Abstract
Introduction
Multidrug resistance (MDR) and emergence of extended-spectrum β-lactamases (ESBLs) that mediate resistance to β-lactam drugs among Escherichia coli and other uropathogens have been reported worldwide. However, there is little information on the detection of ESBLs genes in E. coli from patients with urinary tract infections (UTIs) in the Arab countries using polymerase chain reaction (PCR), and in Libya such information is lacking.
Methods
All patients attending Zawiya Teaching Hospital in Zawiya city between November 2012 and June 2013 suspected of having UTIs and from whom midstream urine samples were taken as part of the clinical workup were included in this prospective study. Samples were examined for uropathogens by standard bacteriological procedures. VITEK-2 automated microbiology system was used to identify the isolated uropathogens and determine the susceptibility of E. coli and Klebsiella spp. isolates to antimicrobials. In addition, phenotypically ESBLs-positive E. coli isolates were tested for ESBLs genes by PCR.
Results
The present study enrolled 1,790 patients with UTIs. Uropathogens were found in 371 (20.7%) urine specimens examined. Mixed pathogens were detected in two specimens with 373 total pathogens isolated. E. coli and Klebsiella spp. were the predominant uropathogens at 55.8% (208/373) and 18.5% (69/373), respectively. Other pathogens were detected in 25.7% (96/373) of urine samples. Of the E. coli and Klebsiella spp. tested, 69.2 and 100% were resistant to ampicillin, 6.7 and 33.3% to ceftriaxone, and 23.1 and 17.4% to ciprofloxacin, respectively. MDR (resistance to ≥3 antimicrobial groups) was found in 69 (33.2%) of E. coli and in 29 (42%) of Klebsiella spp. isolates. ESBLs were detected phenotypically in 14 (6.7%) of E. coli and in 15 (21.7%) of Klebsiella spp. isolates. Thirteen out of the 14 phenotypically ESBL-positive E. coli were positive for ESBL genes by PCR. blaTEM gene was detected in seven isolates, blaOXA gene in 10 isolates and blaCTX-M gene in six isolates. blaSHV gene was not detected in the present study.
Conclusion
The isolation of MDR ESBL-producing uropathogens undoubtedly will limit the choices clinicians have to treat their patients with UTIs. Therefore, there is an urgent need for surveillance studies on antimicrobial resistance and prevalence of ESBLs among uropathogens to guide the clinical treatment of UTIs in Libya in the future.
To access the supplementary material for this article please see Supplementary files under ‘Article Tools’
One of the most common reasons that patients in the community seek medical care and prescribed antibiotics is urinary tract infections (UTIs) (Citation1). Most UTIs are caused by members of the family Enterobacteriaceae mainly Escherichia coli and Klebsiella spp. High resistance rates to antimicrobials used in the treatment of UTIs among E. coli and other uropathogens have been reported worldwide, particularly in developing countries including Libya (Citation2). In recent years, the problem worsened due to the emergence of extended-spectrum β-lactamases (ESBLs) that mediate resistance to β-lactam antimicrobials, especially third generation cephalosporins among these organisms. Genes responsible for ESBLs production arise by point mutation at the active site of the earlier β-lactamases (e.g. TEM-1 and SHV-1 enzymes), and they are usually plasmid mediated (Citation3, Citation4). In addition, ESBLs-positive Gram-negative bacteria often carry genes that confer high levels of resistance to many other antibiotics (e.g. fluoroquinolones and aminoglycosides) (Citation5). Previous use of penicillins, cephalosporins or quinolones is found to be a risk factor, among others, for the development of ESBLs-producing bacteria in non-hospitalized patients with community-acquired UTIs (Citation6).
The epidemiology of antibiotic resistant bacteria, however, varies from region to region, from speciality to speciality, from infection type to infection type, and from year to year (Citation7). Therefore, identification and determination of antimicrobial susceptibility of bacterial pathogens in the local healthcare setting may aid the clinician in selecting the appropriate antimicrobial agent (s) to treat his patients.
Studies on the causative agents of UTIs in Libya and their susceptibility to antimicrobial are few and were mainly carried out in the two main cities in Libya (i.e. Tripoli and Benghazi) (Citation8, Citation9). Furthermore, there is little information on the detection of ESBLs genes in E. coli from patients with UTIs in the Arab countries using molecular methods and in Libya such information is lacking. Therefore, the aim of the present investigation was to determine the prevalence of organisms associated with UTIs in patients attending Zawiya Teaching Hospital (ZTH), their susceptibility to antimicrobial agents, and the presence of ESBLs genes in E. coli isolates using polymerase chain reaction (PCR).
Methods
Patients
All patients attending Zawiya Teaching Hospital in Zawiya city suspected of having UTIs and from whom urine samples were taken as part of the clinical workup were included in this prospective study. The study was carried out between November 2012 and June 2013. Zawiya city is located in northwestern Libya, situated on the Libyan coastline of the Mediterranean Sea about 45 km west of Tripoli with an estimated population of about 200,000 people. ZTH has 400 beds and frequented mainly by inhabitants of Zawiya city and surrounding cities and villages.
In this investigation, urine specimens were collected under approved ethical standards and the study was reviewed and approved by the Academy of Graduate Studies, Janzour, Tripoli, Libya.
Specimen collection
Urine specimens were obtained from all patients included in the study with the assistance of a medical doctor or senior nurse. Midstream urine was collected from patients aged more than 1 year in a sterile container and processed in the laboratory within 2 h of collection. Urine specimens were obtained by suprapubic aspiration from children who are not toilet trained and proceed as above. The consent of the patients or that of their guardians was obtained before urine specimens were collected.
Isolation and identification of uropathogens
Using calibrated loops (10−3 ml, Last Loop Sp, Italiana, Italy), urine specimens were inoculated on to MacConkey and blood agar plates. After 18–24 h incubation at 37°C, the number of colony forming units (cfu) was counted, and urine samples giving ≥105 cfu/ml of urine were considered significant. Isolated colonies from significant plates were identified using the VITEK-2 automated microbiology system (bioMérieux, Inc., Durham, NC, USA) employing the GN and GP colorimetric reagent cards that are incubated and interpreted automatically. The GN card is used for identification of Gram-negative fermenting and non-fermenting bacilli, and the GP card is used for identification of Gram-positive cocci and non-spore forming bacilli.
Reference strains of E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as controls for the Gram-negative bacteria. Staphylococcus aureus ATCC 25923 and Enterococcus faecalis ATCC 929212 were used as Gram-positive control strains.
Antimicrobial susceptibility testing
The VITEK-2 Gram-negative antimicrobial susceptibility testing card (GN AST, bioMérieux, Inc.) was used to determine the susceptibility of E. coli and Klebsiella spp. isolates to different antimicrobial agents. E. coli ATCC 25922 was used as a control organism. VITEK 2 provides AST results for antimicrobials as susceptible, intermediate susceptible, and resistant and is interpreted according to CLSI criteria (Citation10). In the present study, only resistant results were presented.
ESBLs testing
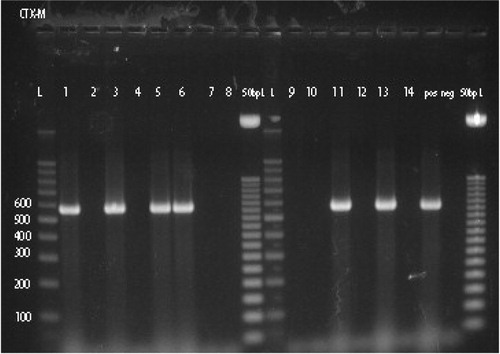
Production of ESBLs by E. coli and Klebsiella spp. isolates was determined phenotypically by VITEK 2 (bioMérieux, Inc.). E. coli isolates positive for ESBLs phenotypically were tested by PCR technique for ESBLs genes as previously reported (Citation11, Citation12). Briefly, whole cell lysate of an overnight bacterial culture was prepared by boiling method as described previously (Citation13). Amplification was done in a 50-ul reaction mixture containing 50 pmol primers (as listed in ), 0.25 mM deoxyribonucleotide, 1.5 mM MgCl2, 1X GoTaq reaction buffer (Promega, Madison, WI, USA), and 0.2 U GoTaq DNA polymerase. Amplification conditions included 30 cycles of 95°C for 30 sec, 55°C for 1 min and 72°C for 1 min. This was followed by a final extension step at 72°C 30 cycles. PCR products were visualized by gel electrophoresis on a 2.0% agarose (Sigma-Aldrich, Steinheim, Germany) using 1X Tris-acetate-EDTA buffer.
Table 1 Polymerase chain reaction (PCR) primer sequences for the detection of extended-spectrum β-lactamases genes in E. coli
Statistical analysis
For statistical analysis, Epi Info 2000 software (Centers for Disease Control and Prevention) was employed. P-values were calculated using the Chi-square test. P<0.05 was considered to be statistically significant.
Results
The present study enrolled 1,790 patients (1,151 females and 639 males) with UTIs. The age of the patients ranged from 3 days to 93 years (mean 25 years). Of these patients, 1,134 were outpatients and 656 inpatients.
Uropathogens were isolated from 371 (20.7%) urine specimens examined. Co-infection was determined in two patient's urine specimens as two different organisms were isolated from each of them with a total of 373 uropathogens being isolated. Predominance of E. coli and Klebsiella spp. among isolated uropathogens was observed at 55.8% (208/373) and 18.5% (69/373), respectively. shows the uropathogens isolated from patients with UTIs attending ZTH.
Table 2 Uropathogens isolates from patients (n=1,790) with UTIs
According to gender and type of hospital admission, E. coli and total uropathogens were significantly detected in female patients compared with male patients and in outpatients compared with inpatients with UTIs. Distribution of E. coli, Klebsiella spp., and other uropathogens isolated from patients with UTIs according to gender and to type of hospital admission is shown in .
Table 3 Distribution of E. coli, Klebsiella spp., and other uropathogens isolated from patients with UTIs according to gender and type of hospital admission
Only E. coli and Klebsiella spp. isolates were tested for antimicrobial susceptibility and ESBL production. Of the E. coli and Klebsiella spp. tested, 69.2 and 100% were resistant to ampicillin, 30.8 and 31.9% to ampicillin/sulbactam, 6.7 and 33.3% to ceftriaxone, 23.1 and 17.4% to ciprofloxacin, and 37 and 24.6% to trimethoprim/sulfamethoxazole, respectively. Nitrofurantoin showed good activity against E. coli isolates (1.9% resistant) but not against Klebsiella spp. (30.4% resistant). shows the antimicrobial resistance patterns of E. coli and Klebsiella spp. from patients with UTIs attending ZTH. From it can be observed that Klebsiella spp. isolates were significantly more resistant to most of the antimicrobial agents tested than E. coli isolates. In addition, multidrug resistance (MDR, resistance to three or more antimicrobial groups) was found in 69 (33.2%) of 208 E. coli isolates and in 29 (42%) of 69 Klebsiella spp. isolates.
Table 4 Antimicrobial resistance patterns of E. coli and Klebsiella spp. from patients with UTIs
ESBLs were detected in 6.7% (14/208) of E. coli and in 21.7% (15/69) of Klebsiella spp. isolates using phenotypic procedure. The detection rate of ESBLs among Klebsiella spp. is significantly higher than the detection rate of ESBLs among E. coli isolates (P<0.0005, OR=3.85). Thirteen of the 14 phenotypically ESBLs-positive E. coli were positive for ESBLs genes by PCR. bla TEM gene was detected in seven isolates, bla OXA gene in 10 isolates and bla CTX-M gene in six isolates (). bla SHV gene was not detected in the present study. Combination of two ESBL genes in a single isolate occurred in four isolates (bla TEM and bla OXA in 1, bla TEM and bla CTX-M in 2, and bla OXA and bla CTX-M in 1) and three ESBL genes combination in three isolates (bla TEM, bla OXA, and bla CTX-M in 3). Distribution of ESBLs genes among 14 phenotypically ESBLs-positive E. coli from patients with UTI is shown in Supplementary Table 1.
Discussion
In the present investigation, uropathogens were detected in more than 20% of patients with UTIs attending ZTH with a predominance of E. coli and Klebsiella spp. Previous studies from different countries reported different rates of uropathogens among patients with UTIs. A study from Kuwait reported uropathogens in 9.2% (4,696/51,562) that cause UTIs in two large teaching hospitals over a period of 1 year (Citation14). The authors reported the predominance of E. coli (47%) followed by Candida spp. (10.8%) and Klebsiella spp. (9.6%). Bercion et al. (Citation15) performed a retrospective analysis of all cases of confirmed UTIs due to Enterobacteriaceae in outpatients attending the Institut Pasteur de Bangui, Central African Republic, between 2004 and 2006. They found bacterial pathogens in 560 of the 5,128 (10.9%) patients with predominance of E. coli (64%) and Klebsiella spp. (10%). Kashef et al. (Citation16) examined 13,333 mid-stream urine samples collected from suspected cases of UTI in Tehran, Iran. Uropathogens were found in 6.3% (840/13,333) with a predominance of E. coli (68.8%, 578/840) followed by Proteus spp. (12.4%, 104/840), and Klebsiella spp. (9.6%, 81/840).
Other uropathogens isolated from ZTH patients include P. mirabilis (5.9%), Enterobacter spp. (4.8%), and Citrobacter spp. (3.2%). A study from Ife, Nigeria reported uropathogens in 36 (12%) of the 301 patients with UTIs (Citation17). The majority of 36 bacterial isolates obtained were E. coli leading to 52.8% of the cases of UTIs, followed by Klebsiella spp. (25%), P. mirabilis (13.9%), Strep.(Enterococcus) faecalis (5.6%), and Ps. aeruginosa (2.8%).
We found E. coli and total uropathogens were significantly isolated from female patients compared to male patients (). Other investigators found no significant difference in the distribution of isolated uropathogens between genders (Citation15, Citation18). However, an Iranian study reported that E. coli occurred significantly more frequently in women (76.5% in women compared to 62.4% in men; P<0.05), whereas the prevalence of UTIs due to K. pneumoniae and Ps. aeruginosa was higher in men than in women (22.6 and 7.5% in men compared to 9.9 and 1.4% in women, respectively; P<0.01) (Citation19). No significant differences (P>0.05) in the distribution of Klebsiella spp. and other uropathogens between genders were observed in this study. Also, E. coli and total uropathogens were significantly isolated from outpatients than from inpatients with UTIs frequenting ZTH. Other studies found no significant difference in the isolation of uropathogens from outpatients and inpatients with UTIs (Citation20).
Differences in the reported isolation rates of uropathogens from different regions may be due to differences in age, gender and number of patients examined, types of uropathogens examined (e.g. some studies did not include Candida spp. in their search as in the present study), and types and number of media used in the isolation of uropathogens. Some studies used blood and MacConkey agars (present study) or blood agar and eosin methylene blue agar plates (Citation16). Other studies used only bromocresol purple agar (Citation15).
In the present study, antimicrobial susceptibility testing showed high resistance rates to ampicillin among E. coli and Klebsiella spp. isolates. However, lower resistance rates than previously reported from Libya to trimethoprim/sulfamethoxazole among E. coli (37%) and Klebsiella spp. (24.6%) were observed. In the mid-1990s, Ghenghesh et al. examined 538 E. coli isolates from patients with UTIs in Tripoli for their susceptibility to antimicrobial agents (Citation8). Of the isolates examined 74% were resistant to ampicillin, 7% to gentamicin, 25% to nitrofurantoin, and 45% to trimethoprim/sulfamethoxazole. Tobgi et al. tested the susceptibility of 148 E. coli isolated from urine specimens from outpatients attending Al-Jalla Hospital in Benghazi to antibiotics (Citation9). They reported that 75% of them were resistant to ampicillin, 18% to gentamicin and 81% to trimethoprim/sulfamethoxazole.
The lower rates observed with trimethoprim/sulfamethoxazole may be due to less use of the drug in treating bacterial infections (including UTIs) in Libya. Newer antibiotics like third generation cephalosporins (e.g. ceftriaxone and cefotaxime) and fluoroquinolones (e.g. ciprofloxacin and norfloxacin) have been more widely used in recent years. This observation can be supported by the findings of the present investigation of high resistance rates to ciprofloxacin among E. coli and Klebsiella spp., and to the cephalosporins such as ceftazidime, ceftriaxone, and cefepime among Klebsiella spp. ().
Studies from other countries also reported high resistance rates to different classes of antimicrobials. A study from Mexico examined 119 E. coli isolates obtained from urine specimens of outpatients with clinical diagnosis of uncomplicated UTIs from 2004 to 2007 (Citation21). Resistance to ampicillin was found in 83.7% of the isolates, to the fluoroquinolone group (i.e. ciprofloxacin, nalidixic acid, ofloxacin, and norfloxacin) in 55.5–60.6%, and to trimethoprim/sulfamethoxazole in 56.4%. In addition, 30.2% of isolates were multidrug-resistant.
MDR was detected in 32.2% of E. coli and 42% of Klebsiella spp. isolated from patients with UTIs attending ZTH. Alzohairy and Khadri reported higher rates of MDR among E. coli (53.6%) and Klebsiella spp. (50%) isolated from community and hospital-acquired UTIs in Qassim province, Saudi Arabia (Citation22).
A previous study from Benghazi examined 119 strains of E. coli isolated from patients with UTIs (14 from HIV-positive patients) for ESBLs production using phenotypic method (Citation23). Only four (3.4%) E. coli strains (two from HIV-negative and two from HIV-positive patients) were found positive for ESBLs. In the present study, ESBLs were detected at a higher rate of 6.7% among E. coli isolates from patients with UTIs attending ZTH using phenotypic techniques.
Other investigators from the Middle East and Africa reported similar findings. A study from Saudi Arabia reported ESBLs in 6.1% (570/9,409) (Citation24). ESBLs were detected in 8.1% (29/357) of E. coli strains isolated from outpatients with suspected uncomplicated UTIs in Central African Republic (Citation15). However, much higher rates of ESBLs have been reported from other countries in Africa. Ejikeugwu et al. examined a total of 83 non-duplicate Escherichia coli isolates from urine specimens of patients suspected of having community-acquired UTIs in Enugu State, Nigeria, from August 2010 to July 2011 (Citation25). They detected ESBLs in 23 (27.7%) of the isolates investigated using the double disk synergy test method. Although the prevalence of ESBLs-producing bacteria is a worldwide problem, the geographical distribution can vary according to countries and institutions (Citation26).
Several studies suggested that urine can be an important source of ESBLs-producing E. coli (Citation27, Citation28). To our knowledge, this the first report of ESBLs detection in E. coli isolates from UTIs in Libya using PCR. Predominance of bla OXA (10/13) followed by bla TEM (7/13) and bla CTX-M (6/13) was observed in this study. Bourjilat et al. detected seven (1.3%) ESBLs-producing E. coli among 535 E. coli isolates from urine specimens of patients with community-acquired urinary infections in Casablanca, Morocco, during July 2004 to July 2007 (Citation29). Employing PCR the investigators detected genes encoding bla CTX-M in most of them (6/7).
The findings of this study showed that E. coli and Klebsiella spp. were the predominant uropathogens causing UTIs in patients frequenting ZTH. High rates of resistance to commonly used and newer antimicrobial agents were observed among these organisms; with Klebsiella spp. isolates being significantly more resistant to these drugs than E. coli isolates. ESBLs and high rates of MDR were observed in E. coli and Klebsiella spp. isolates. In addition, genes coding for ESBLs were detected in E. coli using PCR with a predominance of bla OXA followed by bla TEM and bla CTX-M.
The isolation of MDR ESBLs-producing uropathogens from UTIs patients undoubtedly will limit the choices clinicians have to treat their patients. Of the antimicrobial tested imipenem, meropenem, amikacin, and cefoxitin showed excellent activity against both E. coli and Klebsiella spp. isolates. In addition, third generation cephalosporins and nitrofurantoin demonstrated good activity against E. coli isolates. However, treatment should be guided by the results of antimicrobial susceptibility testing for each individual patient.
Antimicrobial resistance among uropathogens is a serious health problem. Education of clinicians and other healthcare workers on the health risks associated with the problem and on the benefits of prudent use of antimicrobials will help in tackling such a problem. Furthermore, there is an urgent need for surveillance studies on antimicrobial resistance and prevalence of ESBLs among uropathogens to guide the clinical treatment of UTIs in Libya in the future.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
Supplementary Table
Download PDF (149.3 KB)Notes
To access the supplementary material for this article please see Supplementary files under ‘Article Tools’
References
- Mazzulli T. Diagnosis and management of simple and complicated urinary tract infections (UTIs). Can J Urol. 2012; 19: 42–8. [PubMed Abstract].
- Ghenghesh KS, Rahouma A, Tawil K, Zorgani A, Franka E. Antimicrobial resistance in Libya: 1970–2011. Libyan J Med. 2013; 8: 20567. http://dx.doi.org/10.3402/ljm.v8i0.20567.
- Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001; 14: 933–51.
- Pitout JDD, Nordmann P, Laupland KB, Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J Antimicrob Chemother. 2005; 56: 52–9.
- Paterson DL. Recommendation for treatment of severe infections caused by Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs). Clin Microbiol Infect. 2000; 6: 460–3.
- Colodner R, Rock W, Chazan B, Keller N, Guy N, Sakran W, etal. Risk factors for the development of extended-spectrum beta-lactamase-producing bacteria in nonhospitalized patients. Eur J Clin Microbiol Infect Dis. 2004; 23: 163–7.
- Wagenlehner FME, Naber KG. Antibiotics and resistance of uropathogens. EAU Update Ser. 2004; 2: 125–35.
- Ghenghesh KS, Altomi AS, Gashout S, Abouhagar B. High antimicrobial-resistance rates of Escherichia coli from urine specimens in Tripoli-Libya. Garyounis Med J. 2003; 20: 89–93.
- Tobgi RS, Taher IAA, Ali MA. Antibiotic susceptibility of uropathogens in Benghazi, Libya. Jamahiriya Med J. 2001; 1: 46–9.
- Clinical and Laboratory Standards Institute (CLSI). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically . 2009; Wayne, PA: Clinical and Laboratory Standards Institute. Approved standard M07-A8.
- Chen S, Zhao S, White DG, Schroeder CM, Lu R, Yang H, etal. Characterization of multiple-antimicrobial-resistant Salmonella serovars isolated from retail meats. Appl Environ Microbiol. 2004; 70: 1–7.
- Sharma M, Pathak S, Srivastava P. Prevalence and antibiogram of extended-spectrum β-lactamase (ESBL) producing Gram-negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. J Clin Diag Res. 2013; 7: 2173–7.
- Liu PY, Lau YJ, Hu BS, Shyr JM, Shi ZY, Tsai WS, etal. Analysis of clonal relationships among isolates of Shigella sonnei by different molecular typing methods. J Clin Microbiol. 1995; 33: 1779–83. [PubMed Abstract] [PubMed CentralFull Text].
- Al Sweih N, Jamal W, Rotimi VO. Spectrum and antibiotic resistance of uropathogens isolated from hospital and community patients with urinary tract infections in two large hospitals in Kuwait. Med Princ Pract. 2005; 14: 401–7.
- Bercion R, Mossoro-Kpinde D, Manirakiza A, Le Faou A. Increasing prevalence of antimicrobial resistance among Enterobacteriaceae uropathogens in Bangui, Central African Republic. J Infect Dev Ctries. 2009; 3: 187–90.
- Kashef N, Djavid GE, Shahbazi S. Antimicrobial susceptibility patterns of community-acquired uropathogens in Tehran, Iran. J Infect Dev Ctries. 2010; 4: 202–6.
- Aiyegoro OA, Igbinosa OO, Ogunmwonyi IN, Odjadjare EE, Igbinosa OE, Okoh AI. Incidence of urinary tract infections (UTI) among children and adolescents in Ile-Ife, Nigeria. Afr J Microbiol Res. 2007; 1: 13–9.
- Ahmad S, Mubaraki AM. Antimicrobial susceptibility of Gram negative organisms in urine cultures at Armed Forces Hospital in Saudi Arabia. Infect Dis J Pakistan. 2011; 20: 357–60.
- Farajnia S, Alikhani MY, Ghotaslou R, Naghili B, Nakhlband A. Causative agents and antimicrobial susceptibilities of urinary tract infections in the northwest of Iran. Intern J Infect Dis. 2009; 13: 140–4.
- Aboderin OA, Abdu AR, Odetoyin BW, Lamikanra A. Antimicrobial resistance in Escherichia coli strains from urinary tract infections. J Nat Med Assoc. 2009; 101: 1268–73.
- Molina-López J, Aparicio-Ozores G, Ribas-Aparicio RM, Gavilanes-Parra S, Chávez-Berrocal ME, Hernández-Castro R, etal. Drug resistance, serotypes, and phylogenetic groups among uropathogenic Escherichia coli including O25-ST131 in Mexico City. J Infect Dev Ctries. 2011; 5: 840–9.
- Alzohairy M, Khadri H. Frequency and antibiotic susceptibility pattern of uropathogens isolated from community and hospital-acquired infections in Saudi Arabia-a prospective case study. Brit J Med Med Res. 2011; 1: 45–56.
- Buzayan MM, Tobgi RS, Taher I. Detection of extended spectrum ß-lactamases among urinary Escherichia coli and Klebsiella pneumoniae from two centres. Jamahiriya Med J. 2010; 10: 10–6.
- Somily AM, Habib HA, Absar MM, Arshad MZ, Manneh K, Al Subaie SS, etal. ESBL-producing Escherichia coli and Klebsiella pneumoniae at a tertiary care hospital in Saudi Arabia. J Infect Dev Ctries. 2014; 8: 1129–36.
- Ejikeugwu PC, Ikegbunam NM, Ugwu CM, Iroha IR, Esimone CO. Extended-spectrum β-lactamase-producing Escherichia coli isolates from suspected community acquired urinary tract infections. Eur J Sci Res. 2012; 84: 565–71.
- Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 2008; 13: 19044. [PubMed Abstract].
- Harada Y, Morinaga Y, Yamada K, Migiyama Y, Nagaoka K, Migiyama Y, etal. Clinical and molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae and Escherichia Coli in a Japanese Tertiary Hospital. J Med Microbiol Diag. 2013; 2: 127.
- Shigemura K, Tanaka K, Adachi M, Yamashita M, Arakawa S, Fujisawa M. Chronological change of antibiotic use and antibiotic resistance in Escherichia coli causing urinary tract infections. J Infect Chemother. 2011; 17: 646–51.
- Bourjilat F, Bouchrif B, Dersi N, Claude JDPG, Amarouch H, Timinouni M. Emergence of extended-spectrum beta-lactamase-producing Escherichia coli in community-acquired urinary infections in Casablanca, Morocco. J Infect Dev Ctries. 2011; 5: 850–5. [PubMed Abstract].