Abstract
Glyphium encompasses species with erect, carbonaceous ligulate to dolabrate ascomata that are strongly laterally compressed and dehisce along a longitudinal slit. The five currently recognized members of the genus are separated primarily by whether the ascospores disassociate into part-spores within the ascus. Glyphium has traditionally been placed in Mytilinidiaceae (Mytilinidiales, Pleosporomycetidae, Dothideomycetes). The present study, based on freshly collected material of G. elatum and G. grisonense, was initiated to determine the phylogenetic placement of Glyphium. Phylogenies inferred from the analysis of sequences of six gene regions (nuLSU, nuSSU, mtSSU, TEF1, RPB1, RPB2) derived from six accessions indicate that Glyphium belongs to Patellariales (Pleosporomycetidae, Dothideomycetes). Our phylogenies also support the phylogenetic relationship of Patellaria and Hysteropatella within this order. The nomenclatural history of Glyphium is summarized and a key to species is provided.
Introduction
Species of Glyphium Nitschke ex F. Lehm. possess distinctive hatchet-shaped carbonaceous ascomata that dehisce along a longitudinal slit (). Although the ascomata are superficial, their bases are occasionally embedded in the substrate and might be associated with black interwoven hyphal strands that anchor the ascomata to a dense subicular matrix (). The hamathecium consists of trabeculate pseudoparaphyses borne in a gel-matrix that are thin to lacking at maturity (CitationZogg 1962, CitationBarr 1990). The ascospores are filiform, multiseptate, occupy the greater length of the ascus in which they are often spirally arranged and become light brown at maturity (). In some species the ascospores disarticulate into part-spores before maturity while still within the ascus (), while in others the ascospores remain intact even after release (). Glyphium are lignicolous or corticolous and have been recovered from a wide range of hosts, including Alnus, Fagus, Fraxinus, Malus, Populus, Pyrus, Rhamnus, Rhus, Ulmus and Salix; one species is described from Tillandsia (CitationZogg 1962).
Glyphium currently encompasses five species separated primarily by whether the ascospores disassociate into part-spores (). Species with ascospores that remain entire include G. elatum (Grev.: Fr.) H. Zogg (), the type of the genus and G. tillandsiae (E.K. Cash) H. Zogg () (CitationCash 1943). Three species with ascospores that disassociate into part-spores include G. corrugatum (Ellis) Goree (), a species found in the western United States and Canada (CitationSutton 1970, CitationGoree 1974), and two morphologically similar European species, G. grisonense Math. () and G. schizosporum (Maire) H. Zogg () (CitationZogg 1962, CitationSutton 1970). Anamorphs belonging to the form genus Peyronelia have been described for these latter three species (CitationLohman 1933a, CitationSutton 1970, CitationGoree 1974) but are unknown for Glyphium possessing intact ascospores.
The first sequence data for Glyphium was provided by CitationLindemuth et al. (2001) and CitationLumbsch et al. (2005). They sequenced CBS 268.34, the only available culture of G. elatum derived from the rhizoidal strands and subiculum subtending the ascoma of this species. These workers employed sequences from the nuclear small (nuSSU) and large (nuLSU) ribosomal subunits, as well as the mitochondrial small (mtSSU) and large (mtLSU) ribosomal subunits. This demonstrated that Glyphium was not a member of the Mytilinidiaceae but closely associated with Chaetothyriales, in the Eurotiomycetes. The unexpected placement of Glyphium in Chaetothyriales was restated in a number of studies based on CBS 268.34. However, subsequent work by CitationGueidan et al. (2008), CitationTsuneda et al. (2011) and CitationRéblová et al. (2013) positioned this CBS culture within Knufia in Chaetothyriales, resulting in the relabeling of the sequence accessions at GenBank corresponding to CBS 268.34 as “Knufia sp. CBS 268.34”.
Figs 1–8. Ascomatal variation within the genus Glyphium. 1. Glyphium elatum (EB 0329/BPI 892669), Salix caprea, Luxembourg. 2. Glyphium schizosporum (BPI 652567), Salix appendiculata, Switzerland. 3. Glyphium corrugatum (BPI 1107474), decorticated deciduous wood, Oregon, USA. 4. Glyphium grisonense (TROM 4395), Salix caprea ssp. sphacelata, Norway. (photo credit: Jostein Kjærandsen, UiT). 5. Glyphium tillandsiae (labeled as Lophium tillandsiae, BPI 1108420), Tillandsia fasciculata, Florida, USA. 6. Intact ascus containing eight disarticulating ascospores of G. schizosporum (BPI 652567). 7. Intact ascus containing eight filiform ascospores of G. elatum (EB 0342/BPI 892670). 8. Filiform ascospore sending out multiple germ tubes on water agar, recovered from fresh material of G. elatum (EB 0342/BPI 892670). Bars: 1–5 = 0.5 mm, 6–8 = 50 μm.

Based on morphological work, scientists (e.g. CitationBarr 1990) placed Glyphium in what is now Mytilinidiales in Dothideomycetes. The aim of the current study was to determine the phylogenetic placement of this genus based on sequences obtained from fresh, recently collected material of G. elatum and G. grisonense from Luxembourg, France and Norway. DNA sequence data from freshly collected Patellariales, an order with poor representation in the public sequence databases, were included for comparison.
Materials and methods
Taxon sampling.
Fungal specimens, collection data and DNA GenBank accession numbers generated in this study are provided (). Additional accessions used in the analysis are provided (Supplementary table I). Herbarium voucher specimens have been deposited with U.S. National Fungus Collections (BPI) at Beltsville, Maryland. Four fresh specimens of Glyphium elatum, three from Luxembourg and one from France, as well as two specimens of G. grisonense from Norway, one fresh and one dried, were used. We also examined Patellariales collected in Luxembourg.
An attempt was made to obtain cultures from viable ascospores from fresh material. Ascomata of Glyphium elatum affixed to the underside of Petri plate lids with double-sided sticky tape discharged ascospores onto water agar that sent out multiple germ tubes from cells along the length of the intact, non-fragmenting ascospore (). A number of single-ascospore cultures were obtained for G. elatum, but these cultures were lost and we were not able to deposit these into culture collections. Ascospores were not discharged from the ascomata of G. grisonense.
DNA extraction, PCR amplification and sequencing.
Genomic DNA of Glyphium elatum and G. grisonense was recovered from about 100 mg of material (equivalent to about 10 ascomata) using the DNeasy® Plant Mini Kit (QIAGEN Inc.) following the instructions of the manufacturer but employing sterile white quartz sand and a Kontes® battery-powered pestle grinder in 1.5 mL microfuge tubes. DNA from additional Patellariales were obtained by adding Thermo Scientific™ Phire Plant Direct PCR Master Mix to individual Eppendorf tubes that contained ascomata crushed in 20 μL dilution buffer with a glass rod. Efforts to isolate DNA from aged ascomata of Glyphium obtained from BPI were not successful.
The internal transcribed spacers in the nuclear ribosomal cistron (ITS) and the nuSSU were amplified and double-strand sequenced using the primers pairs ITS4 and ITS5 and NS1 and NS4, respectively (CitationWhite et al. 1990). The nuLSU was amplified and sequenced with primers LR0R (CitationRehner and Samuels 1994) and LR7 (CitationVilgalys and Hester 1990) and internal sequencing primers LR3R and LR16 (CitationMoncalvo et al. 1993). Primers and amplification reactions were used as in CitationSchoch et al. (2006) and CitationBoehm et al. (2009b). Primers 983 and 2218R were used to sequence fragments of the translation elongation factor 1 alpha (TEF1) gene. Fragments of the largest subunit of RNA polymerase II (RPB1) were sequenced with primers RPB1-Ac and RPB1-Cr.
Phylogenetic analysis.
We prepared a matrix of aligned nucleotide sequences, consisting of six different phylogenetic markers and 217 taxa representing bitunicate Dothideomycetes and Eurotiomycetes. Sequences initially were downloaded from TreeBASE (www.treebase.org; submission No. 11726) produced by Schoch and Grube (in press) that combined data from CitationSchoch et al. (2009a) and CitationErtz and Tehler (2011). An additional 20 sequences were downloaded from the GenBank nucleotide database and realigned with MAFFT 6.716b (CitationKatoh et al. 2009) using the initial matrix as a seed. Newly generated GenBank accession numbers are indicated (); additional accessions used in this analysis are provided (Supplementary table I). Sequences from the second largest subunit of the RNA polymerase II gene (RPB2) were included to improve this analysis, although no sequences of this gene from Glyphium were obtained. The final alignment consisted of a concatenated matrix from six markers (nuLSU, nuSSU, mtSSU, TEF1, RPB1, RPB2) for 217 taxa with 7085 nucleotide characters of which 54.14% were either missing or gaps.
Phylogenetic analysis was performed at the CIPRES 3.3 webportal (CitationMiller et al. 2010) using RAxML 8.0.9 as part of the RAxML-HPC BlackBox tool (CitationStamatakis 2006, CitationStamatakis et al. 2008) set to find the best tree in maximum likelihood and allowing for boot-stopping with MRE. This was performed applying unique model parameters for each marker and codon (where applicable) with the dataset divided in 12 partitions following CitationSchoch et al. (2009b). A general time reversible model with four gamma rate categories, determining the proportion of invariable sites, was applied and the resulting tree was compressed to indicate major lineages in MEGA 6.0 (CitationTamura et al. 2013). The alignment and the complete phylogeny are deposited in TreeBASE (submission number 16151). The phylogram with values from 250 bootstrap replicates indicated above the branches is presented (). Bayesian inference of maximum likelihood was performed with the MrBayes 3.2.2 tool at the CIPRES 3.3 web portal (CitationMiller et al. 2010). MrBayes was run with similar parameters as RAxML: a general time reversible model with gamma-distributed rate variation across sites (invariance, partitioning across genes and codons). A Markov chain Monte Carlo (MCMC) analysis with two metropolis coupling runs starting from a random tree for 20 000 000 generations, sampling every 2000th cycle. Four chains were run for each separate run simultaneously with the initial 1 500 000 discarded as burn in. Bayesian posterior probabilities of the combined trees from the two separate runs are indicated as percentages below the nodes (). In addition to this two RNA polymerase II subunit sequences (RPB1, RPB2) were obtained from the Patellaria atrata (Hedw.) Fr. genome sequence at JGI (CBS 101060) as part of the 1000 fungal genomes project (CitationGrigoriev et al. 2011) and compared with permission of the original submitters. They revealed a clear affinity to other sequences from P. atrata and P. cf. atrata (data not shown).
Table I. Newly generated sequences for this study, their provenance and GenBank accession numbers
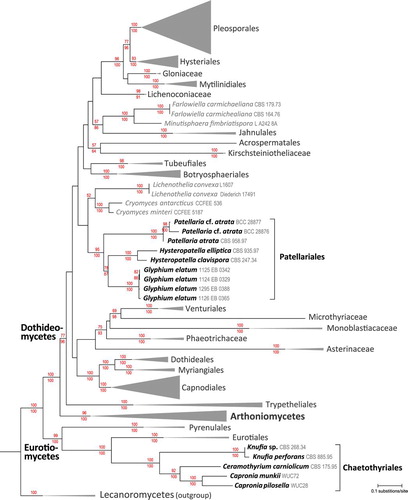
Fig. 9. A RAxML maximum-likelihood tree obtained from a dataset of 217 taxa including representatives of most major orders in Dothideomycetes and representatives of Eurotiomycetes comparing six genes. Two members of Lecanoromycetes, Cladonia caroliniana and Flavoparmelia caperata, were used as outgroups. The set of numbers above the nodes are the percentage bootstraps recovery, and those below represent Bayesian posterior probabilities expressed as percentages.
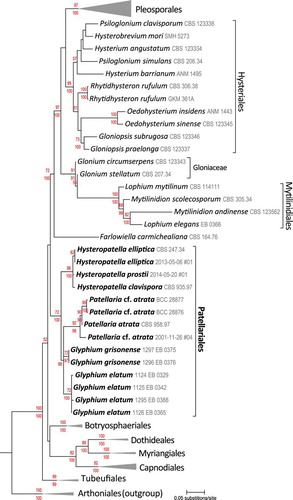
A second dataset of 66 ribosomal nuLSU sequences was chosen for an analysis focused on hysteriaceous species (indicated in Supplementary table I). Sequences from G. grisonense as well as Hysteropatella elliptica (Fr. : Fr.) Rehm, H. prostii (Duby) Rehm and Patellaria cf. atrata were appended and aligned to this template with MAFFT 6.716b (CitationKatoh et al. 2009). This was optimized further for 10 iterations with SATe 2.2.7 (CitationLiu et al. 2009) with MAFFT (CitationKatoh et al. 2009) as the external sequence alignment tool and MUSCLE to merge sub-alignments (CitationEdgar 2004). SATe used FASTTREE for tree estimations with blind mode enabled and decomposition set to centroid. The final alignment consisted of 663 taxa with 1283 characters consisting of 13.35% gaps and undetermined characters. A phylogenetic analysis was performed similarly to the six-marker dataset but without partitioning. The resulting tree is presented (). A Bayesian analysis with similar settings as above was performed for 10 000 000 generations, sampling every 1000th cycle and the initial 1 500 000 generations discarded as burn in. Bayesian posterior probabilities obtained from the combined trees from two independent runs are indicated as percentages below the nodes ().
Results
Phylogenetic analysis.
A RAxML maximum likelihood tree () inferred from an analysis of a concatenated matrix of six marker genes (nuSSU, nuLSU, mtSSU, TEF1, RPB1, RPB2) for 217 fungal taxa, including representatives of all major orders and families of Dothideomycetes. Representative Arthoniales, Trypetheliales and Eurotiomycetes also were included. Two Lecanoromycetes (Cladonia caroliniana Tuck. and Flavoparmelia caperata (L.) Hale) were used as outgroups. In this analysis the genus Glyphium, represented by four accessions of G. elatum, is positioned within Dothideomycetes. The closest related species are in Hysteropatella and Patellaria, taxa that currently are classified in the Patellariales. These results are largely congruent with other recent large-scale phylogenetic analyses of Dothideomycetes (CitationSchoch et al. 2009a, CitationHyde et al. 2013).
The results of an analysis of nuLSU sequences of 66 hysteriaceous taxa is presented (). Three members of Arthoniales (Schismatomma pericleum (Ach.) Branth & Rostr., Dendrographa decolorans (Turner & Borrer) Ertz & Tehler and Roccellographa cretacea J. Steiner) served as outgroups. In this analysis all accessions of Glyphium, representing G. elatum and G. grisonense, were included. We also included newly generated sequences from two Hysteropatella and another species similar to Patellaria atrata. Again, our analysis clearly supports the inclusion of Glyphium within Dothideomycetes and indicates that this genus is most closely related to Hysteropatella and Patellaria ().
Discussion
Glyphium originally was placed by CitationZogg (1962) in Lophiaceae (Citationvon Arx and Müller 1975) a family that also included Actidium, Lophium and Mytilinidion. CitationBarr (1990) reinstated the earlier name Mytilinidiaceae (CitationKirschstein 1924) over Lophiaceae, despite proposals to conserve the latter (CitationHawksworth and Eriksson 1988). Since then a number of genera have been added to the Mytilinidiaceae, including Ostreichnion (CitationBarr 1975, Citation1987, Citation1990), Ostreola (CitationDarker 1963), Quasiconcha (CitationBarr and Blackwell 1980) and Zoggium (CitationVasilyeva 2001). The inclusion of Glyphium within Mytilinidiaceae was based on a suite of presumptive synapomorphic character states, many of which were shared among genera, especially Lophium and Glyphium. These include fragile, yet persistent, carbonaceous, ascomata that are strongly laterally compressed and erect. The lateral walls are more or less converging and extend vertically to a prominent longitudinal apex. They can be clam-, mussel- or hatchet-shaped. These fungi posses a fragile, thin-walled, scleroparenchymatous periderm, that can be composed of cephalothecoid plates. The hamathecium contain narrow trabeculate pseudoparaphyses, borne in a gel matrix, often sparse to lacking at maturity. All genera possess functionally fissitunicate asci, cylindrical with a narrow ocular chamber, and borne on a basal cushion. From a morphological perspective the Mytilinidiaceae form a strikingly homogenous group whose genera are differentiated primarily by ascospore pigmentation and septation (CitationZogg 1962, CitationBarr 1987).
CitationSchoch et al. (2006), using a single isolate of Lophium mytilinum Pers.: Fr., were the first to provide molecular evidence placing Mytilinidiaceae within Pleosporomycetidae. Subsequently CitationBoehm et al. (2009a, Citationb) combined DNA and amino acid sequence data from five genes to reconstruct the phylogeny of Hysteriaceae, Gloniaceae and Mytilinidiaceae. These multigene phylogenies clearly indicated Mytilinidiaceae to be a highly monophyletic lineage for which the new order, Mytilinidiales, was proposed (CitationBoehm et al. 2009a). In this sense the molecular data strongly supported the suite of presumptive synapomorphic character states for the family, mentioned above. However, Ostreichnion was transferred to Hysteriaceae (CitationBoehm et al. 2009a, Citationb). The remaining genera (Ostreola, Zoggium, Glyphium), for which molecular data were lacking, were retained provisionally within Mytilinidiaceae.
Fig. 10. A RAxML maximum likelihood tree obtained from a single marker nuLSU dataset of 66 taxa with a focus on hysteriaceous lineages in Dothideomycetes. Three members of Arthoniales, Arthoniomycetes, Schismatomma pericleum, Dendrographa decolorans and Roccellographa cretacea were used as outgroups. The numbers above the nodes are percentage bootstraps recovery and those below represent Bayesian posterior probabilities expressed as percentages.
Taxonomy
Nomenclatural history and a key to species of Glyphium.
Nomenclatural and taxonomic synonyms and characterization of the members of the genus are summarized below: Glyphium corrugatum (Ellis) Goree Can. J. Bot. 52:1266.1974.
≡ Acrospermum corrugatum Ellis, Bull. Torrey bot. Club 8:124. 1881.
= Lophium leptothecium Earle, in Greene, Plantae Bakerianae 2:11. 1901.
≡ Glyphium leptothecium (Earle) B. Sutton, Trans. Br. Mycol. Soc. 54:256. 1970.
= Acrospermum fultum Harkn., Bull. Calif. Acad. Sci. 1:47. 1884.
CitationZogg (1962) treated Lophium leptothecium Earle as a synonym of Glyphium elatum based on a description by CitationSaccardo (1902). CitationSutton (1970) examined Earle’s original material (CitationEarle 1901), designating one specimen a lectotype. After comparing the lectotype to specimens collected from Populus tremuloides in Manitoba and Saskatchewan CitationSutton (1970) concluded that these were conspecific. Noting that L. leptothecium could be distinguished from G. elatum in possessing a Peyronelia anamorph and ascospores that disarticulated within the ascus before discharge, CitationSutton (1970) transferred this species to Glyphium. CitationGoree (1974) and subsequently treated G. leptothecium as a synonym of G. corrugatum based on the precedence of the name Acrospermum corrugatum (CitationEllis 1881) over Lophium leptothecium (CitationEarle 1901). Glyphium corrugatum occurs on decorticated wood and is known from the western United States and Canada (CitationSutton 1970, CitationGoree 1974).
CitationLohman (1933a) collected a fungus from Salix in Colorado, which he identified as Lophium dolabriforme (Lohman No. 196, MICH) with ascospores that disarticulated within the ascus into one- to three-septate part-spores, 12 × 2 μm. CitationSutton (1970) examined Lohman No. 196 and concluded that it in fact corresponded to G. leptothecium, a species later synonimized by CitationGoree (1974) as G. corrugatum.
CitationLohman (1933a) was unable to induce ascospore germination in G. corrugatum using Lohman No. 196, but he did obtain a culture derived from the rhizoidal strands and subiculum subtending the ascomata from this collection that he deposited as CBS 268.34. CitationSutton (1970) also failed to obtain cultures from the ascospores of G. corrugatum but he did induce conidia to germinate. Although these ceased to grow almost immediately, Sutton was able to demonstrate the connection between the Peyronelia anamorph and the rhizoidal strands subtending the ascomata of this species.
Glyphium elatum (Grev. : Fr.) H. Zogg Beitr. Kryptogamenfl. Schweiz 11:99. 1962.
≡ Lophium elatum Grev. : Fr., Elenchus Fungorum 2:113. 1828.
= Lophium dolabriforme Wallr., Flora cryptogam. germ. 2:433. 1833.
≡ Glyphium dolabriforme (Wallr.) Nitschke ex F. Lehm., Nova Acta Caes. Leop. Carol. Deutsch. Akad. Nat. 50:139. 1886.
CitationGreville (1825) established Lophium, based on L. elatum Grev. and was the first to illustrate the filiform multiseptate ascospores of this species. CitationZogg (1962) transferred three members of the genus, including the type, to Glyphium, noting that the dolabriform nature of the ascomata of these species differed from the conchate ascomata found in Lophium. Glyphium elatum occurs on several angiosperm trees and shrubs in both North (CitationLohman 1933a) and South America (CitationLorenzo and Messuti 2005), Europe (CitationBisby and Ellis 1952, CitationZogg 1962, CitationNordén et al. 1997), the Russian Far East (CitationVasilyeva 2001), China and Taiwan (CitationChen and Hsieh 1996).
Glyphium grisonense Math. Sommerfeltia 20:89. 1993.
Glyphium grisonense resembles G. schizosporum in possessing ascospores that disarticulate within the asci, but it can be distinguished by its wider ascomata that often become obpyriform in outline, considerably wider asci and slightly longer and wider part-spores. In addition grisonense is known only from Salix in northern Norway and on Betula in Switzerland (CitationMathiassen 1993). The conidia are thick-walled, multiseptate, irregularly constricted, verrucose and dark brown and form unbranched chains. They resemble the conidia of the Peyronelia state of G. schizosporum but do not possess longitudinal septa.
Glyphium schizosporum (Maire) H. Zogg Beitr. Kryptogamenfl. Schweiz 11:101. 1962.
≡ Lophium schizosporum Maire, Bull. Soc. Hist. nat. Afr. N. 8:172. 1917.
Glyphium schizosporum is morphologically similar to G. corrugatum and has been reported from Algeria, France, and Switzerland (CitationZogg 1962, CitationSutton 1970). Aside from the differences in the geographical distribution of these species, G. schizosporum is distinguished in possessing part-spores that are larger in diameter and more frequently septate. In addition the verruculose, reddish brown conidia of the Peyronelia state of G. schizosporum typically have up to 50 transverse septa with only an occasional longitudinal septum and produce only straight chains that do not branch; the conidia of G. corrugatum have up to 55 transverse septa, usually possess one or two longitudinal septa, and are prone to producing lateral buds or side branches (CitationSutton 1970).
Glyphium tillandsiae (E.K. Cash) H. Zogg Beitr. Kryptogamenfl. Schweiz 11:103. 1962.
≡ Lophium tillandsiae E.K. Cash, Mycologia 35:596. 1943.
The ascomata of Glyphium tillandsiae are only 0.5 mm tall and contain ascospores comparable in size to other Glyphium that measure nearly the length of the asci. This diminutive Glyphium was collected by Shear in 1943 from Tillandsia fasciculata in Highlands Hammock State Park, west of Sebring, Florida, and described by CitationCash (1943). It is known only from the type locality.
The following dichotomous key to the species of Glyphium is adapted from www.eboehm.com.
Key to species of Glyphium
Based on the analyses presented here it is clearly evident that the genus Glyphium resides within Dothideomycetes. We also included a subset of taxa from Eurotiomycetes in our analysis to provide a context for previous phylogenetic placements of Glyphium based on CBS 268.34, showing its close affinities to Knufia perforans, as supported by CitationGueidan et al. (2008), CitationTsuneda et al. (2011) and CitationRéblová et al. (2013).
Most surprising, the results also do not support the genus Glyphium having an affinity with Mytilinidiales (sensu CitationBoehm et al. 2009b), as proposed by CitationZogg (1962), Citationvon Arx and Müller (1975), CitationBarr (1990) and others, based on presumptive synapomorphic character states. Instead there is strong support for the affinity of Glyphium to two accessions of Hysteropatella, namely H. clavispora (CBS 247.34) and H. elliptica (CBS 975.97). In addition these sequences grouped with good resampling support with four accessions of Patellaria in Patellariales. These including sequences from cultures of the type species of Patellariaceae, P. atrata (CBS 958.97), collected from Luxembourg, and two accessions identified as P. cf. atrata (BCC 28876, BCC 28877), isolated from mangrove wood, collected in Hong Kong and Thailand (CitationSuetrong et al. 2009).
The more focused analysis also supported the placement of Glyphium within Patellariales (). This analysis was based on nuLSU sequences among 66 taxa, including the nuLSU sequences obtained from two accessions of G. grisonense, excluded from the global analysis (). It also should be noted that the representatives of the genus used in our analyses include one species with non-articulating ascospores and no known anamorph (G. elatum) as well as one species with fragmenting ascospores and a Peyronelia anamorph (G. grisonense).
Despite the weight of this molecular evidence, the placement of Glyphium within Patellariales is at odds with what we would have predicted based on the morphology of these taxa. In contrast to Glyphium, taxa traditionally included in Patellariales typically possess black, erumpent or superficial, coriaceous apothecial ascomata that develop from a unilocular stroma beneath the substrate. At maturity these ascomata open at a pore, slit or irregular crack to reveal a flat, cupulate or slightly convex disk that may smooth or minutely roughened but which is normally darkly colored (CitationKutorga and Hawksworth 1997). Patellarioid ascomata have been termed ascostromatic or pseudothecial apothecia or discothecia (CitationKorf 1962) to denote this stromatic ontogeny. The hamathecium is composed of paraphysoids that at first are anchored at both ends to the inner tissue of the closed ascostroma but which elongate and push apart the thin upper stromal layers to reveal a palisade of bitunicate asci borne among paraphysoids. The free ends of the paraphysoids enlarge, branch and become swollen and form the pigmented epithecium characteristic of Patellariales (CitationKutorga and Hawksworth 1997).
From a developmental standpoint this process is distinctive from the ontogeny of the ascomata seen in Glyphium. Although the hamathecia of Glyphium consist of trabeculate pseudoparaphyses, these structures are thin to lacking at maturity (CitationBarr 1987, Citation1990), are not apically swollen, pigmented or encrusted, and they never form a protective epithecium. Furthermore, the thin-walled, fragile, scleroparenchymatous peridium of the ascomata of Glyphium bear little resemblance to the multilayered, corriaceous, pseudoparenchymatous exciple found in Patellariaceae.
The Patellariaceae have long been regarded as a highly heterogenous family (CitationHafellner 1979, CitationSamuels and Müller 1979, CitationBarr 1987, CitationEriksson 1994, CitationKutorga and Hawksworth 1997). We conclude that morphological characters, previously considered as phylogenetically informative synapomorphies within Patellariales, may not be reliable evolutionary markers. Whether Glyphium should be given its own familial status within the Patellariales awaits future studies that include additional members of the genus.
Supplementary text - EXTENDED NOMENCLATURAL HISTORY OF GLYPHIUM
Download MS Word (31.2 KB)Supplementary table I - GenBank accessions used in total analysis
Download MS Word (281 KB)Acknowledgments
We thank Teppo Rämä (University of Tromsø) for a critical reading of the manuscript. CLS acknowledges support from the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Literature cited
- AltschulSFGishWMillerWMyersEWLipmanDJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410.
- BarrME. 1975. The genus Ostreichnion. Mycotaxon 3:81–88.
- BarrME. 1987. Prodromus to class Loculoascomycetes. Hamilton I. Newell Inc., Amherst, Massachusetts: M.E. Barr Bigelow.
- BarrME. 1990. Melanommatales (Loculoascomycetes) II. North American Flora II. 13:1–129.
- BarrMEBlackwellM. 1980. A new genus in the Lophiaceae. Mycologia 72:1224–1227.
- BisbyGREllisMB. 1952. Lophium elatum Grev. Trans Br Mycol Soc 35:299–303.
- BoehmEWASchochCLSpataforaJW. 2009a. On the evolution of the Hysteriaceae and Mytilinidiaceae (Pleosporomycetidae, Dothideomycetes, Ascomycota) using four nuclear genes. Mycol Res 113:461–479.
- BoehmEWAMugambiGKMillerANMMarincowitzSSpataforaJWSchochCL. 2009b. A molecular phylogenetic reappraisal of the Hysteriaceae, Mytilinidiaceae and Gloniaceae (Pleosporomycetidae, Dothideomycetes) with keys to world species. Stud Mycol 64:49–83.
- CashE. 1943. Some new or rare Florida Discomycetes and Hysteriales. Mycologia 35:595–603.
- ChenCYHsiehWH. 1996. Two new species and some new records of ascomycetes from Taiwan. Bot Bull Acad Sin 37:219–227.
- DarkerGD. 1963. A new genus of the Lophiaceae. Can J Bot 41:1383–1388.
- EarleSF. 1901. Fungi from Colorado and New Mexico. Plantae Bakerianae 2:1–30.
- EdgarRC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797.
- EllisJB. 1881. New ascomycetous fungi. Bull Torrey Bot Club 8:123–125.
- ErikssonOE. 1994. Problems in the classification of fissitunicate ascomycetes. In: HawksworthDL, ed. Ascomycete systematics: problems and perspectives in the nineties. NATO ASI series. Vol. 269. New York and London: Plenum Press. p 341–348.
- ErtzDTehlerA. 2011. The phylogeny of Arthoniales (Pezizomycotina) inferred from nucLSU and RPB2 sequences. Fungal Divers 49:47–71.
- GoreeH. 1974. Glyphium in western Canada and United States. Can J Bot 52:1265–1269.
- GrevilleRK. 1825. Scottish Cryptogamic Flora III., Edinburgh.
- GrigorievIVCullenDGoodwinSBHibbettDJeffriesTWKubicekCPKuskeCMagnusonJKMartinFSpataforaJWTsangABakerSE. 2011. Fueling the future with fungal genomics. Mycology 2:192–209.
- GueidanCRuibalCVde HoogGSGorbushinaAAUntereinerWALutzoniF. 2008. A rock-inhabiting ancestor for mutualistic and pathogen-rich fungal lineages. Stud Mycol 61:111–119.
- HafellnerJ. 1979. Karschia: Revision einer Sammelgattung an der Grenze von lichenisierten und nichtlichenisierten Ascomyceten. Beih Nova Hedwigia 62:1–248.
- HawksworthDLErikssonOE. 1988. Proposals to conserve 11 family names in the Ascomycotina (Fungi). Taxon 37:190–193.
- HydeKDJonesEBGLiuJKAriyawansaHBoehmEBoonmeeSBraunUChomnuntiPCrousPWDaiDQDiederichPDissanayakeADoilomMDoveriFHongsananSJayawardenaRLawreyJDLiYMLiuYXLückingRMonkaiJMuggiaLNelsenMPPangKLPhookamsakRSenanayakeICShearerCASuetrongSTanakaKThambugalaKMWijayawardeneNNWikeeSWuHXZhangYAguirre-HudsonBAliasSAAptrootABahkaliAHBezerraJLBhatDJCamporesiEChukeatiroteEGueidanCHawksworthDLHirayamaKDe HoogSKangJCKnudsenKLiWJLiXHLiuZYMapookAMcKenzieEHCMillerANMortimerPEPhillipsAJLRajaHAScheuerCSchummFTaylorJETianQTibprommaSWanasingheDNWangYXuJCYacharoenSYanJYZhangM. 2013. Families of Dothideomycetes. Fungal Divers 63:1–313.
- KatohKAsimenosGTohH. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol 537:39–64.
- KirschsteinW. 1924. Beiträge zur Kenntnis der Ascomyceten. Verh Bot Vereins Prov Brandenburg 66:23–29.
- KorfRP. 1962. A synopsis of the Hemiphacidiaceae, a family of the Helotiales (Discomycetes) causing needle blights of conifers. Mycologia 54:12–33.
- KutorgaEHawksworthDL. 1997. A reassessment of the genera referred to the family Patellariaceae (Ascomycota). Syst Ascomycetum 15:1–110.
- LindemuthRWirtzNLumbschHT. 2001. Phylogenetic analysis of nuclear and mitochondrial rDNA sequences supports the view that loculoascomycetes (Ascomycota) are not monophyletic. Mycol Res 105:1176–1181.
- LiuKRaghavanSNelesenSLinderCRWarnowT. 2009. Rapid and accurate large-scale co-estimation of sequence alignments and phylogenetic trees. Science 324:1561–1564.
- LohmanML. 1933a. Hysteriaceae: Life histories of certain species. Pap Michigan Acad 17:229–288.
- LohmanML. 1933b. Septonema toruloideum: a stage of Mytilidion scolecosporum. Mycologia 25:34–43.
- LorenzoLEMessutiMI.2005. Glyphium elatum (Ascomycota) in Patagonia (Argentina). Bol Soc Argent Bot 40:13–16.
- LumbschHTSchmittILindemuthRMillerAMangoldAFernandoFHuhndorfS. 2005. Performance of four ribosomal DNA regions to infer higher-level phylogenetic relationships of inoperculate euascomycetes (Leotiomyceta). Mol Phylogenet Evol 34:512–524.
- MathiassenG. 1993. Corticolous and lignicolous Pyrenomycetes s. lat. (Ascomycetes) on Salix along a mid-Scandinavian transect. Sommerfeltia 20:1–180.
- MillerMPfeifferWSchwartzT. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway computing environments workshop (GCE), New Orleans, p 1–8.
- MoncalvoJMRehnerSAVilgalysR. 1993. Systematics of Lyophyllum section Difformia based on evidence from culture studies and ribosomal DNA sequences. Mycologia 85:788–794.
- NordénBAppelquistTBarckLLöhmusM. 1997. An ecological field study of wood living pyrenomycetes in a Swedish hardwood forest. Windahlia 22:57–64.
- RéblováMUntereinerWARéblováK. 2013. Novel evolutionary lineages revealed in the Chaetothyriales (Fungi) based on multigene phylogenetic analyses and comparison of ITS secondary structure. PLoS ONE 85:e63547.
- RehnerSASamuelsGJ. 1994. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res 98:625–634.
- SaccardoPA. 1902. Sylloge Fungorum hucusque cognitorum, 16:1–1291.
- SamuelsGJMüllerE. 1979. Life-history studies of Brazilian ascomycetes. 7. Rhytihysteron rufulum and the genus Eutryblidiella. Sydowia 32:277–292.
- SchochCLShoemakerRASeifertKAHambletonSSpataforaJWCrousPW. 2006. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98:1041–1052.
- SchochCLCrousPWGroenewaldJZBoehmEWBurgessTIde GruyterJde HoogGSDixonLJGrubeMGueidanCHaradaYHatakeyamaSHirayamaKHosoyaTHuhndorfSMHydeKDJonesEBKohlmeyerJKruysALiYMLückingRLumbschHTMarvanovaLMbatchouJSMcVayAHMillerANMugambiGKMuggiaLNelsenMPNelsonPOwensbyCAPhillipsAJPhongpaichitSPointingSBPujade-RenaudVRajaHAPlataERRobbertseBRuibalCSakayarojJSanoTSelbmannLShearerCAShirouzuTSlippersBSuetrongSTanakaKVolkmann-KohlmeyerBWingfieldMJWoodARWoudenbergJHYonezawaHZhangYSpataforaJW. 2009a. A class-wide phylogenetic assessment of Dothideomycetes. Stud Mycol 64:1–15.
- SchochCLSungGHLopez-GiraldezFTownsendJPMiadlikowskaJHofstetterVRobbertseBMathenyPBKauffFWangZGueidanCAndrieRMTrippeKCiufettiLMWynnsAFrakerEHodkinsonBPBonitoGGroenewaldJZArzanlouMde HoogGSCrousPWHewittDPfisterDHPetersonKGryzenhoutMWingfieldMJAptrootASuhSOBlackwellMHillisDMGriffithGWCastleburyLARossmanAYLumbschHTLückingRBudelBRauhutADiederichPErtzDGeiserDMHosakaKInderbitzinPKohlmeyerJVolkmann-KohlmeyerBMostertLO’DonnellKSipmanHRogersJDShoemakerRASugiyamaJSummerbellRCUntereinerWJohnstonPRStenroosSZuccaroADyerPSCrittendenPDColeMSHansenKTrappeJMYahrRLutzoniFSpataforaJW. 2009b. The Ascomycota Tree of Life: A phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol 58:224–239.
- SchochCLGrubeM. 2014. Pezizomycotina: Dothideomycetes and Arthoniomycetes In: McLaughlinDJSpataforaJW, eds. The Mycota. Vol. VII, Part B. Berlin, Germany: Springer Verlag, p. 143–178.
- StamatakisA. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690.
- StamatakisAHooverPRougemontJ. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57:758–771.
- SuetrongSSchochCLSpataforaJWKohlmeyerJVolkmann-KohlmeyerBSakayarojJPhongpaichitSTanakaKHirayamaKJonesEB. 2009. Molecular systematics of the marine Dothideomycetes. Stud Mycol 64:155–173.
- SuttonBC. 1970. Glyphium leptothecium (Earle) comb. nov., G. schizosporum (Maire) Zogg, and their imperfect states. Trans Br Mycol Soc 54:255–264.
- TamuraKStecherGPetersonDFilipskiAKumarS. 2013. MEGA 6: molecular evolutionary genetics analysis Mol Biol Evol 30:2725–2729.
- TsunedaADaveyMLCurrahRS. 2011. A new endoconidial black meristematic genus, Atramixtia, associated with declining white spruce and phylogenetically allied to a lineage of dothidealean conifer pathogens. Botany 89:323–338.
- VasilyevaLN. 2001. Hysteriaceous fungi in the Russian Far East IV. Glyphium, Lophium and Mytilinidion. Mikolog Fitopatol 35:15–18.
- VilgalysRHesterM. 1990. Rapid identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246.
- von ArxJAMüllerE. 1975. A re-evaluation of the bitunicate Ascomycetes with keys to families and genera. Stud Mycol 9:1–159.
- WhiteTJBrunsTLeeSTaylorJ. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: InnisMAGelfandDHSninskyJJWhiteTJ, eds. PCR protocols: a guide to methods and applications. New York: Academic Press. p 315–322.
- ZoggH. 1962. Die Hysteriaceae s. str. und Lophiaceae, unter besonderer Berücksichtigung der mitteleuropäischen Formen. Beiträge zur Kryptogamenflora der Schweiz, Band 11:1–190.
