tempting to speculate that differences in the presence of specific strains of inhibitory bacteria may help constrain P. gingivalis and govern the development of periodontitis. Further work will be necessary to test this and other hypotheses regarding the role of these inhibitory strains in health and disease.
Conflict of interest and funding
Tufts Medical Center has filed a provisional patent application for selected Porphyromonas gingivalis-inhibitory bacteria described in this article.
Acknowledgements
The project was supported by NIH grants R21DE016859 (LTH) and 5T32 AI007389-19. The authors wish to thank Dr. Ruben Ovadia, Peter Shin, and Min Seok Kim for their help in collecting clinical samples and Jason Wang for helping with assays.
Authors’ contributions: ELT contributed with the experimental design, laboratory work, data analysis, and preparation and critical review of the manuscript. BAK contributed with the laboratory work, data analysis, and preparation of the manuscript. WSC contributed with the experimental design and collection of patient samples. LTH contributed with the experimental design, data analysis, and preparation and critical review of the manuscript. All authors read and approved the final manuscript.
References
- Cochran DL. Inflammation and bone loss in periodontal disease. J Periodontol. 2008; 79: 1569–76.
- Colombo APV, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al.. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009; 80: 1421–32.
- Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010; 8: 481–90.
- Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007; 71: 653–70.
- Bradshaw DJ, Homer KA, Marsh PD, Beighton D. Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology. 1994; 140: 3407–12.
- Marsh PD. Dental plaque as a biofilm. J Ind Microbiol. 1995; 15: 169–75.
- Wang BY, Deutch A, Hong J, Kuramitsu HK. Proteases of an early colonizer can hinder Streptococcus mutans colonization in vitro. J Dent Res. 2011; 90: 501–5.
- Holt SC, Ebersole J, Felton J, Brunsvold M, Kornman KS. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science. 1988; 239: 55–7.
- Socransky S. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998; 25: 134–44.
- Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999; 20: 168–238.
- Malek R, Fisher JG, Caleca A, Stinson M, van Oss CJ, Lee JY, et al.. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J Bacteriol. 1994; 176: 1052–9.
- Lin L, Li C, Liu J, Zhang D, Zhao J, et al.. Virulence genes of Porphyromonas gingivalis W83 in chronic periodontitis. Acta Odontol Scand. 2009; 67: 258–64.
- Kadowaki T, Nakayama K, Okamoto K, Abe N, Baba A, et al.. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J Biochem. 2000; 128: 153–9.
- Kolenbrander PE, Palmer RJ Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontol ; 42. 2000; 2006: 47–79.
- Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010; 8: 471–80.
- Grenier D. Antagonistic effect of oral bacteria towards Treponema denticola. J Clin Microbiol. 1996; 34: 1249–52.
- Apolonio AC, Carvalho MA, Ribas RN, Sousa-Gaia LG, Santos KV, Lana MA, et al.. Production of antagonistic substance by Eikenella corrodens isolated from the oral cavity of human beings with and without periodontal disease. J Appl Microbiol. 2007; 103: 245–51.
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010; 8: 15–25.
- Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol. 2005; 187: 7193–203.
- Song YL. Clostridium bartletti sp.nov., isolated from human faeces. Anaerobe. 2004; 10: 179–84.
- Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, et al.. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000; 20: 1436–47.
- Shah HN, Gharbia SE, O'Toole CM. Assessment of the relative cytotoxicity of Porphyromonas gingivalis cells, products, and components on human epithelial cell lines. J Periodontol. 1992; 63: 44–51.
- Furuta N, Takeuchi H, Amano A. Entry of Porphyromonas gingivalis outer membrane vesicles into epithelial cells causes cellular functional impairment. Infect Immun. 2009; 77: 4761–70.
- Bakri IM. Douglas CW. Inhibitory effect of garlic extract on oral bacteria. Arch Oral Biol. 2005; 50: 645–51.
- Morioka M, Hinode D, Nagata A, Hayashi H, Ichimiya S, Ueda M, et al.. Cytotoxicity of Porphyromonas gingivalis toward cultured human gingival fibroblasts. Oral Microbiol Immunol. 1993; 8: 203–7.
- Sheets SM, Potempa J, Travis J, Casiano CA, Fletcher HM. Gingipains from Porphyromonas gingivalis W83 induce cell adhesion molecule cleavage and apoptosis in endothelial cells. Infect Immun. 2005; 73: 1543–52.
- Stathopoulou PG, Galicia JC, Benakanakere MR, Garcia CA, Potempa J, Kinane DF. Porphyromonas gingivalis induce apoptosis in human gingival epithelial cells through a gingipain-dependent mechanism. BMC Microbiol. 2009; 9: 107.
- Doran A, Kneist S, Verran J. Ecological control: In vitro inhibition of anaerobic bacteria by oral streptococci. Microb Ecol Health Dis. 2004; 16: 23–7.
- Hillman JD. The relationships between streptococcal species and periodontopathic bacteria in human dental plaque. Arch Oral Biol. 1985; 30: 791–5.
- 30. Tong H, Chen W, Merritt J, Qi F, Shi W, Dong X. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: A possible counteroffensive strategy for interspecies competition. Mol Microbiol. 2007; 63: 872–80.
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005; 43: 5721–32.
- Chen T, Hosogi Y, Nishikawa K, Abbey K, Fleischmann RD, Walling J, et al.. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J Bacteriol. 2004; 186: 5473–9.
- Amornchat C. Rassameemasmaung S, Sripairojthikoon W, Swasdison S. Invasion of Porphyromonas gingivalis into human gingival fibroblasts in vitro. J Int Acad Periodontol. 2003; 5: 98–105.
- Amano A. Bacterial adhesins to host components in periodontitis. Periodontol 2000. 2010; 52: 12–37.
- Brunner J, Wittink FRA, Jonker MJ, De Jong M, Breit TM, Laine ML, et al.. The core genome of the anaerobic oral pathogenic bacterium Porphyromonas gingivalis. BMC Microbiol. 2010; 10: 252.
- Fitzpatrick RE, Wijeyewickrema LC, Pike RN. The gingipains: scissors and glue of the periodontal pathogen, Porphyromonas gingivalis. Future Microbiol. 2009; 4: 471–87.
- O'Brien-Simpson NM, Veith PD, Dashper SG, Reynolds EC. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Protein Pept Sci. 2003; 4: 409.
- Tokuda M. Role of Porphyromonas gingivalis protease activity in colonization of oral surfaces. Infect Immun. 1996; 64: 4067–73.
- Curtis MA, Aduse Opoku J, Rangarajan M, Gallagher A, Sterne JA, Reid CR, et al.. Attenuation of the virulence of Porphyromonas gingivalis by using a specific synthetic Kgp protease inhibitor. Infect Immun. 2002; 70: 6968–75.
- Abe N, Kadowaki T, Okamoto K, Nakayama K, Ohishi M, Yamamoto K. Biochemical and functional properties of lysine-specific cysteine proteinase (lys-gingipain) as a virulence factor of Porphyromonas gingivalis in periodontal disease. J Biochem. 1998; 123: 305–12.
- Zahradnik RT, Magnusson I, Walker C, McDonell E, Hillman CH, Hillman JD. Preliminary assessment of safety and effectiveness in humans of ProBiora3, a probiotic mouthwash. J Appl Microbiol. 2009; 107: 682–90.
- Leke N, Grenier D, Goldner M, Mayrand D. Effects of hydrogen peroxide on growth and selected properties of Porphyromonas gingivalis. FEMS Microbiol Lett. 1999; 174: 347–53.