Abstract
Background: The intestinal microbiota composition varies between healthy and diseased individuals for numerous diseases. Although any cause or effect relationship between the alterations in the gut microbiota and disease is not always clear, targeting the intestinal microbiota might offer new possibilities for prevention and/or treatment of disease.
Objective: Here we review some examples of manipulating the intestinal microbiota by prebiotics, probiotics, and fecal microbial transplants.
Results: Prebiotics are best known for their ability to increase the number of bifidobacteria. However, specific prebiotics could potentially also stimulate other species they can also stimulate other species associated with health, like Akkermansia muciniphila, Ruminococcus bromii, the Roseburia/Enterococcus rectale group, and Faecalibacterium prausnitzii. Probiotics have beneficial health effects for different diseases and digestive symptoms. These effects can be due to the direct effect of the probiotic bacterium or its products itself, as well as effects of the probiotic on the resident microbiota. Probiotics can influence the microbiota composition as well as the activity of the resident microbiota. Fecal microbial transplants are a drastic intervention in the gut microbiota, aiming for total replacement of one microbiota by another. With numerous successful studies related to antibiotic-associated diarrhea and Clostridium difficile infection, the potential of fecal microbial transplants to treat other diseases like inflammatory bowel disease, irritable bowel syndrome, and metabolic and cardiovascular disorders is under investigation.
Conclusions: Improved knowledge on the specific role of gut microbiota in prevention and treatment of disease will help more targeted manipulation of the intestinal microbiota. Further studies are necessary to see the (long term) effects for health of these interventions.
Microbes existed on Earth long before humans; therefore, it is logical that humans have learned to live with them, in fact co-evolved with them. All animals can be looked upon as dualistic ‘superorganisms’, i.e. their selves and their microbiota. Establishment and maintenance of an intestinal microbiota is of utmost importance for health in all mammals.
In the last 2–3 decades, an increasing number of metagenomic analyses have provided us with information about differences in gut microbiota composition between healthy and diseased individuals. Generally, high microbial diversity is thought to be associated with a healthy gut microbiota, while loss of diversity seems to correlate with disease. Nowadays over 25 diseases or syndromes have been linked to an altered intestinal microbiome (Citation1). These diseases range from gastrointestinal diseases like inflammatory bowel diseases (IBDs), irritable bowel syndrome, and colorectal cancer to metabolic diseases and potentially even to diseases like Alzheimer’s disease, autistic spectrum disorders, chronic fatigue syndrome, Parkinson’s disease, and autoimmune diseases like rheumatoid arthritis and multiple sclerosis. The most studied disease conditions in relation to intestinal microbiota are obesity, metabolic syndrome, and type II diabetes on one hand, and bowel diseases (Crohn’s disease, ulcerative colitis, irritable bowel syndrome) on the other hand. Although there is a relationship between the gut microbiota and disease, it is unclear in most cases if alterations in the microbiota are a cause or an effect of the disease, and whether manipulation of the gut microbiota could help to prevent or even treat the disease.
The potential role of the gut microbiota in obesity was first recognized by the group of Jeffrey Gordon. In mice experiments, adult germ-free mice colonized with a normal microbiota of conventionally raised animals had a 60% increase in body fat content and insulin resistance developed within 14 days despite reduced food intake (Citation2). Obese ob/ob mice were found to have a higher Firmicutes/Bacteroidetes ratio compared to lean ob/+ and wild-type mice (Citation3). Although this altered Firmicutes/Bacteroidetes ratio has also been described in some human studies, other studies did not find this correlation, which is still matter of debate (Citation4). It may be that defining the bacterial distribution at a phyla level is not specific enough and differences between obese and lean individuals are better described at genus or even species level. Whether an altered microbiota causes obesity or is caused by the same diet that leads to obesity is still unclear.
In IBDs (Crohn’s disease and ulcerative colitis), the role of the gut microbiota has also been recognized. Numerous culture-independent studies have been carried out recently, comparing the microbiota composition of IBD patients with that of healthy controls (Citation5). In general, an overall decrease in microbial diversity and stability of the intestinal microbiota has been observed in IBD patients. Specific bacterial species, like Faecalibacterium prausnitzii, have been found to have anti-inflammatory properties, as well as a decreased abundance in IBD patients (Citation6). Also the function of the microbiota seems to differ between people with Crohn’s disease compared to healthy controls. People with Crohn’s disease have higher levels of fecal trypsin, an enzyme that is produced by the pancreas and which is normally inactivated by the Bacteroides (Citation7).
In this review, examples of manipulating the gut microbiota by prebiotics, probiotics or fecal transplants are described to give an overview of some of the potential ways to manipulate the gut microbiota and to improve human health. Some probiotic interventions have an impact on disease and digestive symptoms (Citation8, Citation9) and the identification of specific health-promoting bacteria from metagenomic-based studies will provide novel candidates for probiotic intervention. The culture and delivery of such novel probiotics will provide many new challenges. At the same time understanding which bacterial species are present at lower abundance in diseased compared to healthy individuals will enable selective targeting of those bacteria using a prebiotic approach. An alternative to the modulation of specific bacterial species is the transplantation of whole gut microbiota from healthy to diseased individuals. Such a treatment is particularly successful in patients with recurring Clostridium difficile infections.
Manipulating the gut microbiota by prebiotics
The impact of diet on the composition of the gut microbiota is discussed in detail elsewhere in this volume (see the review by Graf et al. in this supplement). Here we will consider the effect of very specific dietary components, prebiotics.
Prebiotics were first described in 1995 (Citation10) and the current, refined, definition states that ‘A prebiotic is a selectively fermented ingredient that results in specific changes in the composition and/or activity of the gastrointestinal microbiota, thus conferring benefit(s) upon host health’ (Citation11). This expanded definition attempts to encompass alterations in other beneficial members of the gut microbiota, rather than focusing solely on the ‘bifidogenic effect’. However prebiotic efficacy is still often stated in terms of the prebiotic index, which relates to the relative increase in bifidobacteria (Citation12), and does not refer to the effect on other members of the gut microbial community.
Prebiotics act to enhance the growth and/or activity of bacteria that are resident in the colon, acting as growth substrates to selectively boost numbers and/or activities of particular bacteria. Data from metagenomic studies comparing the gut microbiota in healthy and diseased individuals (e.g. Metahit and the HMP projects) enables bacterial groups or species that are repressed under specific disease conditions to be identified. Specific growth studies can then be performed under conditions of increasing complexity to identify substrates that can selectively boost the growth or activity of these bacteria, and thus have the potential to redress the dysbiosis associated with the disease when administered as prebiotics.
All food that is indigestible in the upper gastrointestinal tract (GIT) and thus reaches the colon is available for fermentation by the gut anaerobes. The current distinction of a prebiotic is the ‘selective fermentation’, in that not all bacterial species should be able to utilize a specific prebiotic for growth. Substrates that are widely accepted prebiotics include the fructans inulin and fructo-oligosaccharides (FOS), galacto-oligosaccharides (GOS), and lactulose (Citation13, Citation14). Many more, including resistant starches (of which there are many types) and oligosaccharides with a variety of monomeric units, are under investigation and development (Citation12).
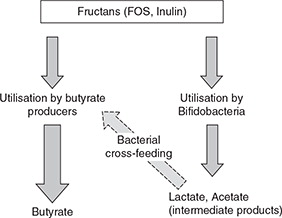
There are many publications demonstrating increased numbers of bifidobacteria in humans following dietary supplementation with fructans of varying chain length. What became apparent from some of these studies was that the level of fecal butyrate also increased following FOS supplementation (Citation15). Since bifidobacteria produce lactate and not butyrate as a fermentation product the effect of the intervention must be more complex. It is likely that at least two mechanisms contribute to the increased detection of butyrate (Fig. 1). Bacteria in the human gut exist within interactive consortia, and the lactate produced by the increased numbers of bifidobacteria probably serves as a growth substrate for lactate-utilizing, butyrate-producing bacteria. The impact of such bacterial cross-feeding on final metabolite detection has been shown in mixed culture work (Citation16–Citation20). In addition some butyrate-producing bacteria are able to use fructans directly for growth (Citation21), and genes for prebiotic degradation were identified in a range of abundant commensal bacteria by functional metagenomic screening (Citation22).
The other important point is that not all bifidobacteria species and strains within a specific species have equal abilities to degrade prebiotic substrates. Detailed work has shown that there are four distinct groups of bifidobacteria with very different abilities to degrade fructan molecules of different chain lengths (Citation23), with the ability to utilize the longer chain length molecules limited to few species. The same research group has identified five, species-independent, clusters of bifidobacteria differing in their ability to utilize arabinoxylan oligosaccharides (Citation24). Although this may seem a trivial difference it is in fact particularly relevant with the increasing use of prebiotics in baby formulae. Most baby formulae milk are supplemented with GOS, and research has shown that this elevates numbers of bifidobacteria compared to babies fed formulae lacking the prebiotic (Citation25). However, the bifidobacterial population is more diverse and less stable in prebiotic supplemented, formula-fed infants (Citation26) compared to breast-fed infants. B. adolescentis, normally found in high numbers in adults, was detected in some formula-fed infants, although it is completely absent in breast-fed babies whose microbiota is dominated by B. longum subspecies infantis (Citation26). It is not clear what impact this distinction between the bifidobacteria species present has on the maturation of the infant gut and immune system.
Pure culture work has revealed that different prebiotics have varying selectivity. FOS was less selective than GOS as a growth substrate for a panel of obligate gut anaerobes tested, while even fewer bacteria were able to use starch and long-chain inulin for growth (Citation21). However, bacterial interactions are key in understanding the true effect of prebiotics, and bacteria that may be able to utilize prebiotics as substrates for growth in pure culture, may not compete well-enough for the substrate in mixed culture. Proper demonstration of the effect of prebiotics requires human supplementation studies, with subsequent analysis of appropriately stored fecal material, analyzing the full microbial content, to at least a genus level.
Bacterial genome sequencing can help to identify those bacteria which have the potential to degrade specific substrates, by enabling the identification of specific genes on the genome, while metagenome sequencing can assess the number of such genes present in entire fecal samples. Many more clones involved in degrading GOS compared to FOS were identified in human intestinal metagenomic libraries (Citation22). Genes for FOS degradation were identified in Bifidobacterium longum and Eubacterium rectale (Citation22) while an inducible fructan utilization operon was previously identified in Roseburia inulinivorans (Citation27). E. rectale and R. inulinivorans both belong to the Roseburia genus (Citation28, Citation29) and, along with Faecalibacterium prausnitzii, are the predominant butyrate producers in the human GIT (Citation30).
Prebiotic stimulation of keystone species
Knowledge of specific bacterial species present at lower abundance in certain disease states offers an opportunity to use prebiotics in a targeted way. Prior to initiating such a strategy, it is essential to have a firm understanding of the role bacteria play in the development of the disease, or at least how they may function to alleviate it, as well as thinking about the wider consequences of increasing numbers of a specific bacterium.
Postulated bacterial targets include:
Akkermansia muciniphila, Lower numbers of this bacterium have been associated with diabetes, obesity, and IBD (Citation31), and supplying a gluten-free diet to mice increased fecal levels of Akkermansia species (Citation32). In contrast however another study found that levels of A. muciniphila were four times higher in patients with colorectal cancer than in healthy controls (Citation33). Thus, it is still unclear whether boosting numbers of this mucin degrading bacterium would actually be beneficial for health.
Ruminococcus bromii has been described as a keystone species for degradation of resistant starch (RS) (Citation34), and numbers of R. bromii clearly responded to increasing the RS content of the diet in human studies (Citation30, Citation35). Co-culture experiments indicate that R. bromii performs the initial degradation of starch externally releasing mono- and oligo-saccharides that can act as substrates for other bacteria (Citation34), and this has also been demonstrated in fermenter models (Citation36). Boosting numbers of the keystone, primary polysaccharide degrader could thus affect the overall composition of the gut microbiota due to bacterial cross-feeding. However, the four types of RS are structurally and chemically different, and many diverse bacteria are able to utilize soluble starch as a growth substrate in vitro. Hence, the type of starch used to selectively increase numbers of specific bacterial species has to be chosen with care. RS2 and RS4 had very different effects on the composition of the microbiota when compared in the same human study (Citation37).
The Roseburia/E. rectale group of butyrate-producing bacteria were also significantly increased on the RS3 diet (Citation30). It was previously shown that reducing the carbohydrate content of the diet had a significant effect on lowering numbers of the Roseburia/E. rectale group, and also resulted in lower butyrate production (Citation38). This linking of bacterial metabolite production and bacterial composition is an important consideration when investigating the potential health effects of prebiotics. Butyrate-producing bacteria are potential targets for prebiotic use to enhance bacterial numbers and elevate butyrate concentrations due to the role of butyrate in causing apoptosis of cancer cells (Citation39, Citation40). Butyrate-producing bacteria were found to be less abundant in fecal samples obtained from colorectal cancer patients compared to healthy controls (Citation41, Citation42).
F. prausnitzii has been shown to respond to prebiotic supplementation using a mixed chain length fructan supplement (Citation43, Citation44). F. prausnitzii is also able to use pectin for growth which may enable a more targeted approach to boosting numbers of this bacterial species (Citation45). Reduced numbers of F. prausnitzii are present in Crohn’s disease patients (Citation44), and since this bacterium has also been shown to have an anti-inflammatory effect (Citation46) it is a strong target for disease therapy.
Oxalobacter formigenes is the key bacterium responsible for the degradation of oxalate in humans (Citation47), and an accumulation of oxalate is the main case of kidney stone formation (Citation48). Patients suffering from calcium oxalate kidney stones are less likely to be colonized by O. formigenes (Citation49). The bacterium is sensitive to many commonly used antibiotics (Citation50) and is less abundant in individuals who have undergone antibiotic treatment at some point in their life (Citation51). Oral recolonisation with Oxalobacter has been successful (Citation52), although it is not permanent. Identification of specific substrates to boost existing numbers of O. formigenes would be a viable alternative therapy, but the preliminary microbiology work has yet to be done.
There are other bacteria whose numbers have been shown to be reduced under certain disease conditions and thus offer targets for prebiotic enhancement, although in many cases the literature is still inconclusive (see review (Citation1)). These include a decreased abundance of Bacteroides species, including B. vulgatus, in pediatric IBS patients (Citation53) and reduced numbers of butyrate-producing bacteria associated with type II diabetes (Citation54).
Manipulating the gut microbiota by probiotics
Lactic acid bacteria were initially used to preserve milk because they occurred spontaneously in the dairy environment. An added bonus was that fermentation of milk by lactobacilli into yogurt improved its digestibility. Yogurt thus provided both a preservable food to eat, and a digestible one. Yogurt was also used to cure diarrhea. Legend states that the French king, François 1st, was cured from chronic diarrhea by a Turkish ‘yogurt’ (Citation55). The concept that some bacteria can provide a health benefit and cure disease, led the way to the development of probiotics. Probiotics are defined as ‘live microorganisms that, when administered in adequate amounts, confer a health benefits on the host’ (Citation56, Citation57). Currently the main groups of probiotic bacteria used for human foods or supplements and/or animal feed or are lactobacilli, streptococci, and bifidobacteria. Furthermore, the yeast Saccharomyces boulardii and one specific strain of E. coli, strain Nissle are commonly used. The European Food Safety Authority (EFSA) recognized the health benefits of different probiotics as components of animal feed for many different bacteria, while for humans (the general population) the only accepted claim is the benefit on lactose digestion, at the present time. However, scientific data are accumulating showing that specific probiotic strains or combination of strains can be beneficial in different diseases (Citation58). Effects can be either a direct effect of the probiotic bacterium itself, or an indirect effect via the interaction with the commensal microbiota.
The first scientific study reporting the capacity of a living bacterium to provide a health benefit to humans, was reported in the 1980s and related to the digestion of lactose. The breath test technique is an easy way to monitor lactose maldigestion, and Levitt & Savaiano’s team demonstrated that live yogurt bacteria (Lactobacillus bulgaricus and Streptococcus thermophilus) were able to compensate for the deficit of lactase in adults (Citation59). This effect has been confirmed by many other groups in different countries, which reported a variable but always significant reduction of lactose malabsorption in lactose malabsorbers consuming yogurt (Citation60). The bacteria themselves possess the enzymatic activity to digest lactose. However, lactase is not the only enzyme involved, as the efficiency of different strains of lactobacilli (as measured by the breath test) is not proportional to their lactose activity per colony forming unit. Probably other enzymes, like a permease activity involved in the influx of lactose into the bacteria, play a role as well (Citation61).
Another recently reported enzymatic activity that a bacterium can provide to improve human health is the degradation of oxalate. The bacterium Oxalobacter formigenes, mentioned earlier, is able to degrade oxalate in the lumen of the intestine, therefore reducing oxalate absorption, oxalate excretion in urine and the risk of kidney stones developing (Citation62). Oxalobacter formigenes is an example of a possible next generation probiotic – a commensal bacteria which is found in the intestine that can confer a defined health benefit to the host. Other examples include those mentioned above, such as Akkermansia muciniphila, Ruminococcus bromii, and Faecalibacterium prausnitzii. Culturing and delivering these (strictly) anaerobic bacteria on an industrial scale is still a technical challenge, and as these bacteria do not have a history of safe use, there also are regulatory hurdles to overcome before these kind of bacteria can be brought to the market.
It has long been known that many microbial strains, including many gut commensals, can produce neurotransmitters and the existence of a gut–brain axis is well established. Many gut commensal bacteria, including clostridial species, Desulfovibrio, Sutterella, B. fragilis, and also bacterial metabolic products, such as propionic acid, have been associated with behavioral alterations including autism spectrum disorders (Citation63–Citation65). The phrase ‘psychobiotic’ has been proposed to describe a class of probiotics of relevance to psychiatry (Citation66).
Other options for next generation probiotics might come from cohorts of people not consuming a ‘westernized’ ‘diet’. Work has started to have such fecal samples stored at the United Nation Global Seed Vault at Svalbard (Citation67). A sample bank like that would be a valuable asset to be used by future generations.
Another multifactorial mechanism of action of some probiotics is to improve human health by influencing the resident microbiota, either by temporarily replacing a missing part of the resident microbiota, or by supplementing the endogenous population, or by stimulating (part of) the resident microbiota. Certain probiotic strains can secrete antibiotic-like factors to prevent localised growth of potential competitors. The ability of a probiotic to efficiently use a niche in the digestive tract to grow and to use available sources of energy may prevent growth of exogenous microbes and lower substrate availability for pathogens. This mechanism of microbial exclusion is likely involved in the effects of certain probiotics to prevent necrotising enterocolitis (NEC) in premature babies (Citation68), and to prevent the occurrence of diarrhea and C. difficile infection during or after the use of antibiotic(s) (Citation69–Citation71). The use of antibiotics is known to have long-term effects on the intestinal microbiota composition and thereby on health (Citation72, Citation73) and certain studies with probiotics have shown lower distortion of the gut microbiota when probiotics were given in parallel with the antibiotics (Citation74–Citation76).
Probiotics can interact with host epithelial cells and other human cells in the body through physico-chemical, or immune signals, in the same way as the commensal gut microbiota. This communication can even reach the brain (Citation77). In the gut, some probiotics can change the composition of the mucus secreted by the colonocytes, by changing the gene expression of the colonocytes (Citation78), while others can strengthen the tight junctions between epithelial cells reducing the deleterious effect on permeability of some pathogens (Citation79, Citation80). Finally, some probiotics are able to modulate the effect, directly or indirectly, of the gut microbiota on the local immune and inflammatory systems, down-regulating over-stimulated inflammatory (Citation81) and/or immune responses (Citation82).
Probiotic stimulation of the intestinal microbiota
Probiotics may also act as ‘prebiotics’ and stimulate the growth of part of the gut microbiota. For example, a probiotic strain of Lactobacillus casei increased the concentration of lactobacilli in the stools of young children (Citation83). A strain of Lactococcus lactis increased the concentration of bifidobacteria and reduced the concentration of Enterococci in human-flora associated rats (Citation84). The levels of a commensal rat ileum bacterium correlated with the positive disease outcome of a prophylactic probiotic therapy in a rat model for acute pancreatitis (Citation85). An additional mechanism by which some probiotics can influence human health is the modification of expressed functions of (part of) the gut microbiota. Some probiotics are able to change the enzymatic activities of the gut microbiota: e.g. the nitrogen metabolism as reflected by urinary concentration of p-cresol, or the glucosidases, or the bile salt hydrolases, or azoreductase (Citation86). The recent genomic and metabolomic tools are able to specifically identify changes, and even without any detectable change in the composition of the fecal microbiota its metabolic activity may be altered by a probiotic (Citation87). It should be emphasized that almost all microbiota studies have used fecal material as a source. It is likely that probiotics mainly act in the small intestine where there is a low concentration of resident microbiota and where an intake of 108 probiotic bacteria per gram (the average concentration of probiotic in fermented milks and in food supplements) is a significant increase. The probiotics are likely to influence the diversity and richness of the microbiota during their transit. In the large intestine, the probiotic bacteria will be outnumbered by the endogenous bacteria, but they can still have direct and indirect effects on health.
Manipulating the gut microbiota by fecal transplants
It is stated that the use of feces to treat a variety of diseases goes back as far as the 4th century AD (Citation88), when according to Merde, Bedouins have used warm camel feces for the treatment of diarrhea (Citation89). However, western medicine was reluctant to use fecal transplant for the treatment of diarrhea, even severe antibiotic-associated diarrhea caused by C. difficile. In a review article, only eight reports utilizing fecal microbiota transplantation (FMT) for relapsing C. difficile infections were published before the year 2000 (Citation90). Since then, a dramatic increase has taken place. By 1 August 2014, there were 613 reports in PubMed, in which FMT had been given for a large variety of disorders.
In principle there are four different types of fecal transplant used in human medicine.
| 1. | Single donor – often to a single recipient. This has been the most common to date, with the donor often a close relative or friend. The donor has to be tested for the absence of pathogens and specific diseases, which is time consuming and often expensive. Case reports demonstrate good results (Citation91, Citation92) but it is difficult – and of little value – to perform microbiomic and metabolomic studies. | ||||
| 2. | Multiple donors – The same preliminary tests of donors have to be done as for 1. ‘Stool banks’ can be established and microbiomic and metabolomic studies are possible. Stool banks are under establishment in the US, although so far there are no published microbiomic and metabolomic data. | ||||
| 3. | Autologous feces transplantation – For this method a fecal sample has to be collected before therapeutic interventions, ideally during a healthy condition, and has to be properly stored until time for usage. The storage may need to be for as long as decades for certain conditions, but this might be more feasible for severely injured patients that most probably will need antibiotic therapy and patients with autologous bone marrow stem cell transplantation. So far the are no publications about this method. | ||||
| 4. | Anaerobically cultivated feces from healthy donor(s) – The benefit of this method is that the selection of donor(s) and screening for pathogens is only necessary once, at the start. This is also probably the cheapest method. Due to the high degree of standardization that is possible, microbiome and metabolomic studies can be performed. Linked to this method are defined collections of mixed commensal anaerobes, which are cultivated separately. Proof of principle studies have already been performed for recurrent C. difficile infection (Citation93). | ||||
Screening of the donor material is of utmost importance to prevent the transfer of a pathogen or a disease from the donor to the recipient. For the screening procedures many protocols are used (Citation94–Citation96). Care also has to be taken with the administration of the feces to the recipient, and different methods have been used, rectally via a catheter of colonoscopy, or oro/nasally, via different protocols using a duodenoscope or a naso-duodenal tube (Citation94–Citation97). Maintaining the viability of the introduced bacteria is extremely important.
At present, antibiotic-associated diarrhea (AAD) and subsequent C. difficile-associated diarrhea (CDAD) are by far the two most common conditions for which fecal transplantation therapy is used. In general, fecal transplants have given very good results for these conditions, superior or equaling the best results obtained by antibiotic therapy (Citation90, Citation94–Citation96). For IBS, especially post-infectious IBS, there are also generally good temporary results published, although there is seldom a complete cure (Citation98, Citation99). Several studies with regard to ulcerative colitis have been performed, and in general remissions are obtained (Citation98, Citation100, Citation101), even in children (Citation102). Results from a very recent study indicated a lack of specific microbes and microbial functions as possible cause(s) in Crohn’s disease, making specific feces transplantation a very interesting prospect (Citation7). For both ulcerative colitis and Crohn’s disease, it should be underlined that well-designed, randomized controlled trials are needed before FMT will become a standard part of therapy for IBD (Citation100).
The most rapidly increasing fields for FMT are the metabolic and cardiovascular disorders. Different case reports have been described, and multiple hypotheses have been raised to explain the observed effects. It is far beyond the scope of this short report to review this field, but it should be kept in mind that a substantial part of the world-wide increase in so-called life-style diseases might be due to diet- or environmental-induced alterations in the intestinal microbiota. If so, FMT might be a therapeutic alternative. Again, well-designed and properly controlled studies are needed. So far, one small but double blind, placebo controlled study, showed temporary effects of fecal transplantation in males with metabolic syndrome (Citation103).
FMT has also been tested in the treatment of many other diseases, including autoimmune and allergic diseases, neurodevelopmental and neurodegenerative disorders, chronic fatigue syndrome, etc. (Citation88, Citation98, Citation104). Most of the reports show promising results, although they are dealing with very few patients and further well-designed, randomized controlled trials are needed to establish the efficacy of FMT for these diseases.
To further understand the mechanisms involved, it would be very helpful if further studies with FMT also analyse the microbial composition before and after FMT therapy. Such studies could also help to identify microbes and their products involved in the pathogenesis of these disorders, and investigate whether so-called intestinal microbiota-associated characteristics are re-established (Citation105). The data generated would also reveal whether FMT has optimal success when the commensal microbiota is decimated, as is the case with CDAD, or whether there is still the potential to replace a disturbed but abundant microbiota with an incoming healthy one.
In general acute adverse effects of FMT are mild and transient and serious adverse effects are extremely rare (Citation97–Citation102, Citation104, Citation106, Citation107). The safety of FMT in immunosuppressed patients has been poorly studied, but a recent report, based on 20 patients stated that there was no health concern (Citation106). The theoretical concern that the transplant may contain microorganisms that might be involved in the development of chronic disorders/diseases in the recipient in later years can only be elucidated by long-term follow–up studies of all recipients. Clearly such documentation is currently lacking.
At present, regulatory agencies have difficulties in establishing rules for FMT. In the USA, their Food and Drug Administration has classified fecal microbiota as a drug, and by this classification physicians have to submit a time-consuming application, although an exception has been made for recurrent C. difficile infections (Citation107). The European Medicine Agency has not yet made a similar classification. In the meantime, the rules published by the European Society for Microbiology and Infectious Diseases are generally followed (Citation97). Accepting human beings as ‘superorganisms’, their intestinal microbiota is an integrated part of themselves, and FMT should follow the same rules as, for instance, blood transfusions.
Concluding remarks
Our intestinal microbiota is an integral part of ourselves and cross-talk between the intestinal microbiota and host leads to life-long epigenetic programming.
The unsuccessful quest for a pathogen for some diseases like Crohn’s disease has triggered the new hypothesis that many so-called ‘life-style’ disorders/diseases may be caused by hitherto not clarified compositional and/or functional ‘weaknesses’ within the intestinal microbiota. Therefore, manipulating the microbiota, either by prebiotics, probiotics or fecal microbial transplantation, seems rational for the prevention and treatment of disease. Whether or not manipulation of the intestinal microbiota is a helpful approach in these and other diseases still needs many more studies. The analyses of missing functions in the intestinal microbiota during disease will help to select the potential prebiotic, probiotic or fecal microbial transplants harbouring the required function.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
Acknowledgements
The authors acknowledge the support of the European Science Foundation (ESF), in the framework of the Research Networking Programme, The European Network for Gastrointestinal Health Research (ENGiHR). The Rowett Institute of Nutrition and Health (KPS) receives support from the Scottish Government (RESAS).
References
- de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev 2012; 70(Suppl 1): S45–56.
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 2004; 101: 15718–23.
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 2005; 102: 11070–5.
- Vrieze A, Holleman F, Zoetendal EG, de Vos WM, Hoekstra JB, Nieuwdorp M. The environment within: how gut micro-biota may influence metabolism and body composition. Diabetologia 2010; 53: 606–13.
- Gerritsen J, Smidt H, Rijkers GT, de Vos WM. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr 2011; 6: 209–40.
- Scaldaferri F, Gerardi V, Lopetuso LR, Del Zompo F, Mangiola F, Boskoski I et al. Gut microbial flora, prebiotics, and probiotics in IBD: their current usage and utility. Biomed Res Int 2013; 2013: 435268.
- Midtvedt T, Zabarovsky E, Norin E, Bark J, Gizatullin R, Kashuba V et al. Increase of faecal tryptic activity relates to changes in the intestinal microbiome: analysis of Crohn's disease with a multidisciplinary platform. PLoS One 2013; 8: e66074.
- Hungin AP, Mulligan C, Pot B, Whorwell P, Agreus L, Fracasso P et al. Systematic review: probiotics in the management of lower gastrointestinal symptoms in clinical practice - an evidence-based international guide. Aliment Pharmacol Ther 2013; 38: 864–86.
- Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB et al. An update on the use and investigation of probiotics in health and disease. Gut 2013; 62: 787–96.
- Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 1995; 125: 1401–12.
- Gibson GR, Scott KP, Rastall RA, Tuohy KM, Hotchkiss A, Dubert-Ferrandon A et al. Dietary prebiotics: current status and new definition. Food Sci Technol Bull Funct Foods 2010; 7: 1–19.
- Roberfroid M. Prebiotics : the concept revisited. J Nutr 2007; 137: 830S–7S.
- Macfarlane G, Steed H, Macfarlane S. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J Appl Microbiol 2008; 104: 305–44.
- Roberfroid MB. Inulin-type fructans: functional food ingredients. J Nutr 2007; 137: 2493S–502S.
- Falony G, De Vuyst L. Ecological interactions of bacteria in the human gut. In: Charlampopoulos D, Rastall RA, eds. Prebiotics, and probiotics Science and Technology. New York, USA: Springer; 2009, p. 641–82.
- Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 2004; 70: 5810–7.
- Falony G, Vlachou A, Verbrugghe K, De Vuyst L. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl Environ Microbiol 2006; 72: 7835–41.
- Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 2006; 72: 3593–9.
- Chassard C, Bernalier-Donadille A. H2 and acetate transfers during xylan fermentation between a butyrate-producing xylanolytic species and hydrogenotrophic microorganisms from the human gut. FEMS Microbiol Lett 2006; 254: 116–22.
- Falony G, Calmeyn T, Leroy F, De Vuyst L. Coculture fermentations of Bifidobacterium species and Bacteroides thetaiotaomicron reveal a mechanistic insight into the pre-biotic effect of inulin-type fructans. Appl Environ Microbiol 2009; 75: 2312–9.
- Scott KP, Martin JC, Duncan SH, Flint HJ. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. Fems Microbiol Ecol 2014; 87: 30–40.
- Cecchini DA, Laville E, Laguerre S, Robe P, Leclerc M, Doré J et al. Functional metagenomics reveals novel pathways of prebiotic breakdown by human gut bacteria. PLoS One 2013; 8: e72766.
- Falony G, Lazidou K, Verschaeren A, Weckx S, Maes D, De Vuyst L. In vitro kinetic analysis of fermentation of pre-biotic inulin-type fructans by Bifidobacterium species reveals four different phenotypes. Appl Environ Microbiol 2009; 75: 454–61.
- Rivière A, Moens F, Selak M, Maes D, Weckx S, De Vuyst L. The ability of bifidobacteria to degrade arabinoxylan oligo-saccharide constituents and derived oligosaccharides is strain dependent. Appl Environ Microbiol 2014; 80: 204–17.
- Sierra C, Bernal M-J, Blasco J, Martinez R, Dalmau J, Ortufio I et al. Prebiotic effect during the first year of life in healthy infants fed formula containing GOS as the only prebiotic: a multicentre, randomised, double-blind and placebo-controlled trial. Eur J Nutr 2014. (in press).
- Klaassens ES, Boesten RJ, Haarman M, Knol J, Schuren FH, Vaughan EE et al. Mixed-species genomic microarray analysis of fecal samples reveals differential transcriptional responses of bifidobacteria in breast-and formula-fed infants. Appl Environ Microbiol 2009; 75: 2668–76.
- Scott KP, Martin JC, Campbell G, Mayer C-D, Flint HJ. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans”. J Bacteriol 2006; 188: 4340–9.
- Aminov RI, Walker AW, Duncan SH, Harmsen HJ, Welling GW, Flint HJ. Molecular diversity, cultivation, and improved detection by fluorescent in situ hybridization of a dominant group of human gut bacteria related to Roseburia spp. or Eubacterium rectale. Appl Environ Microbiol 2006; 72: 6371–6.
- Duncan SH, Aminov RI, Scott KP, Louis P, Stanton TB, Flint HJ. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol 2006; 56: 2437–41.
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 2011; 5: 220–30.
- Rajilie-Stojanovie M, Heilig HG, Tims S, Zoetendal EG, Vos WM. Long-term monitoring of the human intestinal micro-biota composition. Environ Microbiol 2013; 15: 1146–59.
- Marietta EV, Gomez AM, Yeoman C, Tilahun AY, Clark CR, Luckey DH et al. Low incidence of spontaneous type 1 diabetes in non-obese diabetic mice raised on gluten-free diets is associated with changes in the intestinal microbiome. PLoS One 2013; 8: e78687.
- Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan ER Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One 2013; 8: e70803.
- Ze X, Duncan SH, Louis P, Flint HJ. Rum inococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 2012; 6: 1535–43.
- Abell GC, Cooke CM, Bennett CN, Conlon MA, McOrist AL. Phylotypes related to Ruminococcus bromii are abundant in the large bowel of humans and increase in response to a diet high in resistant starch. Fems Microbiol Ecol 2008; 66: 505–15.
- Kovatcheva-Datchary P, Egert M, Maathuis A, Rajilié-Stojanovié M, De Graaf AA, Smidt H et al. Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ Microbiol 2009; 11: 914–26.
- Martinez I, Kim J, Duffy PR, Schlegel VL, Walter J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects PLoS One 2010; 5: e15046.
- Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 2007; 73: 1073–8.
- McIntyre A, Gibson P, Young G. Butyrate production from dietary fibre and protection against large bowel cancer in a rat model. Gut 1993; 34: 386–91.
- Scheppach W Effects of short chain fatty acids on gut morphology and function. Gut 1994; 35: S35–8.
- Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J 2011; 6: 320–9.
- Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 2009; 294: 1–8.
- Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 2009; 101: 541–50.
- Sokol H, Seksik P, Furet J, Firmesse O, Nion-Larmurier I, Beaugerie L et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflam Bowel Dis 2009; 15: 1183–9.
- Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJ, Garcia-Gil LJ, Flint HJ. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol 2012; 78: 420–8.
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermüdez-Humarán LG, Gratadoux J-J et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci 2008; 105: 16731–6.
- Stewart CS, Duncan SH, Cave DR. Oxalobacter formigenes and its role in oxalate metabolism in the human gut. FEMS Microbiol Lett 2004; 230: 1–7.
- Hoppe B. An update on primary hyperoxaluria. Nat Rev Nephrol 2012; 8: 467–75.
- Siener R, Bangen U, Sidhu H, Hönow R, von Unruh G, Hesse A. The role of Oxalobacter formigenes colonization in calcium oxalate stone disease. Kidney Int 2013; 83: 1144–9.
- Lange JN, Wood KD, Wong H, Otto R, Mufarrij PW, Knight J et al. Sensitivity of human strains of Oxalobacter formigenes to commonly prescribed antibiotics. Urology 2012; 79: 1286–9.
- Siva S, Barrack ER, Reddy G, Thamilselvan V, Thamilselvan S, Menon M et al. A critical analysis of the role of gut Oxalobacter formigenes in oxalate stone disease. BJU Int 2009; 103: 18–21.
- Hoppe B, von Unruh G, Laube N, Hesse A, Sidhu H. Oxalate degrading bacteria: new treatment option for patients with primary and secondary hyperoxaluria? Urol Res 2005; 33: 372–5.
- Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 2011; 141: 1782–91.
- Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut 2014; 63: 1513–21. doi: 10.1136/gugn1-2014-306928.
- Deeth H, Tamime A. Yogurt: nutritive and therapeutic aspects. J Food Protect 1981; 44: 78–86.
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 2014; 11: 506–14.
- FAO/WHO Working Group (2001). Report of a joint FAO/ WHO expert consultation on evaluation of health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Córdoba, Argentina: FAO/WHO; 2001.
- Sanders ME. Probiotics: considerations for human health. Nutr Rev 2003; 61: 91–9.
- Savaiano DA, Abou El, Anouar A, Smith DE, Levitt MD. Lactose malabsorption from yogurt, pasteurized yogurt, sweet acidophilus milk, and cultured milk in lactase-deficient individuals. Am J Clin Nutr 1984; 40: 1219–23.
- Rabot S, Rafter J, Rijkers GT, Watzl B, Antoine J-M. Guidance for substantiating the evidence for beneficial effects of probiotics: impact of probiotics on digestive system metabolism. J Nutr 2010; 140: 677S – 89S.
- Savaiano DA, Levitt MD. Milk intolerance and microbe-containing dairy foods. J Dairy Sci 1987; 70: 397–406.
- Okombo J, Liebman M. Probiotic-induced reduction of gastrointestinal oxalate absorption in healthy subjects. Urol Res 2010; 38: 169–78.
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013; 155: 1451–63.
- Midtvedt T. The gut: a triggering place for autism-possibilities and challenges. Microb Ecol Health Dis 2012; 23. doi: 10.3402/mehd.v23i0.18982.
- MacFabe DF. Short-chain fatty acid fermentation products of the gut microbiome: implications in autism spectrum disorders. Microb Ecol Health Dis 2012; 23. doi: 10.3402/mehd.v23i0.19260.
- Dinan TG, Stanton C, Cryan JF. Psychobiotics: a novel class of psychotropic. Biol Psychiatry 2013; 74: 720–6.
- Shenderov BA, Midtvedt T. Epigenomic programing: a future way to health? Microb Ecol Health Dis 2014; 25.
- Alfaleh K, Anabrees J, Bassler D, Al-Kharfi T. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev 2011: CD005496.
- Hempel S, Newberry SJ, Maher AR, Wang Z, Miles JN, Shanman R et al. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: a systematic review and meta-analysis. JAMA 2012; 307: 1959–69.
- Goldenberg JZ, Ma SS, Saxton JD, Martzen MR, Vandvik PO, Thorlund K et al. Probiotics for the prevention of Clostridium difficde-associated diarrhea in adults and children. Cochrane Database Syst Rev 2013: CD006095.
- Hell M, Bernhofer C, Stalzer P, Kern JM, Claassen E. Probiotics in Clostridium difficile infection: reviewing the need for a multistrain probiotic. Benef Microbes 2013; 4: 39–51.
- Greenwood C, Morrow AL, Lagomarcino AJ, Altaye M, Taft DH, Yu Z et al. Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of enterobacter. J Pediatr 2014; 165: 23–9.
- Blaser M. Antibiotic overuse: stop the killing of beneficial bacteria. Nature 2011; 476: 393–4.
- Koning CJ, Jonkers DM, Stobberingh EE, Mulder L, Rombouts FM, Stockbrugger RW. The effect of a multi-species probiotic on the intestinal microbiota and bowel movements in healthy volunteers taking the antibiotic amoxycillin. Am J Gastroenterol 2008; 103: 178–89.
- Engelbrektson A, Korzenik JR, Pittler A, Sanders ME, Klaenhammer TR, Leyer G et al. Probiotics to minimize the disruption of faecal microbiota in healthy subjects undergoing antibiotic therapy. J Med Microbiol 2009; 58: 663–70.
- Rehman A, Heinsen FA, Koenen ME, Venema K, Knecht H, Hellmig S et al. Effects of probiotics and antibiotics on the intestinal homeostasis in a computer controlled model of the large intestine. BMC Microbiol 2012; 12: 47.
- Tillisch K, Labus J, Kilpatrick L, Jiang Z, Stains J, Ebrat B et al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology 2013; 144: 1394–401, 401e1–4.
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science 2005; 307: 1915–20.
- Putaala H, Salusjärvi T, Nordstrom M, Saarinen M, Ouwehand AC, Bech Hansen E et al. Effect of four probiotic strains and Escherichia coli 0157: H7 on tight junction integrity and cyclo-oxygenase expression. Res Microbiol 2008; 159: 692–8.
- Van Hemert S, Verwer J, Schuetz B. Clinical studies evaluat-ing effects of probiotics on parameters of intestinal barrier function. Adv Microbiol 2013; 3: 212–21.
- Sang L-X, Chang B, Zhang W-L, Wu X-M, Li X-H, Jiang M. Remission induction and maintenance effect of probiotics on ulcerative colitis: a meta-analysis. World J Gastroenterol 2010; 16: 1908.
- Klaenhammer TR, Kleerebezem M, Kopp MV, Rescigno M. The impact of probiotics and prebiotics on the immune system. Nat Rev Immunol 2012; 12: 728–34.
- Guerin-Danan C, Chabanet C, Pedone C, Popot F, Vaissade P, Bouley C et al. Milk fermented with yogurt cultures and Lactobacillus casei compared with yogurt and gelled milk: influence on intestinal microflora in healthy infants. Am J Clin Nutr 1998; 67: 111–7.
- Bernbom N, Licht TR, Brogren CH, Jelle B, Johansen AH, Badiola I et al. Effects of Lactococcus lactis on composition of intestinal microbiota: role of nisin. Appl Environ Microbiol 2006; 72: 239–44.
- Gerritsen J, Timmerman HM, Fuentes S, van Minnen LP, Panneman H, Konstantinov SR et al. Correlation between protection against sepsis by probiotic therapy and stimulation of a novel bacterial phylotype. Appl Environ Microbiol 2011; 77: 7749–56.
- Ouwehand AC, Lagstrom H, Suomalainen T, Salminen S. Effect of probiotics on constipation, fecal azoreductase activity and fecal mucin content in the elderly. Ann Nutr Metab 2002; 46: 159–62.
- McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med 2011; 3: 106ra06.
- Zhang F, Luo W, Shi Y, Fan Z, Ji G. Should we standardize the 1,700-year-old fecal microbiota transplantation? Am J Gastroenterol 2012; 107: 1755; author reply p. 55–6.
- Merde LA. Excursions in scientific, cultural, and socio-cultural coprology. New York: Randon House; 1999.
- Bakken JS. Fecal bacteriotherapy for recurrent Clostridium difficile infection. Anaerobe 2009; 15: 285–9.
- Swaminath A. The power of poop: patients getting ahead of their doctors using self-administered fecal transplants. Am J Gastroenterol 2014; 109: 777–8.
- Brandt LJ, Reddy SS. Fecal microbiota transplantation for recurrent Clostridium difficile infection. J Clin Gastroenterol 2011; 45(Suppl): S159–67.
- Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome 2013; 1: 3.
- Midtvedt T, Norin E, Benno P, Dahlgren A-L. Response to Surawicz et al. Am J Gastroenterol 2013; 108: 1931–2.
- Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol 2011; 9: 1044–9.
- Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol 2013; 108: 478–98; quiz 99.
- Debast SB, Bauer MP, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect 2014; 20\(Suppl 2): 1–26.
- Smits LP, Bouter KE, de Vos WM, Borody TJ, Nieuwdorp M. Therapeutic potential of fecal microbiota transplantation. Gastroenterology 2013; 145: 946–53.
- Morken MH, Valeur J, Norin E, Midtvedt T, Nysaeter G, Berstad A. Antibiotic or bacterial therapy in post-giardiasis irritable bowel syndrome. Scand J Gastroenterol 2009; 44: 1296–303.
- Anderson JL, Edney RJ, Whelan K. Systematic review: faecal microbiota transplantation in the management of inflammatory bowel disease. Aliment Pharmacol Ther 2012; 36: 503–16.
- Allegretti JR, Hamilton M.I. Restoring the gut microbiome for the treatment of inflammatory bowel diseases. World J Gastroenterol 2014; 20: 3468–74.
- Kunde S, Pham A, Bonczyk S, Crumb T, Duba M, Conrad H, Jr., et al. Safety, tolerability, and clinical response after fecal transplantation in children and young adults with ulcerative colitis. J Pediatr Gastroenterol Nutr 2013; 56: 597–601.
- Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012; 143: 913–16-e7.
- Aroniadis OC, Brandt U. Fecal microbiota transplantation: past, present and future. Curr Opin Gastroenterol 2013; 29: 79–84.
- Gustafsson A, Berstad A, Lund-Tonnesen S, Midtvedt T, Norin E. The effect of faecal enema on five microflora-associated characteristics in patients with antibiotic-associated diarrhoea. Scand J Gastroenterol 1999; 34: 580–6.
- Brandt LJ, Aroniadis OC. An overview of fecal microbiota transplantation: techniques, indications, and outcomes. Gastrointest Endosc 2013; 78: 240–9.
- Smith MB, Kelly C, Alm EJ. Policy: how to regulate faecal transplants. Nature 2014; 506: 290–1.