Abstract
Two new species of Hypocrea are added here to the European funga. Hypocrea britdaniae, a fungus with unknown anamorph and large, conspicuous stromata resembling basidiomata of a corticiaceous fungus, is a sister species to the Longibrachiatum clade, while H. foliicola, a leaf-dwelling species that forms pulvinate stromata, is recognized as an additional member of the pachybasium core group. Hypocrea foliicola sporulates in culture in a reduced verticillium-like manner, while it produces a white, typical pachybasium-like anamorph in nature. Ecologically H. foliicola is remarkable in inhabiting leaves, a substrate rarely recorded for Hypocrea. All relevant morphological teleomorphic and anamorphic traits are given. The phylogenetic placement of the new species within Hypocrea/Trichoderma was determined with combined analyses of rpb2 and tef1 exon sequences.
Introduction
Extensive field work, followed by molecular data and morphological studies, enabled CitationJaklitsch (2009, Citation2011) to present and illustrate 75 Trichoderma species that form Hypocrea teleomorphs in Europe. This diversity exceeded all expectations. CitationJaklitsch (2011, p 202) mentioned additional species in Europe that could not be included in his monograph. One of these species has been known as a single collection in the Kew fungarium. Although this species belongs among those that produce most conspicuous stromata, it has not been described. This might be because mycologists are deterred by the numerous names in the genus worldwide, which have been re-examined only partly in a modern context. This species is described here together with another one, based on material that was received after the submission of part II of the monograph. The latter species forms small pulvinate stromata on oak leaves.
Materials and methods
Isolates and specimens
Ascospore and conidial isolates were prepared as described by CitationJaklitsch (2009). Specimen information is provided for each species after its description. Representative isolates have been deposited at the Centraalbureau voor Schimmelcultures, Utrecht, the Netherlands (CBS). Recently collected specimens have been deposited in the Herbarium of the Institute of Botany, University of Vienna (WU).
Growth, morphology, DNA extraction, PCR and sequencing
Growth rate experiments were carried out and teleomorph and anamorph morphology was determined as described by CitationJaklitsch (2009). Genomic DNA extraction, PCR and sequencing was performed as described in that work, except that before DNA extraction by the DNeasy Plant Minikit (QIAGEN GmbH, Hilden, Germany) mycelium was grown in 2% liquid malt extract culture, harvested by filtration, freeze-dried and ground according to CitationVoglmayr and Jaklitsch (2008). For H. britdaniae, which could not be isolated in pure culture, DNA was extracted directly from stromata with a modified CTAB protocol described in CitationVoglmayr and Jaklitsch (2011). GenBank accession numbers of sequences retrieved from GenBank and those generated in this study are provided ().
Table I Strain numbers and GenBank accessions of sequences used for phylogenetic analyses
Analysis of sequence data
To reveal the phylogenetic affiliation of the new species, a rough phylogenetic analysis was performed on an extensive matrix of the second largest subunit of RNA polymerase II (rpb2) containing 278 sequences covering all lineages of Hypocrea/Trichoderma for which rpb2 sequences were available. Alignments were produced with MUSCLE 3.6 (CitationEdgar 2004). After exclusion of excessive leading/trailing gap regions, the matrix contained 1072 characters. Maximum parsimony (MP) analyses were performed with PAUP* 4.0 b10 (CitationSwofford 2002), using 1000 replicates of a heuristic search with random addition of sequences and subsequent tbr branch swapping (Multrees option in effect, collapse = maxbrlen, steepest descent option not in effect), each replicate limited to 1 000 000 rearrangements. All molecular characters were unordered and given equal weight; analyses were performed with gaps treated as missing data. According to this analysis, Hypocrea britdaniae was found to be affiliated with the Longibrachiatum clade and H. foliicola with the pachybasium core group (data not shown; see CitationJaklitsch 2009, Citation2011 for clade circumscriptions).
Based on this tree, ITS, tef1 (translation elongation factor 1-alpha) and rpb2 sequences of a smaller selection of species, in particular all available species of the Longibrachiatum clade and the pachybasium core group, were downloaded from GenBank and combined with those used by CitationJaklitsch (2009, Citation2011). For ITS, only a sequence alignment was produced with MUSCLE to enable sequence comparisons, but no further phylogenetic analyses were performed due to insufficient phylogenetic information (i.e. high sequence similarity of numerous species).
For combined analyses of tef1 and rpb2 sequences a single representative sequence was selected for each species, preferably from the types. Because several sequences of ex-type strains were short, these were replaced by identical or highly similar sequences from other authentic sources where available or by those determined for newly isolated strains. For the new species, several representative sequences were included in the matrix. Sequence alignments for final phylogenetic analyses were produced with MAFFT 6.847 (CitationKatoh et al. 2002, CitationKatoh and Toh 2008) implemented in UGENE 1.10.0 (http://ugene.unipro.ru), with a maximum of 100 iterative refinements and gap-opening penalties of 1.53 for rpb2 and 0.53 for tef1 respectively. The resulting alignments were checked with BioEdit 7.0.9.0 (CitationHall 1999). For the subsequent analyses tef1 introns were excluded because the latter are alignable only within clades of Trichoderma while our analyses span a range of several clades. After exclusion of excessive leading/trailing gap regions, the combined matrix contained 955 and 1075 characters from tef1 and rpb2 respectively. Before phylogenetic analyses, the approach of CitationWiens (1998) was applied to test for significant localized incongruence between the two gene partitions, using the level of bootstrap support (CitationSung et al. 2007). For this we compared the 60% maximum parsimony (MP) bootstrap trees of the individual gene regions, which were calculated with the same parameters as for the combined analysis given below. No topological conflicts were observed between these bootstrap trees of tef1 and rpb2, indicating the absence of significant incongruence and combinability of both matrices (CitationWiens 1998).
Maximum parsimony (MP), maximum likelihood (ML) and Bayesian analyses, including selection of models of sequence substitution for the latter two, were performed as described for tef1 and rpb2 in CitationJaklitsch and Voglmayr (2011), with these differences: Before MP analyses of the combined matrix, the transition/transversion (Ti/Tv) bias R was calculated with Mega 5.05 (CitationTamura et al. 2011) with the maximum composite likelihood (MCL) method and the substitution model of CitationTamura and Nei (1993). Because these values suggested a significant Ti/Tv bias (R = 2.5 for rpb2 and R = 1.9 for tef1), the transversions of the rpb2 and tef1 genes were weighted according to the R values in the subsequent MP analyses (CitationBirksa and Edwards 2002). For ML analyses, 1000 rounds of random addition of sequences as well as 1000 thorough bootstrap replicates were computed with RAxML (CitationStamatakis 2006) as implemented in raxml-GUI 0.95 (CitationSilvestro and Michalak 2011) using respectively the GTRGAMMAI and GTRCATI algorithms. For Bayesian analyses using MrBayes 3.1.2 (CitationHuelsenbeck and Ronquist 2001), three parallel runs of four incrementally heated simultaneous Markov chains were performed over 5 000 000 generations from which every 500th tree was sampled in each run. The first 1000 trees were discarded and a 90% majority rule consensus of the remaining trees was computed to obtain estimates for the probabilities that groups are monophyletic based on the sequence data (posterior probabilities). To test convergence of runs, the results were analyzed with AWTY (CitationNylander et al. 2008); no indication of lack of convergence was detected. The sequence alignment files were deposited in TreeBASE and are available at http://purl.org/phylo/treebase/phylows/study/TB2:S12541.
In addition to the combined analyses, separate phylogenetic analyses were performed for the Longibrachiatum clade and the pachybasium core group on tef1 alignments that included the introns. However, because they did not provide significant additional information concerning the phylogenetic position of the new species, the results are not shown.
Results
Molecular phylogenetic analyses
Of the 2030 characters included in the combined matrix, 544 were parsimony informative (179 in tef1, 365 in rpb2). MP analyses revealed one MP tree with a score of 3609.5 (not shown). The best ML tree (lnL = −15254.3383) () is similar to the MP tree except for minor topological differences in few nodes with low or insignificant MP/ML support. In the MP analyses, H. luteffusa was placed as sister clade to the rest of the pachybasium core group, H. pachypallida was sister clade to the H. alutacea/minutispora/atlantica clade and H. gelatinosa was sister clade to the H. estonica/parestonica/strictipilosa/longipilosa clade (not shown). Tree topologies of the Bayesian analyses were fully congruent with the ML tree. The three Bayesian runs revealed almost identical posterior probabilities (PP). MP and ML bootstrap support above 60% and Bayesian posterior probabilities above 90% are illustrated () at first, second and third position respectively, above or below the branches.
Fig. 1 Phylogram of the best ML tree (lnL = −15254.3383) revealed by RAxML from an analysis of the combined tef1-rpb2 alignment, showing the phylogenetic position of the two new Hypocrea species (in boldface). MP and ML bootstrap support above 60 and Bayesian posterior probabilities above 90% are given respectively at first, second and third position, above or below the branches.
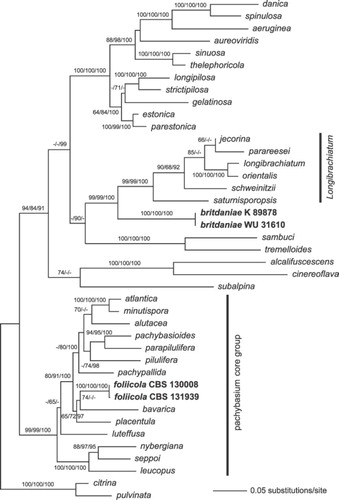
In all analyses, H. foliicola was confirmed as a member of the highly supported pachybasium core group; the sister group relationship to H. bavarica () is not well supported. Hypocrea britdaniae formed a sister group of the Longibrachiatum clade with high support.
Taxonomy
Hypocrea britdaniae Jaklitsch & Voglmayr, sp. nov.
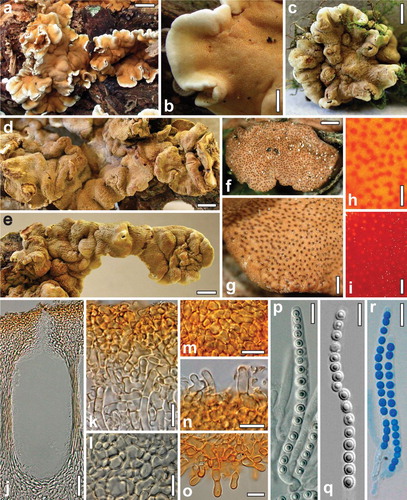
Fig. 2 Teleomorph of Hypocrea britdaniae. a, b. Fresh stromata. c–g. Dry stromata. h. Surface of rehydrated stroma. i. Surface of rehydrated stroma in 3% KOH. j. Perithecium in section. k. Cortex and subcortical tissue in section. l. Subperithecial tissue in section. m. Cortex in face view. n. Hairs on the stroma surface. o. Hairs at the stroma base. p–r. Asci (r. in cottonblue/lactic acid). a, b. Hampshire 28 Aug 2008 (images by Stuart Skeates). c–e, g–q. K(M) 89878. f, r. WU 31610. Bars: a = 15 mm; b, d = 2 mm; c, e = 3 mm; f = 0.6 mm; g = 0.3 mm; h, i = 0.15 mm; j = 30 μm; k, m, n, r = 10 μm; l, o = 15 μm; p, q = 7 μm.
MycoBank MB 564137
Etymology: occurring in Great Britain and Denmark
Stromata large, undulate, up to ca. 10 cm long and 3 mm thick when fresh, scattered or forming dense clusters extending up to 80 cm, becoming variably disintegrated into smaller parts. Stromata hard when dry, variably lobed or undulate, individual parts 1.3–7 mm diam, 0.5–1 mm thick; narrowly attached (i.e. margin for a large part free) sterile, concolorous, white when young. Surface smooth and even, except for coarse tubercles or folds, distinctly folded or rugose when immature, covered by a farinose to floccose covering layer; ostioles erumpent through this layer, ostiolar dots fine but distinct, densely disposed, circular or oblong in outline, (23–)27–51(–95) μm wide (n = 50), flat to distinctly convex, dark brown or reddish brown with light center. Stroma surface light to medium brown, sometimes reddish brown, 5CD4–7, 6–7CD4–6, 7D6–8, 5E5–8, lighter or more orange-brown under stereomicroscope; pale brownish inside. Spore deposits white or yellow. Stromata after rehydration 2 mm or more thick, more orange by reddish ostiolar dots on a bright yellow stroma, white inside; in 3% KOH turning orange-red, finally brown.
Stroma anatomy: Cortical layer (14–)18–27(–36) μm thick (n = 30), comprising a dense textura angularis of small thin-walled cells (3–)4–8(–12) × (2–)3–5(–6.5) μm (n = 65) in section and in face view, at the surface individually projecting and becoming detached, forming amorphous matter, yellow in lactic acid, golden yellow around the ostiole; surface with scant projecting cells or short hairs (6–)7–14(–17) × (2.5–)3.0–4.0(–4.5) μm (n = 20), 1–3-celled, thin-walled, (sub)hyaline. Subcortical tissue comprising textura intricata of short-celled, thin-walled hyaline hyphae (3.0–)3.5–5.0(–5.5) μm wide (n = 15). Subperithecial tissue a dense hyaline textura angularis-epidermoidea of cells (4–)5–26(–41) × 4–12(–25) μm (n = 30) with walls up to 2 μm; toward the base predominantly comprising wide, thick-walled hyphae. Stroma base of golden yellow to orange-brown thick-walled cells (up to 14 μm wide) and hyphae projecting in non-attached regions as smooth or warted, elongate cells or up to six-celled, terminally rounded hairs (13–)18–50(–77) × (5–)6–8(–11) μm (n = 33). In section perithecial layer comprising a small fraction of the whole stroma. Perithecia (174–)200–235(–255) μm high, (60–)80–125(–165) μm wide (n = 30), mostly cylindrical or ellipsoid to subglobose, crowded, up to 11 per mm; peridium (16–)17–25(–28) μm wide at the base, (5–)6–12(–17) μm at the sides (n = 30), yellow. Ostioles cylindrical, (46–)54–68(–73) μm long, not projecting, (11–)15–32(–43) μm wide at the apex inside. Asci (52–)60–75(–86) × (3.7–)4.0–5.0(–5.5) μm, including a stipe (1–)7–16(–23) μm long (n = 40), cylindrical; apex truncate, thickened to 1.5 μm, with a flat ring below the thickening. Ascospores hyaline, cells monomorphic, (sub)globose, less commonly ellipsoid, (2.0–)2.5–3.0(–3.8) × (2.0–)2.5–3.0(–3.5) μm, l/w 0.9–1.1(–1.4) (n = 120), smooth to finely spinulose, yellow to pale orange when old, rarely dimorphic, then proximal cells ellipsoid or wedge-shaped and 3–3.5 μm long.
Distribution: Europe (Denmark, England).
Habitat: on wood and bark of Salix spp.
Holotype: UK, ENGLAND, WEST SUSSEX, Petworth, The Mens and The Cut Nat. Reserve, on branch of Salix sp., stromata on bark and green moss, 15 Sep 2001, N.W. Legon, comm. B.M. Spooner (K(M) 89878).
Other known material: DENMARK, S. JUTLAND, West of Ribe, Munkesø, in wet lakeside willow carr, on partly moss-covered branches of Salix sp., 6 Aug 2010, S.A. Elborne, comm T. Laessoe (WU 31610). ENGLAND, HAMPSHIRE, Romsey Baddesley Common, grid SU38812140, 50:59: 26.510N 1:26:54.277W, boggy area, on log of Salix sp., 28 Aug 2008, S.J. Skeates (Hampshire Fungus Recording Group), FRDBI record No. 1476775; same area and host, grid SU3821, 8 Sep 2008, A. Lucas (K(M)161869), FRDBI record No. 1500707.
Notes: Stromata of H. britdaniae look like basidiomata of a corticiaceous fungus. Large effused stromata of Hypocrea occur in the section Hypocreanum, while similar compact and firm stromata with densely disposed ostiolar dots are reminiscent of members of the Longibrachiatum clade, to which it is closely related. In stroma characteristics some similarity with species of that clade forming brown stromata such as Hypocrea orientalis is apparent. In the latter stromata are much smaller and ostioles turn green in lactic acid. The perithecial layer in H. britdaniae comprises only a small fraction of the whole stroma thickness. Remarkable also are the orange-brown hair-like hyphae on the non-attached stroma base that have not been seen in any other species of Hypocrea.
No anamorph is known for this conspicuous and unusual species. Ascospores of fresh material from Denmark did not germinate. A green Trichoderma accompanying stromata was determined as T. viridescens.
Hypocrea foliicola Jaklitsch & Voglmayr, sp. nov.
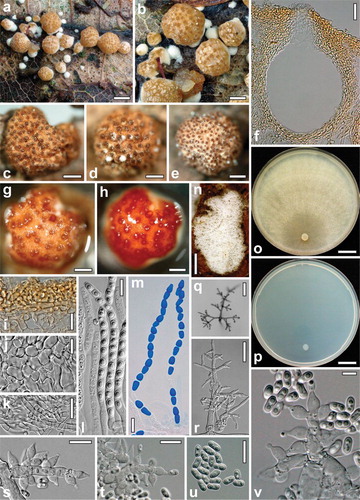
Fig. 3 Hypocrea foliicola. a–m. Teleomorph. a, b. Fresh stromata. c–e. Dry stromata. f. Perithecium in section. g. Rehydrated stroma. h. Rehydrated stroma in 3% KOH. i. Cortex and subcortical tissue in section. j. Subperithecial tissue in section. k. Stroma base in section. l, m. Asci (m. in cotton blue/lactic acid). n–v. Cultures and anamorph. n. Colony on the natural substrate. o, p. Cultures after 21 d at 25 C (o. on PDA. p. on SNA). q, r. Conidiophores on SNA. s–v. Conidiophores with phialides and conidia on the natural substrate. a, b. WU 31612 (Images by Jens H. Petersen). c–v. WU 31611/CBS 130008. Bars: a, n = 1 mm; b, e = 0.5 mm; c, d, g, h = 0.3 mm; f = 30 μm; i–k = 15 μm; l, m, s–u = 10 μm; o, p = 15 mm; q = 50 μm; r = 25 μm; v = 5 μm.
MycoBank MB 564138
Etymology: occurring on leaves
Stromata scattered on the leaf surface, when dry (0.4–)0.5–1.4(–2) mm diam, (0.3–)0.4–0.8(–1.2) mm high (n = 20), starting as small white mycelial tufts, center becoming compact and pigmented, pulvinate to semiglobose with circular or angular outline and free margin when mature, often elevated on a compact whitish sterile mycelial base; surface smooth when fresh or with a whitish covering layer splitting into peeling flakes, tubercular or ragged when dry. Ostiolar dots distinct, (40–)60–102(–118) μm diam (n = 50), slightly projecting, convex or bluntly conical, pale reddish brown or dark brown; surface cream, yellowish to light or medium brown. Rehydrated stromata with smooth surface, slightly convex ostiolar dots, orange with more orange or reddish brown dots in 3% KOH.
Stroma anatomy: Cortical layer (20–)21–30(–37) μm thick (n = 30), comprising a yellow textura angularis of thin-walled cells (3–)4–11(–14) × (2.7–)3–7(–9) μm in section (n = 30). Subcortical textura intricata of hyaline, thin-walled hyphae (2.7–)3–5(–6) μm wide (n = 30), often strongly reduced. Tissue below perithecia of a hyaline textura angularis-epidermoidea comprising thin-walled cells (6.5–)8–25(–35) × (5–)6–14(–18) μm (n = 33). Compressed base layer of thin-walled hyaline hyphae (2.5–)4–7(–8.5) μm wide (n = 30), partly appearing as textura angularis-globulosa, similar hyphae present also at the sterile stroma margin, yellow upward. Perithecia (190–)200–235(–250) μm high, (140–)160–235(–270) μm wide (n = 21), subglobose, globose or ellipsoid; peridium (15–)17–22(–25) μm wide at the base, (11–)13–18(–19) μm at the sides (n = 21), yellow. Ostiolar canal (50–)60–82(–95) μm long, plane with the surface or projecting to ca. 20 μm, apically (20–)25–40(–47) μm wide inside (n = 21), conical or cylindrical, periphysate, apical marginal cells cylindrical or narrowly clavate. Asci (84–)94–116(–128) × (5.0–)5.5–6.5(–7.0) μm, stipe (5–)9–24(–34) μm long (n = 45), apex thickened. Ascospores hyaline, verruculose; cells dimorphic, large, multiguttulate, distal cell (4.5–)5.0–6.3(–7.5) × (3.2–)3.8–4.5(–4.7) μm, l/w (1.1–)1.2–1.5(–1.8), subglobose, wedge-shaped or ellipsoid, proximal cell (4.8–)5.3–6.5(–7.5) × (3.0–)3.3–3.8(–4.2), l/w (1.3–)1.5–1.9(–2.3) (n = 60), oblong or wedge-shaped.
Anamorph on the natural substrate: Colonies up to 11 mm long, white, flat, roundish longish, loose, granulose within due to conidial masses, surface hairy. Conidiophores typically pachybasium-like, with short stout branches at right angles, mostly 6–8 μm wide in lower regions, branching points up to 10 μm, phialide origins 4–5 μm, ends 3–5 μm wide, ends sterile when young, becoming fertile; no differentiated elongations present. Phialides produced in whorls of 2–5, bluntly lageniform to ampulliform, (6.0–)6.5–9.0(–11) × (3.2–)3.5–4.2(–5.0) μm, (1.5–)2.3–3.4(–3.7) μm wide at the base, l/w (1.5–)1.7–2.4(–2.8) (n = 32). Conidia formed in minute heads, oblong, (4.7–)5.2–6.0(–6.3) × 2.5–3.0 μm, l/w (1.7–)1.9–2.2(–2.4) (n = 30), hyaline, smooth, with two or more guttules.
Cultures and anamorph: Optimal growth at 25 C on all media, slower on SNA than on CMD and PDA, slow and restricted growth at 30 C, no growth at 35 C. On CMD after 72 h 9–12 mm at 15 C, 27–31 mm at 25 C, 2–3 mm at 30 C; mycelium covering the plate after 1 wk at 25 C. Colony hyaline, thin, dense, not zonate, circular; margin well defined, mycelium radial, without conspicuous differences in hyphal width; surface hyphae soon degenerating from the center, appearing empty. Aerial hyphae inconspicuous. No conidiation, no chlamydospores, autolytic excretions, coilings, pigment or distinct odor noticeable within 21 d.
On PDA after 72 h 12–14 mm at 15 C, 24–27 mm at 25 C, 1–4 mm at 30 C; mycelium covering the plate after 9–10 d at 25 C. Colony circular, dense, margin wavy, aerial hyphae forming ill defined concentric zones of fine whitish floccules on the surface. No conidiation appearing within 21 d; autolytic excretions and coilings inconspicuous; diffusing pigment lacking, odor indistinct or slightly unpleasant.
On SNA after 72 h 6–8 mm at 15 C, 11–15 mm at 25 C, 1–3 mm at 30 C; mycelium covering the plate after 9 d at 25 C. Colony hyaline, dense, mycelium radial, not zonate, margin well defined, aerial hyphae scant, more frequent at the distal margin after ca. 10 d. No chlamydospores, pigment or distinct odor noticeable within 3 wk. Autolytic excretions and coilings scant. Conidiation lacking or scant, effuse, appearing at the distal margin earliest after 9–14 d, often only after agar injury. Conidiophores formed on surface or aerial hyphae, mostly less than 0.5 mm long, (2.5–)2–4(–5) μm wide, simple, verticillium-like, with mostly asymmetrical branching, forming conidia in variable wet heads up to ca. 40 μm diam on phialides in whorls of 3–5, divergent to nearly parallel. Phialides (8–)10–15(–19) × (2.5–)3.0–3.5(–3.8) μm, (1.7–)2.0–2.5(–3.0) μm wide at the base, l/w (2.5–)3.1–4.9(–6.8) (n = 30), lageniform, straight, curved or sigmoid. Conidia (4.3–)5.0–6.5(–7.2) × (2.0–)2.3–2.8(–3.2) μm, l/w (1.5–)2.0–2.6(–2.8) (n = 38), hyaline, oblong, sides often slightly pinched, smooth, with few minute guttules, scar indistinct.
Distribution: Europe (Denmark, Germany).
Habitat: On leaves of Quercus robur lying on the ground.
Holotype: GERMANY, BRANDENBURG, Potsdam-Mittel-mark, 2 km NW Krielow, grid square 3542/43, on leaves of Quercus robur in a stand of Rubus caesius, soc. white anamorph, Mycosphaerella punctiformis and rhizomorphs, 31 Oct 2010, V. Kummer (WU 31611; culture Hypo 645 = CBS 130008)
Paratype: DENMARK, Jutland, Aalborg, SE Vestbjerg, 57°07′25″ N, 9017658′58″ E, 20 m, deciduous forest, on leaves of Quercus robur, soc. Mycosphaerella punctiformis, teleomorph, 23 Aug 2011, J.H. Petersen JHP-11.324 (WU 31612, culture Hypo 650 = CBS 131939).
Possibly also representing this species due to similar but immature stromata occurring on a leaf: FINLAND, Mekrijärvi, 28 Aug 1996, C. Lange, comm. T. Læssøe & J.H. Petersen (JHP-96.091).
Notes: Foliicolous habit, an often persistent whitish base and a strongly reduced, verticillium-like, colorless anamorph on SNA are diagnostic for H. foliicola. The solitary or scattered stromata do not permit a standardized color determination. A conspicuously uneven stroma surface, the hyphal base, but also the oblong conidia of the anamorph on the natural substrate and in culture are reminiscent of H. longipilosa. The latter however differs in many traits (e.g. lignicolous habit, green ascospores and conidia and elongations on conidiation pustules). In addition, H. longipilosa is closely related to H. strictipilosa with high support () while being phylogenetically distant from H. foliicola.
Discussion
With the two species described here, Hypocrea species add up to 77 in central and northern Europe. Hypocrea britdaniae seems to be rare. It was known only from a single specimen in the Kew fungarium until recently. It appears to be highly substrate specific, because it has been collected only on corticated dead Salix trunks from damp sites. Another hypocrealean fungus forming large conspicuous stromata on Salix is Hypocreopsis lichenioides, which differs for example by radiating finger-like lobes or ridges and large one-septate, non-disarticulating ascospores and is often associated with Hymenochaete tabacina.
The phylogenetic placement of H. britdaniae was determined with DNA extracted from stromata, because ascospores did not germinate on artificial media. A putative anamorph could not be studied for the same reasons. CitationDruzhinina et al. (2012) reassessed the phylogenetic structure of the Longibrachiatum clade, while CitationSamuels et al. (2012) reviewed the taxonomy of the clade and added eight species. The tef1 alignment used in the former work, not published at this time, was taken to compare and ascertain clear distinction of sequences of H. britdaniae from all other species in the clade. However, the final alignment used to compute the tree () is reduced, because H. britdaniae turned out to be situated close to but outside the Longibrachiatum clade. Teleomorph habit of H. britdaniae (viz. large brown stromata with densely set ostiolar dots, which are typical for several species of the Longibrachiatum clade) correlates well with this phylogenetic placement.
The most obvious reason why H. foliicola was not detected earlier is that leaves were not regarded and examined as potential substrates of Hypocrea. Only few species were recorded from leaves, nearly exclusively of monocotyledonous plants.
The anamorph of H. foliicola was found to be pachybasium-like in nature, while in culture it resembles effused conidiation of species such as H. pachypallida or H. bavarica (CitationJaklitsch 2011) but is reduced substantially, scant and present only at the distal margin of the Petri plate or after agar injury of SNA plates covered by mycelium. Based on the light pulvinate stromata devoid of hairs and the white pachybasium-like conidiation in nature, Hypocrea foliicola fits perfectly in the pachybasium core group. A whitish scurf, albeit less conspicuous, often occurs also on stromata of H. minutispora, the most common species of this clade.
Finally we want to alert readers to the importance of the sophisticated skills necessary for the collectors of the species described in this paper. These skills include the careful examination of biological material, alertness and the ability to correctly recognize fungi and place them within their taxonomic context. This is crucial to expand sampling of the world's fungal biodiversity, to spot new bio-resources, as well as to increase the number of well validated fungal DNA sequences.
Acknowledgments
We thank the collectors and communicators of Hypocrea specimens, Volker Kummer, Thomas Laessoe, Jens H. Petersen, Stuart Skeates; Brian Spooner and Begoña Aguirre-Hudson at the fungarium Kew and Walter Till at WU for sending and managing collections.
Literature cited
- BirksaSMEdwardsSV. 2002. A phylogeny of the megapodes (Aves: Megapodiidae) based on nuclear and mitochondrial DNA sequences. Mol Phylogenet Evol 23:408–421, doi:10.1016/S1055-7903(02)00002-7
- DruzhininaISKomoń-ZelazowskaMIsmaielAJaklitschWMMulawTSamuelsGJKubicekCP. 2012. Molecular phylogeny and species delimitation in the Longibrachiatum clade of Trichoderma. Fungal Genet Biol 49:358–368, doi:10.1016/j.fgb.2012.02.004
- EdgarRC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797, doi:10.1093/nar/gkh340
- HallTA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98.
- HuelsenbeckJPRonquistF. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755, doi:10.1093/bioinformatics/17.8.754
- JaklitschWVoglmayrH. 2011. Nectria eustromatica sp. nov, an exceptional species with a hypocreaceous stroma. Mycologia 103:209–218, doi:10.3852/10-178
- JaklitschWM. 2009. European species of Hypocrea I. The green-spored species. Stud Mycol 63:1–91, doi:10.3114/sim.2009.63.01
- JaklitschWM. 2011. European species of Hypocrea II. Species with hyaline ascospores. Fungal Divers 48:1–250, doi:10.1007/s13225-011-0088-y
- KatohKMisawaKKumaKMiyataT. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066, doi:10.1093/nar/gkf436
- KatohKTohH. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298, doi:10.1093/bib/bbn013
- NylanderJAWilgenbuschJCWarrenDLSwoffordDL. 2008. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24:581–583, doi:10.1093/bioinformatics/btm388
- SamuelsGJIsmaielAMulawTBSzakacsGDruzhininaISKubicekCPJaklitschWM. 2012. The Longibrachiatum clade of Trichoderma: a revision with new species. Fungal Divers 55:77–108 doi:10.1007/s13225-012-0152-2
- SilvestroDMichalakI. 2011. raxmlGUI: a graphical front-end for RAxML. Org Div Evol doi:10.1007/s13127-011-0056-0.
- StamatakisE. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 doi:10.1093/bioinformatics/btl446
- SungGHSungJMHywel-JonesNLSpataforaJW. 2007. A multigene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Mol Phylogenet Evol 44:1204–1223, doi:10.1016/j.ympev.2007.03.011
- SwoffordDL. 2002. PAUP* 4.0b10: phylogenetic analysis using parsimony (*and other methods). Sunderland, Massachusetts: Sinauer Associates.
- TamuraKNeiM. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526.
- TamuraKPetersonDPetersonNStecherGNeiMKumarS. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol doi:10.1093/molbev/msr121
- VoglmayrHJaklitschWM. 2008. Prosthecium species with Stegonsporium anamorphs on Acer. Mycol Res 112:885–905, doi:10.1016/j.mycres.2008.01.020
- VoglmayrHJaklitschWM. 2011. Molecular data reveal high host specificity in the phylogenetically isolated genus Massaria (Ascomycota, Massariaceae). Fungal Divers 46:133–170, doi:10.1007/s13225-010-0078-5
- WiensJJ. 1998. Combining datasets with different phylogenetic histories. Syst Biol 47:568–581, doi:10.1080/106351598260581