Abstract
Background
Over 700 bacterial species reside in human oral cavity, many of which are associated with local or distant site infections. Extensive characterization of the oral microbiome depends on the technologies used to determine the presence and proportions of specific bacterial species in various oral sites.
Objective
The objective of this study was to compare the microbial composition of dental plaque at baseline using Human Oral Microbe Identification Microarray (HOMIM) and Human Oral Microbe Identification using Next Generation Sequencing (HOMINGS) technologies, which are based on 16S rRNA.
Methods
Dental plaque samples were collected from 96 patients at baseline prior to a dental procedure involving manipulation of gingival tissues. The samples were surveyed for 293 and 597 oral bacterial species via HOMIM and HOMINGS, respectively, based on 16S rRNA gene sequences. We determined the concordance between the two technologies for common species. Genus level analysis was performed using HOMINGS-specific genus identification capabilities.
Results
HOMINGS detected twice the number of species in the same dental plaque samples compared to HOMIM. For the species detected by both HOMIM and HOMINGS, there was no difference in relative proportions of overall bacterial composition at the species, genus or phylum levels. Additionally, there was no difference in relative proportion for total species per patient between the two technologies.
Conclusion
HOMINGS significantly expanded oral bacterial species identification compared to HOMIM. The genus and species probes, combined in HOMINGS, provided a more comprehensive representation of oral bacterial community, critical for future characterization of oral microbes in distant site infections.
The oral microbiome encompasses over 700 predominant bacterial species, many of which have not been formally named or cultivated (Citation1, Citation2). These bacterial species reside on various oral surfaces including tongue, buccal mucosa, gingiva, hard palate, and supra- and sub-gingival dental plaque (Citation3, Citation4).
Numerous oral bacterial species have been associated with oral diseases including dental caries, gingivitis, and periodontitis (Citation5, Citation6). Some oral bacterial species have increasingly been shown to be associated with systemic diseases such as infective endocarditis (IE) and prosthetic joint infection (PJI) (Citation7). These species are thought to gain access to internal tissues following bacteremia that may result from various manipulations of the oral mucosa, some of which are highly invasive (e.g. tooth extraction) and others are considered less invasive (e.g. tooth brushing or chewing) (Citation8). Oral bacterial niche characterization is, therefore, fundamental for determining predictors of bacteremia and associated systemic disease. Proper characterization of the oral microbiome is dependent upon the technologies used to determine the relative proportions of specific bacterial species in various oral sites.
This study used two semi-quantitative technologies for oral bacterial species identification: the Human Oral Microbe Identification Microarray (HOMIM) and Human Oral Microbe Identification using Next Generation Sequencing (HOMINGS) (Citation9). Both technologies use 16S ribosomal RNA (rRNA) gene sequences for species identification. While HOMIM relies on an in vitro hybridization procedure similar to DNA microarray technology, HOMINGS follows an in silico hybridization process, in effect processing of unique electronic or ‘e’-hybridization events referred to as ‘hits’.
Prior to the introduction of HOMIM in 2008, other sequence-based methods, while providing an abundance of 16S rRNA gene data, typically identified taxa at the genus or higher levels (Citation10, Citation11). HOMIM, for the first time, provided 379 species-level probes capable of identifying 293 predominant oral bacterial species.
HOMINGS, introduced in 2014, provides species-level identification via species probes and genus level identification via genus probes for those sequences not uniquely e-hybridized to a species probe. Genus probes are useful for detection of species at the genus level that are highly conserved and for which design of specific probe sequences used for e-hybridization is difficult, e.g. Streptococcus and Fusobacterium species. These probes are also useful to conduct effective genus level analyses. Also, unknown species may be captured by genus probes, providing further opportunities for species probe development. The 672 species probes and 93 genus probes used for HOMINGS are capable of identifying 597 species and 83 genera (due to multiple probes for certain species or genera), respectively. Determination of total hits for a given genus is thereby achieved by adding hits from corresponding species probes to the genus probe hits.
Our objective was to compare baseline oral microbiome of dental plaque using these two technologies.
Materials and methods
Study population
Patients (n=96) were recruited from our hospital-based dental clinic as a subset of a previous clinical bacteremia study (Citation8, Citation12). All patients had the need for extraction of at least one erupted tooth. Patients were excluded from the study if they had fewer than 10 teeth, had taken a systemic antibiotic within 2 weeks prior to the study, or were immunocompromised. Patient demographics and clinical characteristics are summarized in (Citation8). As changes of clinical parameters were not the focus of this study, we kept the description here as basic.
Table 1. Demographic and clinical characteristics of study cohort
IRB approval and patients’ signed consents were obtained for the study.
Sample collection and DNA extraction
Dental plaque samples were collected at least 1 h before any dental manipulation, such as eating, tooth brushing, or tooth extraction. They were acquired by scraping a dental scaler across the supra-and superficial sub-gingival aspect of 1–4 teeth, with the deepest probing depths, and pooled. The sample was suspended in TE buffer (50 mM Tris HCL, 1 mM EDTA pH 7.6), transferred on ice, and stored at −80°C until furthzer analysis. Bacterial DNA extraction was performed using a modification of QIAamp DNA Mini Kit (QIAGEN, Valencia, CA), as described previously (Citation12) and used for simultaneous analysis by HOMIM and HOMINGS.
HOMIM and HOMINGS platforms
Bacterial DNA samples were used to survey bacterial identification utilizing HOMIM and HOMINGS technologies. HOMIM uses in vitro microarray hybridization technology (Citation13, Citation14). Briefly, 16S rRNA-based probes were printed on aldehyde-coated slides. 16S rRNA gene sequences were polymerase chain reaction (PCR) amplified and labeled with Cy3-dCTP in a second nested PCR. Hybridization was performed overnight followed by washing. Microarray plate probes were scanned and the resulting fluorescence intensities translated into a ‘barcode’ format (Citation10). Individual intensity signals were normalized by comparison to the average signals from universal 16S rRNA gene probes (Citation10). Band intensity was scored by approximation using a 0–5 discrete scale, with 0 corresponding to no detection and 5 to highest presence (relative abundance) of a bacterial species. Species were identified based on a BLAST search of the Human Oral Microbiome Database (Citation2).
HOMINGS employs a ProbeSeq program for species detection with modifications as previously described (Citation15). Briefly, 50 ng of genomic DNA was used for each initial PCR. Amplification of the 16S rRNA gene (V3–V4 region) was followed by purification and processing using a modified next generation sequencing method as described by Caporaso et al. (Citation16) using a Miseq (Illumina, Inc., San Diego, CA). ProbeSeq sequence identification used rRNA-based in silico probes in a BLAST program to determine the species taxa and frequency (Citation15). HOMINGS follows an in silico hybridization process. Species-specific 16S rRNA-based oligonucleotide probes, many of which had originally been designed for HOMIM, were used in a BLAST program, called ProbeSeq for HOMINGS, to identify the frequency of oral bacterial targets. ProbeSeq loads the raw sequence files into a cell array and then loops through the array, one sequence at a time, looking for a ‘string’ (segment of text) that matches one of the oligomers. When a match is found, a running counter starts to give the total number of probe ‘hits’. Partial matches are not considered as a match.
The ProbeSeq process was iterative, that is, the sequences that had not been detected by a single species probe were subsequently processed against genus probes, which consist of two or more species within the genus. All hits were accumulated by species/genus by patient, with a higher hit total interpreted as representing a higher presence of a given species. Modifications of the protocol include the following: DNA was amplified directly in the initial PCR—two rounds of PCR were used in the previous study (Citation15), and chimeric sequences were not removed from analyses for this study. However, in a separate analysis, chimeric sequences ranged from 10 to 15% of the total reads. The relative proportions of detected taxa did not vary significantly (data not shown).
Statistical analysis
HOMIM intensity data (0–5) and HOMINGS hits data (0–300,000 range) were provided as Excel spreadsheets for downstream statistical analysis. HOMIM in vitro signal intensity estimations and HOMINGS e-hybridization hits are not directly comparable. Therefore, the data from both technologies were converted into relative proportions to allow semi-quantitative comparison of their outcomes. Common species total abundance per patient between the two technologies was determined based on species probes information. For the common species, Wilcoxon signed-rank test was used to determine the statistical significance of differences found between HOMIM and HOMINGS relative proportions at phylum, genus, and species levels, using a significance level of <0.05 (FDR [false discovery rate] adjusted p-value). Statistical analyses including outlier determination were performed using XLSTAT-Pro (version 2014.4.06) and/or SAS Enterprise Guide® (version 6.1).
Results
HOMIM and HOMINGS capacities and bacterial identification in dental plaque samples
HOMINGS and HOMIM data were obtained from dental plaque samples of a patient cohort scheduled for single tooth extraction. Demographics and gingival characteristics of these patients are shown in . HOMINGS doubles the capacity for identification of bacterial species and increases the number of identifiable genera by nearly 85% compared to HOMIM, based on use of species and genus probes counts (). In our study of 96 pla que samples, HOMINGS detected 489 species compared to 244 species by HOMIM (). This is consistent with the doubled capacity provided by HOMINGS technology. A greater number of genera were also detected by HOMINGS (i.e. 129) than HOMIM (i.e. 84), corresponding to 53% increase (). Detailed examination of the 129 genera revealed that 59 were represented by both species and genus probes, while 68 (i.e. 105 species) were represented by species probes alone. Unaccounted reads, that is, sequence reads that did not uniquely e-hybridize to species or genus probes, ranged from 4.5 to 36.7% [Mean% (SD): 14% (4.9)] of total sequences per sample and were over 20% for nine of the 96 samples (data not shown).
Table 2. HOMINGS and HOMIM capacities based on Human Oral Taxonomy (HOT) designation.
Table 3. Dental plaque microbiome composition by HOMIM and HOMINGS
Analysis of the common taxa detected by both HOMIM and HOMINGS
Because HOMINGS detected double the number of species, full dataset statistical comparisons at any taxonomy level would be deemed meaningless, as it would simply highlight the obvious. To assess the concordance between the two technologies, we adjusted the HOMINGS dataset to match those species contained in HOMIM dataset to enable relative proportion of global comparisons at the phylum, genus, and species levels. Both HOMIM and HOMINGS identified 198 species in common, corresponding to 74 genera and 10 phyla (). Comparison of the relative proportions at three taxonomic levels (e.g. species, genera, and phyla) obtained by HOMIM and HOMINGS showed no significant overall differences when FDR correction was applied for the multiple comparisons (FDR>0.05) ().
In addition, we compared the total common species (i.e. 198 spp.) relative proportion per patient with regard to total HOMIM intensities or HOMINGS hits for all 96 patients. No significant difference was found (p=0.617) (a). These results did not change when removing patient 26 (Pt26), the most prominent outlier according to HOMINGS total species/genus relative proportions per patient (p=0.805) (b). Pt26 had the highest number of total hits by HOMINGS (116,385, i.e. 2.75% per total 96 patients) compared to a relatively lower total intensity by HOMIM (i.e. 149 total intensity, i.e. 1.09% per total 96 patients). Further analysis showed that Prevotella melaninogenica was responsible for this discrepancy compared to the other 197 common species in Pt26. P. melaninogenica had 60,505 hits representing 52% of the total hits of 116,385 (data not shown).
Fig. 1. HOMINGS versus HOMIM comparison of total taxa relative proportion per patient. Total taxa (i.e. species common to both technologies) relative proportion for a patient was calculated by dividing the sum of the intensity scores (HOMIM) or hits (HOMINGS) of all bacterial taxa of the patient by the total number of these values for all 96 patients. (a) Differences in relative taxa proportions are illustrated by patient to patient line chart (HOMINGS [blue]; HOMIM [red]). These differences were not statistically significant overall (p=0.617, Wilcoxon signed-rank test). (b) Grubbs test identified six patients as outliers, with patient 26 (Pt26) representing the most prominent outlier. Removing Pt26 from the analysis also showed that differences in taxa relative proportions were overall not statistically significant (p=0.805).
![Fig. 1. HOMINGS versus HOMIM comparison of total taxa relative proportion per patient. Total taxa (i.e. species common to both technologies) relative proportion for a patient was calculated by dividing the sum of the intensity scores (HOMIM) or hits (HOMINGS) of all bacterial taxa of the patient by the total number of these values for all 96 patients. (a) Differences in relative taxa proportions are illustrated by patient to patient line chart (HOMINGS [blue]; HOMIM [red]). These differences were not statistically significant overall (p=0.617, Wilcoxon signed-rank test). (b) Grubbs test identified six patients as outliers, with patient 26 (Pt26) representing the most prominent outlier. Removing Pt26 from the analysis also showed that differences in taxa relative proportions were overall not statistically significant (p=0.805).](/cms/asset/6cc79749-6d23-4147-aa91-95d9a1547b57/zjom_a_11821315_f0001_ob.jpg)
Analysis of the HOMINGS added taxa identification capacity compared to HOMIM
Descriptive analysis of HOMINGS added taxa identification capacity is summarized in , and and . At the species level, Porphyromonas gingivalis was the most represented species by HOMINGS (). The proportion of this species was nearly twice that of next most represented species, Dialister invisus. The opposite ratio was observed for the proportions of these two species detected by HOMIM ().
Fig. 2. Dental plaque bacterial genera composition by HOMINGS with added taxa identification capacity compared to HOMIM. *HOMINGS microbiome profiling of dental plaque samples from a patient cohort (n=96) shows a large increase (>100%) in the detection of Actinomyces, Fusobacterium, Leptotrichia, and Prevotella genera compared to HOMIM, likely due to additional genera probes. Relative proportions per genus were calculated based on total hits by HOMINGS or total intensity scores by HOMIM per 96 patients. Genera accounting for 96% of total hits by HOMINGS are represented.
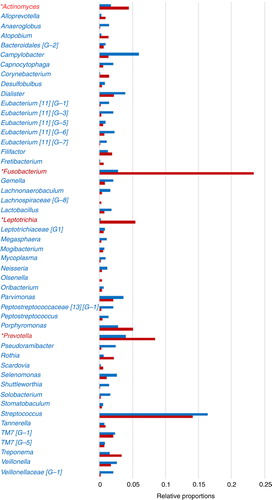
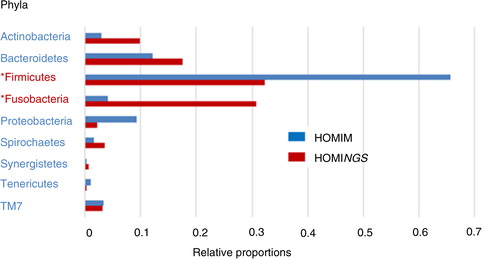
Fig. 3. Dental plaque bacterial phyla composition by HOMINGS with added taxa identification capacity compared to HOMIM. *HOMINGS microbiome profiling of dental plaque samples from a 96 patients cohort shows a large shift in relative proportions for Fusobacteria (3.8% HOMIM vs. 29.4% HOMINGS) and Firmicutes (64.4% HOMIM vs. 32.6% HOMINGS). Relative proportions per phylum were calculated based on total hits by HOMINGS or total intensity score by HOMIM per 96 patients. Chloroflexi, GN02 and SR1 were omitted due to negligible representation.
At the genus level, HOMINGS genus probes accounted for 38.7% of total hits (). Due to this added capacity, larger relative proportions compared to HOMIM were apparent for several genera including Fusobacterium, Prevotella, and Leptotrichia (, ). The highest relative proportion was observed for Fusobacterium, that is, 23.4% (including 2.1% species probes derived genera) by HOMINGS, compared to 2.8% detected by HOMIM (). The next four genera with the highest relative proportion detected by HOMINGS were Streptococcus (14.1% including 6.2% species probes derived genera) followed by Prevotella, Leptotrichia, and Porphyromonas (). Of these genera, Streptococcus yielded similar proportions by HOMIM (16.4%).
At the phylum level, Firmicutes represented twice the proportion by HOMIM (64.4%) compared to HOMINGS (32.6%) (, ). Firmicutes and Fusobacteria were represented in different proportions by HOMIM alone (64.4 and 3.8%), whereas HOMINGS yielded nearly equal representation of these two phyla (32.6 and 29.4%). Additionally, Fusobacteria were detected at a larger proportion by HOMINGS (29.4%) compared to HOMIM (3.8%) (, ). Similarly, Bacteroidetes and Actinobacteria phyla proportions differed to a large extent using HOMIM (11.8 and 3.5%) and were in similar range (17 and 10.9%) using HOMINGS.
Discussion
Identification of bacterial species from various sites in the mouth and quantification of the relative abundance of each species is a daunting task, given the wide diversity of species known to inhabit the human oral cavity. Both HOMIM and HOMINGS provide semi-quantitative identification of oral microbiome bacterial species.
Using the same dental plaque samples in this study, HOMINGS technology expanded the detection of the number of species (via species-level probes) by 100% and the number of genera (via species probes and genus probes) by nearly 85%. This represents a significant improvement toward achieving full knowledge of oral bacterial composition at the species level.
Incorporation of genus probes by HOMINGS represents a significant improvement. This capability gives recognition to the fact that bacterial species do not all provide equal unique e-hybridization potential based on the degree of conservation of the V3–V4 region of the 16S rRNA gene. Examples of highly conserved species that are not effectively uniquely e-hybridized to species-level probes are found in several genera including Fusobacterium and Streptococcus. By using an iterative process to combine species with genus probe e-hybridization events, HOMINGS technology maximizes reads identification. However, for our dental plaque dataset, there was an average of 14% unaccounted reads per patient. These correspond to sequences which either do not e-hybridize to a target probe or e-hybridize to multiple target probes. These sequences provide opportunities for further species probe refinement and new probe development in order to identify all bacterial species present in oral samples to the lowest detection limit possible.
Identification of species-level information is preferable, but in the absence of this, genus level information is useful and informative. In fact, genus probes altogether accounted for nearly 40% of total hits in this study. Therefore, for genera that have species probes but no genus probes, it is unclear whether there are species remaining unaccounted for within the genus. In addition, within the bulk of genus probes total hits, Fusobacterium represented ~20% of hits, followed by, but to a significantly smaller extent, Streptococcus (~8%). Therefore, the largest contribution by HOMINGS with the addition of genus probes is the added capacity of detecting more Fusobacterium compared to HOMIM. Based on analyses of the samples tested, it seems clear that some key taxa of Fusobacterium, Leptotrichia, and/or Actinomyces are not differentiated at the species level. Additionally, similar proportions for Firmicutes with Fusobacteria and Bacteroidetes with Actinobacteria were determined by HOMINGS, but not by HOMIM, as probes for many of these taxa were difficult to construct within proper hybridization parameters, for example, GC content and Tm. Nevertheless, HOMIM phylum level data were in overall agreement with the literature (Citation4), namely, Firmicutes was the most predominant phylum. Results obtained by HOMINGS suggest a more balanced representation of several phyla of dental plaque. However, they might only be representative of the patient population in need for tooth extraction used in our study. Therefore, the results may not be extrapolated to the general population with a better oral hygiene and less dental disease, which clearly may impact the composition of the dental plaque microbiome.
In addition, the introduction of unique e-hybridization events (hit counting) to determine the relative abundance of species and genera is a more precise semi-quantitative process as compared to HOMIM. The fluorescence-based quantification employed in HOMIM provides only a rough estimate of relative proportions on a narrow discrete scale basis (0–5). In contrast, HOMINGS in silico matching of target sequence to probe must be exact for a hit to be counted, resulting in no ambiguity of presence or absence of a species. Thus, a low number of hits determined by HOMINGS would most likely correspond to background intensity or no detection by HOMIM. Taking into account the fact that there is no practical limitation to the number of hits that can be counted, hit counting intrinsically provides a more precise and comprehensive representation of the species relative abundance.
These fundamental changes make direct comparisons of HOMINGS semi-quantitative data with those of HOMIM difficult. A fair comparison cannot be achieved because there is no method to convert in vitro hybridization intensities accurately for each species into e-hybridization hits. Nonetheless, in the present study, a comparison of relative proportions limited to those species found in both HOMIM and HOMINGS demonstrated concordance between the two technologies in that there were no statistically significant differences noted in the overall results at the phylum, genus, or species taxonomic levels. Concordance between the two technologies was also found regarding the total species relative proportion by patient. In essence, HOMINGS partially confirmed HOMIM data obtained from the same dental plaque sample collection, while differences largely out of range might have been anticipated. In this respect, previous studies that have used HOMIM have been helpful to the extent that differences between disease and control groups can be understood on a global microbial community at the species level to the limit of technology accuracy.
In conclusion, the determination of bacterial species composition in the oral cavity will likely move forward in a significant way with the adoption of HOMINGS. Assuming that further improvements of HOMINGS or other bioinformatic technology would enable detection of all the species present in various oral sites, true quantification may be achieved based on the calibration of bacterial DNA input and knowledge of the bacterial genome sizes for the species detected. Refinement of HOMINGS will provide a better identification tool for the oral microbiome and will be beneficial to the understanding of distant site infections originating from the oral cavity, such as IE and PJIs.
Conflict of interest and funding
The authors declare no conflict of interest.
Author contributions
J-L.C. Mougeot designed the study and wrote the manuscript. C.B. Stevens contributed to data analysis and writing of the manuscript. S.L. Cotton and K. Krishnan performed ProbeSeq/HOMINGS analysis. D.S. Morton performed sample processing. M.T. Brennan directed the clinical staff for patient recruitment and samples collection. P.B. Lockhart designed the bacteremia study and conducted the clinical study. B.J. Paster designed and led ProbeSeq/HOMINGS analysis and reviewed the manuscript. F. B. Mougeot designed the study and wrote the manuscript.
Acknowledgements
This study was supported by National Institute of Dental and Craniofacial Research/National Institute of Health grants # R01 DE13559-01, R01 DE021565, and Carolinas HealthCare System (CHS) Research Fund. We thank the residents and clinical coordinators at CMC Dental Clinic for sample collection, Megan Templin for her help in statistical analysis, and P. Hanjra for her help with the database.
References
- Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010; 2010: baq013.
- HOMD database: Human Oral Microbiome Database, All Human Microbial Taxa. Available from: http://www.homd.org/modules.php?op=modload&name=HOMD&file=index [cited 26 June 2015]..
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu WH, etal. The human oral microbiome. J Bacteriol. 2010; 192: 5002–17.
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, etal. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001; 183: 3770–83.
- Belstrøm D, Fiehn NE, Nielsen CH, Klepac-Ceraj V, Paster BJ, Twetman S, etal. Differentiation of salivary bacterial profiles of subjects with periodontitis and dental caries. J Oral Microbiol. 2015; 7: 27429. doi: http://dx.doi.org/10.3402/jom.v7.27429.
- Shaddox LM, Huang H, Lin T, Hou W, Harrison PL, Aukhil I, etal. Microbiological characterization in children with aggressive periodontitis. J Dent Res. 2012; 91: 927–33.
- Mougeot FB, Saunders SE, Brennan MT, Lockhart PB. Associations between bacteremia from oral sources and distant-site infections: tooth brushing versus single tooth extraction. Oral Surg Oral Med Oral Pathol Oral Radiol. 2015; 119: 430–5.
- Lockhart PB, Brennan MT, Sasser HC, Fox PC, Paster BJ, Bahrani-Mougeot FK. Bacteremia associated with toothbrushing and dental extraction. Circulation. 2008; 117: 3118–25.
- HOMINGS, Forsyth Institute. Available from: http://homings.forsyth.org/index2.html [cited 26 June 2015]..
- Cotton SL, Klepac-Ceraj V, Murphy CM, Kokaras AS, Paster BJ. Species level determination of high-throughput sequencing data using HOMIM Probes. J Dent Res (AADR). 2013; 92: 3828.
- Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, etal. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012; 13: R42.
- Bahrani-Mougeot FK, Paster BJ, Coleman S, Ashar J, Barbuto S, Lockhart PB. Diverse and novel oral bacterial species in blood following dental procedures. J Clin Microbiol. 2008; 46: 2129–32.
- Colombo APV, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, etal. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis and periodontal health using the Human Oral Microbe Identification Microarray (HOMIM). J Periodontol. 2009; 80: 1421–32.
- Colombo APV, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, etal. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the Human Oral Microbe Identification Microarray. J Periodontol. 2012; 83: 1279–87.
- Gomes BP, Berber VB, Kokaras AS, Chen T, Paster BJ. Microbiomes of endodontic–periodontal lesions before and after chemomechanical preparation. J Endod. 2015; 41: 1975–84. doi: http://dx.doi.org/10.1016/j.joen.2015.08.022 [PubMed Abstract].
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, etal. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS. 2011; 108(Suppl 1): 4516–22.
