Abstract
Fructophilic lactic acid bacteria (FLAB) are a special group of lactic acid bacteria (LAB), which prefer fructose but not glucose as growth substrate. They are found in fructose-rich niches, e.g. flowers, fruits, and fermented foods made from fruits. Quite recently, they were found in the gastrointestinal tracts of animals consuming fructose, which were bumblebees, tropical fruit flies, and Camponotus ants. These suggest that all natural sources that are rich in fructose are possible their habitats. Fructobacillus spp., formerly classified as Leuconostoc spp., are representatives of these microorganisms, and Lactobacillus kunkeei has also been classified as FLAB. They share several unique biochemical characteristics, which have not been found in LAB inhabited in other niches. FLAB grow well on fructose but very poor on glucose. These organisms grow well on glucose only when external electron accepters, e.g. pyruvate or oxygen, are available. LAB have been shown to have specific evolution to adapt to their niches and have several niche-specific characteristics. FLAB must have fructophilic evolution during adaptation to fructose-rich niches. FLAB are unique food-related LAB, suggesting a great potential for future food and feed applications.
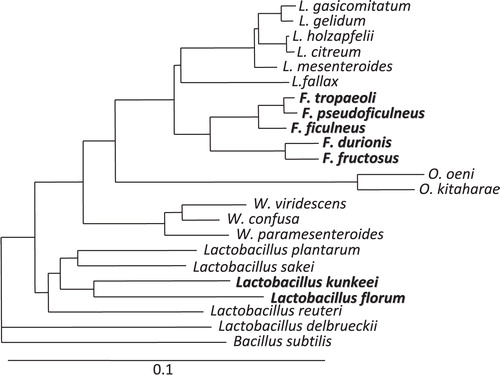
Lactic acid bacteria (LAB) can be found in various environments including fermented foods and gastrointestinal tracts of human and animals. Fructophilic lactic acid bacteria (FLAB) are a special group of LAB, which like fructose as a growth substrate Citation1. They are found in fructose-rich niches, e.g. flowers, fruits, and fermented foods made from fruits. Fructobacillus spp. are representatives of these microorganisms. They had been originally classified as Leuconostoc species Citation2–Citation4 and later reclassified as Fructobacillus species based on their phylogenetic positions and biochemical and morphological characteristics (; 5). Lactobacillus kunkeei is only the fructophilic species classified in the genus Lactobacillus at present (). This species was originally isolated from wine, and at the time of proposal of the species, its fructophilic characteristics had not been discussed Citation6. The species was later characterized as FLAB Citation7. Lactobacillus florum shares similar characteristics with FLAB, but it is differentiated from FLAB based on several biochemical characteristics. Thus, this organism is now classified as facultatively FLAB Citation1 Citation8.
Niche
Fructophilic lactic acid bacteria have been found in various fructose-rich niches until now. Fructobacillus fructosus, type species of the genus Fructobacillus, was originally isolated from a flower in Japan in 1956 Citation9, and isolation of the species had not been reported for over 50 years. The species has been isolated from flower, wine, and taberna, a beverage produced in Mexico Citation1 Citation10 Citation11. Fructobacillus pseudoficulneus is the most frequently seen Fructobacillus species in natural sources, and it has been isolated from figs, bananas, flowers, and taberna Citation1 Citation3 Citation11. Fructobacillus ficulneus and Fructobacillus durionis have been isolated only from figs and tempoyak (fermented durian condiment), respectively Citation2 Citation4. Fructobacillus tropaeoli is a recently described species from flowers Citation12. Recently, a few studies have suggested that cocoa bean fermentation is a rich source of Fructobacillus species Citation13–Citation15. L. kunkeei has been isolated from wine, honey, and flowers Citation1 Citation7. Interestingly, these FLAB species have also been found in the gastrointestinal tracts of several insects, e.g. bumblebees, tropical fruit flies, and Camponotus ants Citation16–Citation18. Bumblebees and tropical fruit flies are flower-related insects, and their diets are rich in fructose. Camponotus ants consume honeydew containing fructose obtained from aphids. These findings suggest that the gastrointestinal tracts of insects consuming large amounts of fructose can be colonized by various FLAB. Such findings are of interest as FLAB prefer aerobic conditions over anaerobic conditions for their growth. The organisms cannot grow under anaerobic conditions if available carbon source is glucose and thus possibly colonize the intestinal tract of vegetarian subjects.
Biochemical characteristics
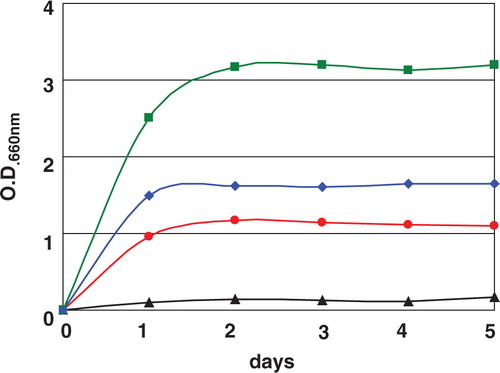
Fructophilic lactic acid bacteria have very unique biochemical characteristics when compared to other LAB. FLAB grow very well on fructose, but growth is very poor on glucose (), although glucose is the most general substrate for almost all LAB. FLAB produce mannitol from fructose, and they grow well on glucose in the presence of pyruvate or under aerobic conditions (). These results mean that FLAB need electron acceptor for glucose metabolism, and pyruvate and oxygen are used as electron acceptor. These organisms can grow well on agar medium containing glucose as sole carbon source under aerobic conditions but hardly grow under anaerobic conditions Citation1 Citation12. These characteristics easily distinguish FLAB from other LAB. Facultatively FLAB species, L. florum, grows well on fructose and on glucose in the presence of electron acceptor as same as other FLAB. However, this organism is differentiated from FLAB based on its growth characteristics on glucose without electron acceptor, meaning that L. florum can grow on glucose but at delayed growth rate. Based on the carbohydrate fermentation test, FLAB can metabolize only a limited number of carbohydrates Citation1. They are only two to five. Most of the Fructobacillus species can metabolize only fructose, glucose, and mannitol. Metabolism of fructose was done within 1 day, and that of glucose took 2–4 days Citation1.
Fig. 2. Growth characteristics of Fructobacillus tropaeoli F214-1T on fructose (![]()
All FLAB produce gas from glucose, indicating that they are obligately heterofermentative LAB. However, FLAB are differentiated from other obligately heterofermentative LAB by production of acetate instead of ethanol Citation1 Citation5 Citation7. Obligately heterofermentative LAB normally use phosphoketolase pathway for glucose metabolism. Acetyl phosphate arisen from cleaving of xylulose-5-phosphate by phosphoketolase is converted to ethanol via acetyl-CoA and acetaldehyde in the pathway. AdhE protein coded on adhE gene is needed for the conversion Citation19. However, Fructobacillus spp. do not have adhE gene (unpublished data). Because of this absence, Fructobacillus spp. produce acetic acid instead of ethanol. Conversion of acetyl phosphate to ethanol is an important step to oxidize NADH to NAD, for which NADH is produced in upper stream of the phosphoketolase pathway. Thus, the missing step leads to shortage of NADH for glucose metabolism in the pathway. Therefore, FLAB would need electron acceptors for glucose metabolism. This finding correlates well with their growth characteristics.
Niche-specific characteristics of LAB
Lactic acid bacteria can be found in various niches, and several studies suggested that LAB have adapted to their niches for their lives Citation20 Citation21. Diary LAB, e.g. Lactobacillus delbrueckii subsp. bulgaricus, Lactobacillus helveticus, Streptococcus thermophilus, prefer lactose over glucose as growth substrate and are well known to have proteolytic activity Citation22. Proteolysis is a very important characteristics for dairy LAB as free amino acids are scarce in milk. Gastrointestinal LAB are usually bile tolerant and produce various bacteriocins to compete with other bacteria Citation23. As FLAB inhabit in fructose-rich niches, they might have lost ability to metabolize glucose during their adaptation to their niches. Genome sequencing of FLAB might clear the evidences of their evolution.
Fructophilic lactic acid bacteria are unique food-related LAB, as described above, suggesting a great potential for future food and feed applications.
Conflict of interest and funding
Most of the research was supported by postdoctoral grant from the Claude Leon Foundation, Cape Town, South Africa.
References
- Endo A, Futagawa-Endo Y, Dicks LMT. Isolation and characterization of fructophilic lactic acid bacteria from fructose-rich niches. Syst Appl Microbiol. 2009; 32: 593–600. 10.3402/mehd.v23i0.18563.
- Antunes A, Rainey FA, Nobre MF, Schumann P, Ferreira AM, Ramos A, et al. Leuconostoc ficulneum sp. nov., a novel lactic acid bacterium isolated from a ripe fig, and reclassification of Lactobacillus fructosus as Leuconostoc fructosum comb. nov. Int J Syst Evol Microbiol. 2002; 52: 647–55.
- Chambel L, Chelo IM, Zé-Zé L, Pedro LG, Santos MA, Tenreiro R. Leuconostoc pseudoficulneum sp. nov., isolated from a ripe fig. Int J Syst Evol Microbiol. 2006; 56: 1375–81. 10.3402/mehd.v23i0.18563.
- Leisner JJ, Vancanneyt M, van der Meulen R, Lefebvre K, Engelbeen K, Hoste B, et al. Leuconostoc durionis sp. nov., a heterofermenter with no detectable gas production from glucose. Int J Syst Evol Microbiol. 2005; 55: 1267–70. 10.3402/mehd.v23i0.18563.
- Endo A, Okada S. Reclassification of the genus Leuconostoc and proposals of Fructobacillus fructosus gen. nov., comb. nov., Fructobacillus durionis comb. nov., Fructobacillus ficulneus comb. nov. and Fructobacillus pseudoficulneus comb. nov. Int J Syst Evol Microbiol. 2008; 58: 2195–205. 10.3402/mehd.v23i0.18563.
- Edwards CG, Haag KM, Collins MD, Hutson RA, Huang YC. Lactobacillus kunkeei sp. nov.: a spoilage organism associated with grape juice fermentations. J Appl Microbiol. 1998; 84: 698–702. 10.3402/mehd.v23i0.18563.
- Endo A, Irisawa T, Futagawa-Endo Y, Takano K, du Toit M, Okada S, et al. Characterization and emended description of Lactobacillus kunkeei as a fructophilic lactic acid bacterium. Int J Syst Evol Microbiol. 2012; 62: 500–4. 10.3402/mehd.v23i0.18563.
- Endo A, Futagawa-Endo Y, Sakamoto M, Kitahara M, Dicks LMT. Lactobacillus florum sp. nov., a novel fructophilic species isolated from flowers. Int J Syst Evol Microbiol. 2010; 60: 2478–82. 10.3402/mehd.v23i0.18563.
- Kodama R. Studies on the nutrition of lactic acid bacteria. Part IV. Lactobacillus fructosus nov. sp., a new species of lactic acid bacteria. J Agric Chem Soc Jpn. 1956; 30: 705–8.
- Alcántara-Hernández RJ, Rodríguez-Álvarez JA, Valenzuela-Encinas C, Gutiérrez-Miceli FA, Castañón-González H, Marsch R, et al. The bacterial community in ‘taberna’ a traditional beverage of Southern Mexico. Lett Appl Microbiol. 2010; 51: 558–63. 10.3402/mehd.v23i0.18563.
- Mesas JM, Rodríguez MC, Alegre MT. Characterization of lactic acid bacteria from musts and wines of three consecutive vintages of Ribeira Sacra. Lett Appl Microbiol. 2011; 52: 258–68. 10.3402/mehd.v23i0.18563.
- Endo A, Irisawa T, Futagawa-Endo Y, Sonomoto K, Itoh K, Takano K, et al. Fructobacillus tropaeoli sp. nov., a novel fructophilic lactic acid bacterium isolated from a flower. Int J Syst Evol Microbiol. 2011; 61: 898–902. 10.3402/mehd.v23i0.18563.
- Lefeber T, Gobert W, Vrancken G, Camu N, De Vuyst L. Dynamics and species diversity of communities of lactic acid bacteria and acetic acid bacteria during spontaneous cocoa bean fermentation in vessels. Food Microbiol. 2011; 28: 457–64. 10.3402/mehd.v23i0.18563.
- Nielsen DS, Teniola OD, Ban-Koffi L, Owusu M, Andersson TS, Holzapfel WH. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int J Food Microbiol. 2007; 114: 168–86. 10.3402/mehd.v23i0.18563.
- Papalexandratou Z, Falony G, Romanens E, Jimenez JC, Amores F, Daniel HM, et al. Species diversity, community dynamics, and metabolite kinetics of the microbiota associated with traditional ecuadorian spontaneous cocoa bean fermentations. Appl Environ Microbiol. 2011; 77: 7698–714. 10.3402/mehd.v23i0.18563.
- He H, Chen Y, Zhang Y, Wei C. Bacteria associated with gut lumen of Camponotus japonicus Mayr. Environ Entomol. 2011; 40: 1405–9. 10.3402/mehd.v23i0.18563.
- Koch H, Schmid-Hempel P. Bacterial communities in central European bumblebees: low diversity and high specificity. Microb Ecol. 2011; 62: 121–33. 10.3402/mehd.v23i0.18563.
- Thaochan N, Drew RA, Hughes JM, Vijaysegaran S, Chinajariyawong A. Alimentary tract bacteria isolated and identified with API-20E and molecular cloning techniques from Australian tropical fruit flies, Bactrocera cacuminata and B. tryoni. J Insect Sci. 2010; 10: 131. 10.3402/mehd.v23i0.18563.
- Koo OK, Jeong DW, Lee JM, Kim MJ, Lee JH, Chang HC, et al. Cloning and characterization of the bifunctional alcohol/acetaldehyde dehydrogenase gene (adhE) in Leuconostoc mesenteroides isolated from kimchi. Biotechnol Lett. 2005; 27: 505–10. 10.3402/mehd.v23i0.18563.
- Makarova KS, Koonin EV. Evolutionary genomics of lactic acid bacteria. J Bacteriol. 2007; 189: 1199–208. 10.3402/mehd.v23i0.18563.
- Marri PR, Hao W, Golding GB. Gene gain and gene loss in streptococcus: is it driven by habitat?. Mol Biol Evol. 2006; 23: 2379–91. 10.3402/mehd.v23i0.18563.
- Kunji ER, Mierau I, Hagting A, Poolman B, Konings WN. The proteolytic systems of lactic acid bacteria. Antonie Van Leeuwenhoek. 1996; 70: 187–221. 10.3402/mehd.v23i0.18563.
- Klaenhammer TR, Altermann E, Pfeiler E, Buck BL, Goh YJ, O'Flaherty S, et al. Functional genomics of probiotic Lactobacilli. J Clin Gastroenterol. 2008; 42: S160–2. 10.3402/mehd.v23i0.18563.