Abstract
During a global investigation of fungi in house dust, we isolated six novel arthroconidial fungi. Phylogenies from combined analysis of nuc rDNA 18S, 28S and internal transcribed spacers sequences demonstrated that these fungi and two species preserved in culture collections represent undescribed species of Spiromastigaceae, Onygenales. Seven of the eight species lacked sexual states and only characters of asexual states and growth rates on different media could be used to characterize them. The eighth species produced ascomata only on water agar. We introduce six new species and one new combination in Spiromastix and validate the recently proposed family Spiromastigaceae, genus Pseudospiromastix and combination Ps. tentaculata. The new genus Sigleria is proposed for two new species that differ from Spiromastix by conidiophore branching patterns, slower growth and a limited ability to utilize nitrate as a sole N source. A key to the three genera of Spiromastigaceae, Spiromastix, Pseudospiromastix and Sigleria, is provided. Phylogenetic analyses support the placement of Spiromastigaceae within Onygenales.
Introduction
The number of species of Onygenales is increasing as new habitats are explored and sequence data accumulates (e.g. CitationGuarro et al. 2002, CitationRizzo et al. 2014). Many are dermatophytes or agents of other diseases of vertebrates and some are keratinolytic; others are saprobes in soil or dung. The order is characterized by smooth or appendiculate, brightly colored ascomata with walls either of loosely interwoven or anastomosing hyphae or of textura angularis, unitunicate asci that are subglobose to turbinate and one-celled, hyaline ascospores (CitationCurrah 1994; CitationSugiyama et al. 1999, Citation2002). Asexual states feature thallic conidium ontogeny and arthro- and/or aleurioconidia. The order includes six families and about 50 teleomorph genera (CitationLumbsch and Huhndorf 2007, CitationKirk et al. 2008), distinguished by morphological, physiological and molecular phylogenetic characters (CitationCurrah 1985; CitationSugiyama et al. 1999, Citation2002; CitationUntereiner et al. 2004). Nineteen anamorph genera are associated with the order (CitationSigler 2002, CitationSeifert et al. 2011), including significant human pathogens such as Blastomyces Costantin & Rolland, Coccidioides G.W. Stiles, Histoplasma Darling and Paracoccidioides F.P. Almeida. Morphological patterns represented by the concepts of the anamorph genera Chrysosporium, with terminal or lateral aleurioconidia, and Malbranchea, with chains of rhexolytically seceding arthroconidia, are broadly distributed in the order.
Spiromastix Kuehn & G.F. Orr was based on the gymnothecial species Sp. warcupii Kuehn & G.F. Orr (CitationKuehn and Orr 1962), commonly isolated from wheat field soils in Adelaide, Australia (CitationWarcup 1957, as Myxotrichum sp.). Spiromastix now includes four species, Sp. asexualis D.A. Sutton, L. Rizzo, Stchigel & Guarro, Sp. saturnispora Uchiy., Kamiya & Udagawa, Sp. sphaerospora Udagawa & Uchiy. and Sp. warcupii. Three of these lack an asexual state, unusual for onygenalean fungi, and absence of conidia initially was considered a generic character. The first evidence that Spiromastix might not be exclusively sexual was the discovery of a strain lacking ascomata but with a malbranchea-like asexual state, which seemed to be a Spiromastix species based on molecular data (CitationSugiyama and Mikawa 2001). CitationRizzo et al. (2014) described Sp. asexualis, the first named species lacking a mature sexual state, although it produced sterile ascomata with loosely coiled appendages.
Several species attributed to Spiromastix are classified elsewhere. CitationUdagawa and Uchiyama (1999) described Sp. princeps Udagawa & Uchiy. but later placed it in a new genus and renamed it Acanthogymnomyces princeps (Udagawa & Uchiy.) Udagawa & Uchiy. (CitationUdagawa and Uchiyama 2000). Spiromastix grisea Currah & Locq.-Lin. (CitationCurrah and Locquin-Linard 1988) was transferred to Ajellomyces McDonough & A.L. Lewis as A. grisea (Currah & Locq.-Lin) Unter. & J.A. Scott based on partial nuc 28S rDNA subunit and small subunit rDNA sequences (CitationUntereiner et al. 2002). Spiromastix tentaculata Guarro, Gené & de Vroey (CitationGuarro et al. 1993) was classified by CitationRizzo et al. (2014) in the invalidly published, monotypic “Pseudospiromastix Guarro, Stchigel & Cano”, differing from Spiromastix by undulate ascomata appendages with swollen tips and ascospores with irregular furrows.
Spiromastix initially was classified in the family Gymnoascaceae and distinguished from similar genera by helical ascomata appendages (CitationBenny and Kimbrough 1980, Citationvon Arx 1987, CitationGuarro et al. 1993). CitationCurrah (1985) classified it as Onygenaceae based on teleomorph morphology but later questioned this because the Spiromastix species then known lacked a conidial state and were non- or only weakly keratinolytic (CitationCurrah 1994). In the absence of keratinolysis and presence of a Q10 (H2) ubiquinone system, CitationUdagawa (1997) tentatively placed Spiromastix in Amauroascaceae. Early phylogenetic studies of rDNA sequences supported Spiromastix as a monophyletic genus of Onygenaceae (CitationSugiyama and Mikawa 2001; CitationSugiyama et al. 2002; CitationUntereiner et al. 2002, Citation2004), placing Spiromastix as a basal lineage sister to Ajellomycetaceae, a family including nonkeratinolytic saprobic or pathogenic species. This phylogenetic relationship correlates with the weak or absent keratinolysis and lack of pathogenicity in Spiromastix (CitationScott and Untereiner 2004). Based on analyses of D1–D2 domains of nuc 28S rDNA sequences, CitationRizzo et al. (2014) proposed the family “Spiromastixaceae” for species with or without conidia and ascomata with slender, curved or twisted ascomata appendages and oblate to globose ascospores, often with an equatorial rim and several wall ornamentation patterns, including reticulate, punctate or irregular furrows.
During our investigation of the mycota of settled house dust, six novel chrysosporium- or malbranchea-like fungi were isolated from samples from several continents. Two additional species, identified only as Spiromastix sp., were obtained from culture collections. Phylogenetic trees inferred from analyses of nuc rDNA internal transcribed spacers 1 and 2, including the 5.8S (ITS) and combined nuc 18S and 28S rDNA sequences demonstrated that these fungi represent new species in Spiromastix clade. Here we describe and illustrate six new species of Spiromastix. Two species are relatively distant from the core of Spiromastix, subtended by a long, well-supported branch and we interpret these to represent a distinct genus, described here as Sigleria. The family name Spiromastigaceae, the genus name Pseudospiromastix and a new combination in Pseudospiromastix are validated.
Materials and methods
Culture isolation
House dust samples used in this study were collected by CitationAmend et al. (2010). About 8000 cultures were isolated by dilution to extinction as described by CitationVisagie et al. (2014), of which ca. 50 were relevant to this study. Samples were serially diluted and dispensed into panels of 96 capped 1.5 mL microtubes containing 1 mL isolation medium, with dilutions calibrated to deliver ~ one particle/tube. This method favors isolation of slow-growing fungi by eliminating competition from fast-growing or antibiotic-producing microbes. Isolation media used were MEA and 20SMEA described below, with 100 mg/L chloramphenicol added to retard bacterial growth.
Morphological studies
For colony descriptions and growth rates, we used single-point inoculations on malt extract agar (MEA: 20 g Bacto malt extract (Difco, Sparks, Maryland), 15 g agar (EMD, Gibbstown, New Jersey), 1000 mL distilled water) and oatmeal agar (OA, CitationSamson et al. 2010), standard media used in taxonomic studies of Onygenales, and 20% Sucrose MEA [20SMEA: 200 g sucrose, 20 g Bacto malt extract, 15 g agar, 1000 mL distilled water) because our house-dust strains were isolated with that medium. Other media were included to attempt induction of ascomata, using 6 cm Petri dishes of Sabouraud’s glucose agar (SGA, CitationSabouraud 1910), modified Leonian’s agar (MLA, CitationMalloch 1981), freezing agar (FA, CitationKuehn et al. 1961), or 1.5% water agar (WA). Three-point inoculations were made on 9 cm Petri dishes containing creatine sucrose agar (CREA, CitationFrivsad 1985) to assess production of organic acids and bases by growing fungal colonies. Cultures were incubated at 25 C in the dark, and colony morphologies and diameter were recorded seven and 14 d after inoculation. Growth experiments were conducted in triplicate. Capitalized colony and alphanumeric codes refer to CitationKornerup and Wanscher (1978). Micrographs were taken from material mounted in 85% lactic acid from ca. 1 mo old cultures grown on MEA, 20SMEA or OA. An Evolution MP digital microscope camera paired with ImagePro 6.0 (Media Cybernetics, Rockville, Maryland) mounted on an Olympus BX50 compound microscope (Olympus, Tokyo, Japan) was used for photomicrographs. ImageJ (rsbweb.nih.gov/ij) was used for scale calibration and size measurements. For measurements of ascospores and conidia, the means, standard deviations and 95% confidence intervals were calculated with Systat 10 (Systat Software, San Jose, California), based on 50 measurements.
Mating tests were undertaken for the one species for which we had multiple strains, described below as Sigleria carmichaelii. Petri dishes, 9 cm diam with MEA, were inoculated with agar plugs two strains ~2 cm apart and incubated at 25 C in the dark. Six strains (DAOMC 250071, 250073–250077) were inoculated in all possible pairwise combinations. Plates were examined periodically for ascomata with the dissecting microscope.
For scanning electron microscopy of ascomata and ascospores of Sp. sugiyamae, fungal tissue on WA was excised with a scalpel and fixed overnight at 4 C in 2.5% glutaraldehyde in 0.1 M sodium cacodylate pH 7.2. Specimens were rinsed and dehydrated in an ethanol series and dried with CO2 as the transitional fluid. Specimens were mounted with double-sided tape, grounded with silver paint and gold coated (Emitech K550X, Kent, UK) to ~10 nm thickness. A Quanta 600 FEI SEM (Hillsboro, Oregon) operating at 5, 10 and 20 kV was used to examine the specimens.
Physiological studies
Keratinolytic ability was determined with a keratin azure assay modified from CitationScott and Untereiner (2004). A modified basal CZA medium (lacking sucrose) was dispensed into 25 mL screw cap tubes, autoclaved at 15 psi for 20 min and cooled in an upright position. Finely chopped keratin azure (Sigma, St Louis, Missouri) was suspended in modified basal CZA at 4 mg/mL and autoclaved; 1 mL was dispensed onto the solidified basal medium and cooled before inoculation. Inoculum was grown on modified basal CZA amended with 1% sucrose for 4 wk. Tubes were inoculated with a 10 mm disk and incubated at 25 C in the dark 3 mo, then monitored weekly for release and diffusion of azure dye into the lower layer that would indicate keratin degradation. Each strain was tested in triplicate, and the experiment was repeated. An uninoculated negative control and positive control (Aphanoascus fulvescens [Coole] Apinis, CBS 111.58) were included.
Czapek’s-Dox agar (CZA, CitationMalloch 1981), a diagnostic medium used for taxonomic studies of Penicillium and Aspergillus, was used to test the ability of strains to utilize nitrate as a sole N source.
DNA extraction, PCR and sequencing
Mycelial mats were harvested with sterilized needles from colonies grown on MEA or 20SMEA in 6 cm diam Petri plates ~1 mo (or from the dried agar specimen of Sp. sphaerospora UAMH 10013). Genomic DNA extractions were performed with the Ultra-Clean Microbial DNA Isolation (MO BIO, Carlsbad, California) or the NucleoSpin® 96 Plant II kits (Macherey-Nagel, Germany). PCR amplification and sequencing of ca. 1100 bp of the 5′ end of the 18S gene and ca. 1000 bp of the 28S gene followed the protocols of CitationNguyen et al. (2013). PCR success was confirmed by agarose gel electrophoresis. Amplicons were sequenced with Big Dye Terminator (Applied Biosystems, Foster City, California) in 10 μL reactions with the protocol of Citationde Cock and Levesque (2004). All sequences were deposited in GenBank (Supplementary table I).
Phylogenetic analyses
We downloaded available data for all species of Spiromastix and relatives from GenBank and UNITE. Twenty-two cultures of Spiromastix and relatives were sequenced for phylogenetic analysis and assembled and edited with SeqMan II 8.0 (DNA STAR Inc., Madison, Wisconsin). Sequences of each gene were aligned with MAFFT 7.122b (CitationKatoh and Standley 2013) and trimmed with BioEdit (CitationHall 1999). Alignments are deposited in TreeBASE (www.treebase.org/treebase) as study number S16079. The identities of all 56 cultures of Sigleria carmichaelii were confirmed by ITS barcoding.
Molecular phylogenies were constructed with maximum likelihood (ML) and Bayesian inference (BI). For the ITS tree, Aspergillus montevidensis Talice & J.A. Mackinnon was used as outgroup. Outgroups for the 28S + 18S tree were Aleuria aurantia (Pers.) Fuckel and Ascobolus crenulatus P. Karst. To select the most appropriate model of sequence evolution, jmodeltest 2.1.1 (CitationDarriba et al. 2012) was applied to each alignment and the model chosen according to the Akaike information criterion (AIC). The gtr+i+g model was selected for ITS, 18S and 28S. Before ML and BI analysis the 18S and 28S matrices were concatenated with SeaView (CitationGouy et al. 2010).
ML analyses were performed on the ITS and concatenated 18S + 28S data with Garli 2.1 (CitationZwickl 2006). ML bootstrap (ML-BS) analyses with 1000 replicates were conducted with GARLI web service (CitationBazinet et al. 2014). The bootstrap consensus tree for combined datasets was calculated in PAUP*4.0b10 (CitationSwofford 2002). A threshold of ≥80% was used as the cut-off for significantly supported nodes.
BI analyses were performed with MrBayes 3.2 (CitationRonquist and Huelsenbeck 2003). Three independent Markov chain Monte Carlo (MCMC) runs were run simultaneously. Each MCMC ran for 3.0 × 106 generations, sampling every 500 generations until convergence (standard deviation of split frequency < 0.01). The first 25% trees were discarded as burn-in, and the remaining 13 503 trees were combined into one tree with 50% majority rule consensus. Consensus trees in Newick format were imported into FigTree 1.3.1 (tree.bio.ed.ac.uk/software/figtree) and exported as SVG vector graphics for figure assembly. BI posterior probabilities (BI-PP) above 0.95 were considered significant.
To calculate pairwise distance between species, out-group taxon Aspergillus montevidensis was removed from the ITS alignment. The dataset was re-aligned with MUSCLE (Edgar 2004) and imported into MEGA 6 (CitationTamura et al. 2013), where a pairwise distances were calculated with the maximum composite likelihood model (CitationTamura et al. 2004).
Results
Morphological studies
None of the arthroconidial strains that we isolated from house dust produced ascomata, despite incubation for periods of up to a year on nine different agar media. One species that was not part of our house dust study, described below as Spiromastix sugiyamae, produced mature ascomata sparsely only on WA. In contrast all house dust strains produced moderate to abundant arthroconidia on at least some media. Each putative species had a distinct conidiophore branching pattern. Dimensions of terminal and single lateral conidia are also used for distinguishing species ().
Physiological studies
Keratin azure assays showed that seven of our eight new species had no keratinolytic activity. Only Sp. pyramidalis had weak keratinolytic activity. Strains behaved identically in the two replicated experiments, with positive and negative controls performing as expected.
Growth rates on MEA and ability to grow using nitrate as a sole N source were useful characters for distinguishing the new genus Sigleria, described below, from Spiromastix. None of the strains in this study produced acids or bases on CREA.
Phylogenetic studies
In our 18S + 28S phylogenetic tree, 71 sequences representing eight families of Onygenales and 38 other fungi were included to estimate phylogenetic placement of Spiromastix and its relatives (Supplementary table I, Fig. 4). After removing ambiguously aligned regions, the 18S and 28S alignments were both 1300 bp long. In the 18S alignment there were 273 parsimony informative characters (21%), with 431 (33%) in the 28S alignment. The topology of the BI analysis was congruent with the best scoring ML tree for the concatenated two-locus dataset. The BI tree () places Spiromastigaceae basal to Onygenales as a monophyletic clade with high BI-PP (1.00) and moderate ML-BS (72%) support. Within Spiromastigaceae we interpret the three main clades as genera, namely Spiromastix, Sigleria (both BI-PP 1.00, ML-BP 100%) and Pseudospiromastix (1.00 BI-PP, 76% ML-BP support). Onygenaceae sensu CitationCurrah (1985, Citation1994) is polyphyletic and distributed among three clades.
To estimate species concepts, we used phylogenetic analysis of ITS including 85 strains representing seven families (supplementary table I, Fig. 5). After removing ambiguously aligned regions, ITS alignments were 1106 bp long with 526 (48%) parsimony informative characters. Spiromastigaceae included the same three clades identified as genera in the 18S + 28S analysis, Spiromastix (BI-PP 1.00, weak ML-BS), Sigleria (BI-PP 1.00, ML-BP 100%) and Pseudospiromastix (BI-PP 1.00, ML-BP 99%). Based on pairwise analyses, the genetic distance separating Spiromastix and Sigleria was 0.134 ± 0.001, while the average genetic distance between other accepted genera of the Onygenales was 0.137 ± 0.001. Onygenaceae was divided into four clades.
Sequence data for Sp. saturnispora and Sp. sphaerospora are unavailable because ex-type cultures are missing (S. Uchiyama pers comm). We failed to obtain PCR products or sequences of any rDNA markers from an authentic dried specimen of the ex-type of Sp. sphaerospora (UAMH 10013).
Taxonomy
Spiromastigaceae Hirooka, Tanney & Seifert, fam. nov. MycoBank MB811851
Typification: Spiromastix Kuehn & G.F. Orr, Mycologia 54:160. 1962.
Ascomata present in some species; when present, discrete or confluent, globose to subglobose, yellowish white to dark brown, sometimes irregular in shape, with appendages. Peridial hyphae irregularly branched, often curved or contorted, forming a loose network; appendages arising from basal swollen cells of peridial hyphae and extending outward, pale brown to brown, curved or straight, thick-walled, smooth or rough, sinuous, loosely coiled or swollen at the apex. Asci eight-spored, globose to ovoid or pyriform, protunicate, short-stipitate, evanescent. Ascospores globose to subglobose or oblate, hyaline, yellowish to pale brown, reticulate, punctate or sulcate ornamentation, sometimes with an equatorial rim. Asexual state present in some species; conidiophores well-developed in some species, hardly differentiated from hyphae in other species; conidia dry, thallic arthric, terminal, lateral and/or intercalary, mostly subglobose, broadly ellipsoidal, broadly clavate or ovoid; secession rhexolytic. Chlamydospores sometimes observed.
Notes: CitationRizzo et al. (2014) proposed new family and order names based on Spiromastix Kuehn & G.F. Orr, namely “Spiromastixaceae” Guarro, Cano & Stchigel and “Spiromastixales” Guarro, Cano & Stchigel. The two names were given only a single description that was not explicitly linked to either of the two names and therefore fail to fulfill Art. 38.11 of the Melbourne Code (CitationMcNeill et al. 2012). Further the authors also used only one identifier, a single MycoBank number, for the two names, failing to comply with Art. 42.1. We validated the family name Spiromastigaceae above, replacing the original “x” used in the name by CitationRizzo et al. (2014) with a “g” to reflect the genitive of the Greek suffix “mastix” (i.e. “mastiggos”). Our concept of the family varies slightly from that of CitationRizzo et al. (2014), primarily by the addition of anamorph characters and to disassociate the name from the concept of the “Spiromastixales”, which we do not accept, we validated the family name with our own authority above. The family includes three genera and 14 species.
In the descriptions and key below, we refer to asexual states as “malbranchea-like” when they form usually sparingly branched conidiophores that turn into chains of rhexolytically seceding arthroconidia that alternate with conspicuous “separating cells” (e.g. , cf. CitationSigler and Carmichael 1976). We use the term “chrysosporium-like” for sparingly branched conidiophores with solitary terminal or lateral conidia on fertile hyphae or in short chains, and sometimes single or short chains of intercalary conidia on some of the branches or the axis of the conidiophore (e.g. in CitationRizzo et al. 2014, cf. also CitationCarmichael 1962, CitationSigler 1997, Citationde Hoog et al. 2000). “Pyramidal conidiophores” have multiple layers of branching at more or less right angles, generally with longer branches toward the base of the conidiophore and shorter branches toward the apex (e.g. ). Conidiophores with a few basal layers of more or less right-angled branching and distinctly short conidiogenous branches (e.g. ) are referred to as “bush-like”. All microscopic structures hyaline with smooth walls unless indicated.
Sigleria Hirooka, Tanney & Seifert, gen. nov. MycoBank MB811852.
Typification: Sigleria carmichaelii Hirooka, Tanney & Seifert
Etymology: In honor of Prof Lynne Sigler, University of Alberta, Edmonton, Canada, in recognition of her many contributions to the taxonomy of Onygenales.
Colonies relatively slow growing, whitish, with velvet-like aerial mycelia, strong or moderate arthroconidial production, reverse pale to grayish yellow on MEA. Somatic hyphae septate, branched, thin-walled. Conidiophores common on aerial hyphae, erect or repent, with pyramidal branching, branches tending to be opposite, many terminating in a single conidium. Conidiogenous hyphae sparingly branched or laterally branched, thick-walled, septa produced basipetally to form arthroconidia. Terminal and single lateral conidia unicellular, borne on the main fertile hypha or on short lateral branches, subglobose to clavate or broadly ellipsoidal with a truncate base, thick-walled, secession rhexolytic. Arthroconidia 0(–1)-septate, cylindrical, cuboid, or doliiform, thick-walled, secession rhexolytic. Chlamydospores terminal or intercalary, solitary, unicellular, subglobose, thick-walled, sometimes appearing as swollen cells in hyphae when immature. Growth poor on CZA indicating a limited ability to utilize nitrate as the sole N source. Sexual state unknown.
Notes: Sigleria is morphologically similar to Spiromastix, but its conidiophores tend to have opposite branching whereas most Spiromastix species have a predominantly unilateral branching pattern (although infrequent opposite branching occurs in Sp. pyramidalis and Sp. frutex). Growth rates are useful to distinguish the genera. The two species of Sigleria grow slowly on the eight different media tested, with diameters < 5 mm on MEA after 7 d, in contrast to growth of > 5 mm for Spiromastix species (). The poor growth of Sigleria species on CZA, indicating a limited ability to utilize nitrate as a N source, is a clear physiological character distinguishing it from Spiromastix. These characters correspond well with the phylogenetic distance between Sigleria and Spiromastix (, ), which in both analyses exceeds or is equal to that separating most other pairs of genera in related families.
Fig. 1. Growth on different media and temperatures. A. Growth on MEA in the dark at 5 C intervals 5–40 C after 7 d. B. Growth on eight different media in the dark at 25 C after 14 d. Bars = standard errors.
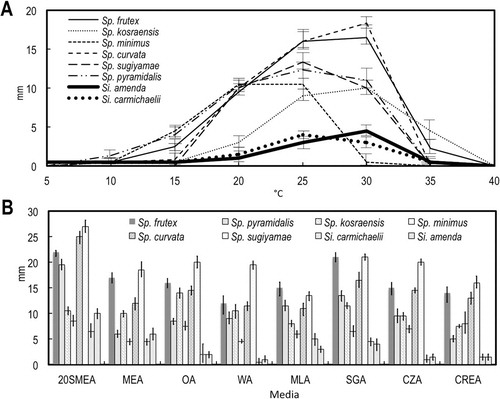
Ascomata were not produced by either species of Sigleria.
Sigleria amenda Hirooka, Tanney & Seifert, sp. nov. ,
Fig. 2. Colonies of six new species of Spiromastix and two new species of Sigleria on four media after 14 d in the dark at 25 C. In each pair of photographs the top is the obverse and the bottom is the reverse.
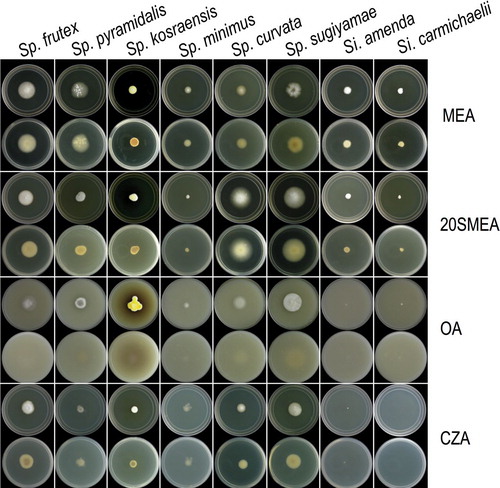
Fig. 3. Graphs of 95% confidence intervals of lengths (open shapes) and widths (gray shapes) of conidia, with means indicated by the constriction. A. Terminal and single lateral conidia. B. Arthroconidia. Bars = standard errors.
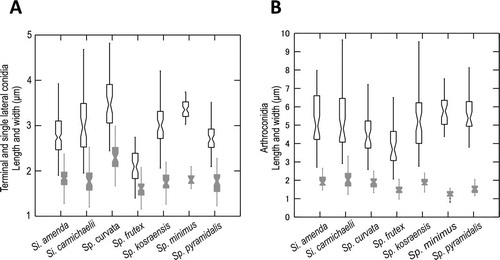
Fig. 4. Bayesian tree based on 18S + 28S sequences showing the relationship between the new genus Sigleria and new species of Spiromastix, their placement in the family Spiromastigaceae among other taxa of Onygenales and Eurotiomycetes. Clades with 1.00 Bayesian inference posterior probabilities (BI-PP) and 100% maximum likelihood bootstrap branch support (ML-BS) are indicated by thick black lines. Clades with > 0.95 BPP and 80% ML-BS are indicated by thicker gray lines, with the corresponding support values indicated; the BI-PP value is to the left and the ML-BS to the right. Asterisks indicate Bayesian posterior probabilities of 1.00 or maximum likelihood bootstrap branch support of 100%. Dashes indicate support values lower than 0.96 BPP or 80% ML-BS. Some support values for terminal branches were removed to reduce clutter. GT indicates that the strain represents the type species of its genus and ET indicates ex-type cultures. GenBank accession numbers for all sequences are provided (Supplementary table I).
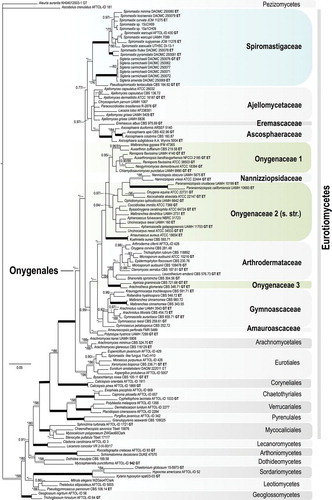
Fig. 5. Bayesian tree based on ITS sequences of the relationship between the new genus Sigleria and new species of Spiromastix, their placement in the family Spiromastigaceae among other families of Onygenales. Symbols and coding the same as for . GenBank accession numbers for all sequences are provided (Supplementary table I).
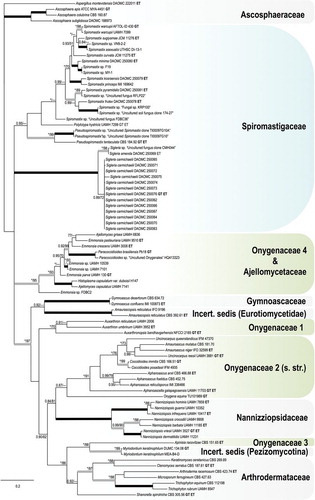
Fig. 6. Sigleria amenda. Ex-type culture on MEA. A. Sporodochial and aerial conidiophores. B–F. Conidiophores and conidia. G, H. Terminal and single lateral conidia and arthroconidia. I. arthroconidia. Bars: A = 500 μm, B–J = 10 μm.
MycoBank MB811853
Typification: MEXICO: Nayarit, Casa Xocotla, Sayulita, Mariposa 2, dried culture isolated from settled house dust, 9 Jan 2009, A. Amend (holotype DAOM 588476). Ex-type culture DAOMC 250069 = CBS 138257.
Etymology: amenda, named in honor of Prof Anthony Amend, University of Hawaii at Manoa, in recognition of his contributions to the metagenomic analysis of indoor fungi.
Colony diameter (mm) at 7 d, 25 C: 20SMEA 3; MEA 1–2; OA < 1; WA < 1; MLA 1; SGA 1; CZA 1; CREA < 1; 14 d: 20SMEA 10; MEA 5–6; OA 2; WA 1; MLA 3; SGA 4; CZA 1–2; CREA 1–2. Colonies after 14 d at 25 C: On 20SMEA slightly convex, white, aerial mycelia woolly, margin white, diffuse, reverse pale yellow (4A3); on MEA convex, white, aerial mycelia velvet-like, margin entire, white, reverse grayish yellow (4B5); on OA convex, white, granular because of arthroconidial production, aerial mycelia woolly in center, margin diffuse, colorless, reverse white. Arthroconidial production profuse on MLA, OA, moderate on 20SMEA, CREA, MEA, SGA, sparse on CZ, absent on WA.
Somatic hyphae on MEA septate, branched, thin-walled, 1.0–1.5 μm diam. Conidiophores on aerial hyphae abundant, erect or repent, up to 200 μm long, with pyramidal branching, most branches opposite, many branches terminating with a single conidium. Conidiogenous hyphae sparingly branched or laterally branched, 4.5–22.5 μm long, 0.5–1.5 μm wide at base, thick-walled, septa developing basipetally to form arthroconidia. Terminal and single lateral conidia unicellular, borne on the main fertile hypha or on short lateral branches, subglobose to clavate or broadly ellipsoidal with a truncate base, straight, (2.0–)2.5–3.7(–4.7) × (1.2–)1.4–2.2(–3.2) μm, thick-walled. Arthroconidia 0(–1)-septate, cylindrical, cuboid, or doliiform, straight, (2.7–)3.9–6.9(–8.0) × (1.5–)1.6–2.4(–3.1) μm, thick-walled. Chlamydospores terminal or intercalary, solitary, unicellular, subglobose, 3.0–9.0 μm, thick-walled.
Cardinal temperatures: optimum 25–30 C, minimum 10 C, maximum 35 C.
Notes: Sigleria amenda and Si. carmichaelii are morphologically very similar (, , ). Sigleria amenda grew faster than Si. carmichaelii on most media (, ) and its optimal growth occurred at 30 C rather than 25 C (). The phylogenetic distinction between the two species is clear from the ITS sequences, with high ML-BP and BI-PP support. However, the calculated pairwise distance of 0.023 between these two species is the closest of any sibling species pair in our analysis, which otherwise varies from ~0.033–0.057. Our ITS phylogeny also showed Si. amenda clustering with the sequence of an uncultured Spiromastix sp. from house dust in Missouri, USA (CitationRittenour et al. 2014) (). It seems unlikely that Si. amenda and this uncultured fungus are the same species because there are 6 bp differences between them in their ITS sequences (99% similarity).
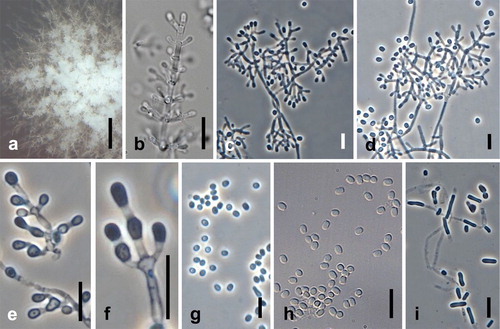
Fig. 7. Sigleria carmichaelii. Ex-type culture on MEA. A. Sporodochial and aerial conidiophores. B–E. Conidiophores and conidia. F–H Conidia. I, J. Chlamydospores. Bars: A = 500 μm, B–J = 10 μm.
Sigleria carmichaelii Hirooka, Tanney & Seifert, sp. nov. ,
MycoBank MB811854
Typification: FEDERATED STATES OF MICRONESIA: Kosrae; Malem, dried culture isolated from settled house dust in a private residence, N05.29507 E163.02364, 15 Mar 2009, W. Law (holotype DAOM 571575). Ex-type culture DAOMC 250076 = CBS 138264.
Etymology: carmichaelii, in honor of the late Prof John W. Carmichael, University of Alberta, Edmonton, Canada, in recognition of his contributions to Chrysosporium taxonomy. He was a mentor to Prof Sigler.
Colony diameter (mm) at 7 d, 25 C: 20SMEA 2; MEA 1; OA 1; WA < 1; MLA 1; SGA 1; CZA < 1; CREA < 1; 14 d: 20SMEA 6–7; MEA 4–5; OA 2; WA < 1; MLA 5; SGA 4–5; CZA 1; CREA 1–2. Colonies after 14 d at 25 C: On 20SMEA slightly convex, white, aerial mycelia woolly, margin diffuse, reverse pastel yellow (3A4); on MEA convex, white, aerial mycelia velvet-like, margin entire, white, reverse pale yellow (4A3); on OA flat, white, aerial mycelia velvet-like in center, margin entire, colorless, reverse pale yellow (4A3). Arthroconidial production profuse on MEA, MLA, OA, SGA, moderate on CREA, 20SMEA, WA, moderate to sparse on CZA.
Somatic hyphae on MEA, septate, branched, thin-walled, 1.1–1.5 μm diam. Conidiophores abundant on aerial hyphae, erect or repent, up to 180 μm long, with pyramidal branching, most branches opposite, many branches terminating with a single conidium. Conidiogenous hyphae sparingly branched or laterally branched, 5.1–20.0 μm long, 1.0–2.0 μm wide at base, thick-walled, septa produced basipetally to form arthroconidia. Terminal and single lateral conidia unicellular, borne on the main fertile hypha or on short lateral branches, subglobose to clavate or broadly ellipsoidal with a truncate base, straight, (1.9–)2.4–3.2(–4.2) × (1.3–)1.6–2.0(–2.4) μm, thick-walled. Arthroconidia 0(–1)-septate, cylindrical, cuboid, or doliiform, straight, (2.9–)3.6–7.2(–9.6) × (1.2–)1.6–2.6(–3.3) μm, thick-walled. Chlamydospores terminal or intercalary, solitary, unicellular, subglobose, 4.5–10.0 μm, thick-walled.
Cardinal temperatures: optimum 25–30 C, minimum 10 C, maximum 35 C.
Distribution: Federated States of Micronesia, Mexico, Thailand.
Habitat: House dust, often in association with Wallemia spp.
Additional specimens and cultures examined: FEDERATED STATES OF MICRONESIA: Kosrae; Yela, Tofol, Lelu, N05.32840 E163.00861, from settled dust in an office, 31 Mar 2009, W. Law, DAOM 588478, culture DAOMC 250071 = CBS 138259; ibid, from settled dust in private residence, N05.35748 E163.01065, 1 Apr 2009, W. Law, DAOM 571571, culture DAOMC 250072 = CBS 138260; ibid., DAOM 571573, culture DAOMC 250074 = CBS 138262; ibid., DAOM 571572, culture DAOMC 250073 = CBS 138261; ibid., DAOM 571574, culture DAOMC 250075 = CBS 138263. MEXICO: Nayarit; Casa Xocotla, Mariposa 2, Sayulita, from settled dust in a studio, 9 Jan 2009, A. Amend, DAOM 588469, culture DAOMC 250062 = CBS 138250; ibid., DAOM 588473, culture DAOMC 250066 CBS 138254; ibid., DAOM 588475, culture DAOMC 250068 = CBS 138256; ibid., DAOM 588470, culture DAOMC 250063 = CBS 138251; ibid., DAOM 588474, culture DAOMC 250067 = CBS 138255; ibid., DAOM 588472, culture DAOMC 250065 = CBS 138253; ibid., DAOM 588477, culture DAOMC 250070 = CBS 138258; ibid., DAOM 588471, culture DAOMC 250064 = CBS 138252. THAILAND: Surat Thani; 31 M006, Makhamtia, Muang Surat Thani, from settled dust in a private residence, 21 Apr 2009, P. Noonim, DAOM 571576, culture DAOMC 250077 = CBS 138265.
Notes: The differences between Si. carmichaelii and Si. amenda are discussed in Notes under the former species. We conducted mating experiments with six isolates of Si. carmichaelii but no ascomata were observed after 6 mo on MEA.
In 38 cultures isolated from Micronesian house dust with 20SMEA, Si. carmichaelii grew directly on colonies of Wallemia species. This frequent co-occurrence could be coincidental or may indicate mycoparasitism of Wallemia by Si. carmichaelii.
Pseudospiromastix Guarro, Stchigel & Cano, gen. nov. MycoBank MB805790
Typification: Pseudospiromastix tentaculata (Guarro, Géne & de Vroey) Guarro, Stchigel & Cano
Etymology: Pseudo- (Latin), meaning similar to Spiromastix
Diagnosis: See CitationRizzo et al. (2014).
Notes: CitationRizzo et al. (2014) proposed “Pseudospiromastix” and the new combination “P. tentaculata”. Unfortunately the name Pseudospiromastix and the combination P. tentaculata are nomenclaturally invalid. The designated type for “Pseudospiromastix” was the “species” labeled “Pseudospiromastix tentaculata”, but that name was not validly published because no basionym was fully cited (Art. 40.3, CitationMcNeill et al. 2012). Further only one MycoBank identifier was used for both the “new” generic name and the “new” combination, which does not fulfill Art. 42.1. We validate the genus name above and the new combination below, in the name of and with the permission of the original authors.
CitationRizzo et al. (2014) treated Pseudospiromastix as incertae sedis at family and order ranks because of its phylogenetic distance from their concept of “Spiromastixaceae” and “Spiromastixales” clades in their phylogenetic analysis. Our 28S + 18S phylogenetic tree places the genus in Spiromastigaceae clade supported by high BI-PP (1.00) and moderate ML-BP (72%) and we suggest that Pseudospiromastix be classified in that family ().
The most distinctive sexual character for Pseudospiromastix is the undulate ascomata appendages with swollen ends.
Pseudospiromastix tentaculata (Guarro, Géne & De Vroey) Guarro, Stchigel & Cano, comb. nov.
MycoBank MB805912
≡ Spiromastix tentaculata Guarro, Géne & De Vroey, Mycotaxon 46:308. 1993. (basionym, as tentaculatum).
Typification: SOMALIA: Province of Hiran; from soil, Mar 1966, J. Bosmans (holotype IMI 351264). Ex-type culture RV 24167 = CBS 184.92 = FMR 3842.
Diagnosis: CitationGuarro et al. (1993).
Spiromastix Kuehn & G.F. Orr, Mycologia 54:160. 1962.
Typification: Spiromastix warcupii Kuehn & G.F. Orr, Mycologia 54:160. 1962.
Ascomata absent for some species; when present, discrete or confluent, globose to subglobose, yellowish white to dark brown, sometimes shaped irregularly, appendages present. Peridial hyphae irregularly branched, forming a loose network, often curved or contorted near the ascomata wall but more straight toward the end; appendages arising from the basal swollen cells of peridial hyphae and extending outward, pale brown to brown, curved, helical or straight, thick-walled, smooth or rough walled, not swollen at the apex. Asci eight-spored, globose to ovoid or pyriform, short-stipitate, evanescent. Ascospores hyaline, yellowish or pale brown, reticulate, sulcate or punctate ornamentation, sometimes with an equatorial rim. Somatic hyphae on MEA septate, branched, thin-walled. Conidiophores on aerial hyphae common, with several branching patterns, branches generally unilateral or infrequently opposite in some species, many branches terminating with a single conidiogenous, erect or repent. Conidiogenous hyphae sparingly branched or laterally branched, thick-walled, septa produced basipetally to form arthroconidia. Terminal and single lateral conidia unicellular, borne on the main fertile hypha or on short lateral branches, subglobose to clavate or broadly ellipsoidal with a truncate base, thick-walled. Arthroconidia 0(–1)-septate, cylindrical, cuboid or doliiform, thick-walled. Chlamydospores when present terminal or intercalary, solitary, unicellular, subglobose, thick-walled, sometimes appearing as swollen cells in hyphae when immature. Growth moderate on CZA indicating an ability to utilize nitrate as the sole N source.
Notes: CitationDoweld (2013) renamed Spiromastix Kuehn & G.F. Orr as Spiromastigoides Doweld citing the priority of Spiromastix CitationPerfiliev (1929) for an alga, 33 y before the fungal genus was named. The nomenclatural arguments for conserving Spiromastix Kuehn & G.F. Orr require consultation with phycologists and will be formally presented elsewhere.
Our treatment of Spiromastix includes 11 species, five lacking a sexual state, six lacking an asexual state and no species having both.
Spiromastix curvata Hirooka, Tanney & Seifert, sp. nov. ,
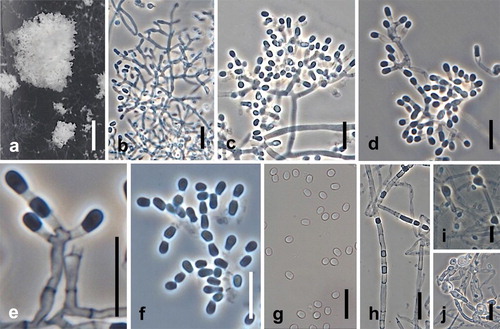
Fig. 8. Spiromastix curvata. Ex-type culture on MEA. A. Aerial conidiophores. B–D. Conidiophores and conidia. E–G. Terminal and single lateral conidia and arthroconidia. H. Chlamydospores. Bars: A = 500 μm, B–H = 10 μm.
MycoBank MB811856
Typification: MEXICO: Guerrero or Morelos, dried culture isolated as a contaminant in a strain of Histoplasma capsulatum Darling (EH366) that originally was isolated from a bat intestine, s.d. but probably 1999, T. Kasuga and M.L. Taylor (holotype DAOM 571582). Ex-type culture JCM 11275, originally identified as Malbranchea sp. (=CBS 140477).
Etymology: curvata (Latin), referring to its curved conidiophores.
Colony diameter (mm) at 7 d, 25 C: 20SMEA 8–9; MEA 4–5; OA 6–7; WA 5; MLA 2–5; SGA 7; CZA 5–6; CREA 5–6; 14 d: 20SMEA 25; MEA 12; OA 14–15; WA 11–12; MLA 10–12; SGA 16–17; CZA 14–15; CREA 11–14. Colonies after 14 d at 25 C: on 20SMEA convex, white, aerial mycelia woolly, margin diffuse, colorless, reverse white with reddish yellow (4A6) center; on MEA flat to slightly convex, white, aerial mycelia woolly, margin diffuse, colorless, reverse grayish yellow (4B5); on OA flat to slightly convex, white, aerial mycelia velvet-like, margin diffuse, reverse white. Arthroconidial production profuse on OA, moderate on SGA, sparse on 20SMEA, MEA, MLA, SGA, absent on CREA, WA.
Somatic hyphae on WA septate, branched, thin-walled, 1.0–2.0 μm diam. Conidiophores on submerged or aerial hyphae, unbranched or loosely branched, sinuate to curved, up to 28 μm long. Conidiogenous hyphae sparingly branched or laterally branched, 7.5–23.5 μm long, 1.0–2.5 μm wide at base, thick-walled, septa produced basipetally to form arthroconidia. Terminal and single lateral conidia unicellular, borne on the main fertile hypha or on short lateral branches, subglobose to clavate or broadly ellipsoidal with a truncate base, straight, (2.4–)2.9–4.1 (–4.8) × (1.7–)2.0–2.6(–3.0) μm, thick-walled. Arthroconidia 0(–1)-septate, cylindrical, cuboid, or doliiform, straight, (2.6–)3.6–5.6(–7.2) × (1.3–)1.6–2.2(–2.5) μm, thick-walled. Chlamydospores terminal or intercalary, solitary, 0-septate, subglobose, 3.0–6.0 μm, thick-walled.
Cardinal temperatures: optimum 30 C, minimum 15 C, maximum 35 C.
Habitat: Uncertain. The ex-type culture was a contaminant in a culture originally isolated from a bat intestine.
Notes: Spiromastix curvata was identified previously as Malbranchea sp. by CitationSugiyama and Mikawa (2001) based on morphological and phylogenetic results. They did not name the species in Spiromastix because it lacked ascomata, then an essential character for this genus.
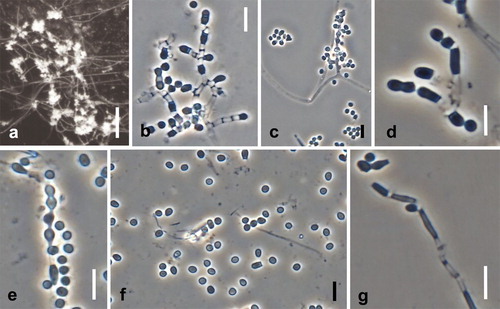
Spiromastix frutex Hirooka, Tanney & Seifert, sp. nov. ,
Fig. 9. Spiromastix frutex. Ex-type cultures on MEA. A. Aerial conidiophores. B, C. Conidiophores and conidia. D–G. Terminal and single lateral conidia and arthroconidia. Bars: A = 500 μm, B–G = 5 μm.
MycoBank MB811857
Typification: MEXICO: Nayarit; Sayulita, Casa Xocotla, Mariposa 2, dried culture from settled dust in a studio, 9 Jan 2009, A. Amend (holotype DAOM 571577). Ex-type culture DAOMC 250078 = CBS 138266.
Etymology: frutex (Latin), describing the bush-like conidiophores.
Colony diameter (mm) at 7 d, 25 C: 20SMEA 6–7; MEA 8–9; OA 6–7; WA 4; MLA 7–8; SGA 9; CZA 6; CREA 2–4; 14 d: 20SMEA 21–22; MEA 17; OA 16–17; WA 12–15; MLA 15–16; SGA 20–21; CZA 15; CREA 9–13. Colonies after 14 d at 25 C: On 20SMEA slightly convex, white, aerial mycelia wooly, margin diffuse, white, reverse pastel yellow (3A4); on MEA convex, white, aerial mycelia velvet-like, margin entire, reverse pale yellow (4A3); on OA flat, white, aerial mycelia velvet-like in center, margin entire, colorless, reverse pale yellow (4A3). Arthroconidia abundant on all media.
Somatic hyphae on MEA septate, branched, thin-walled, 1.0–1.5 μm diam. Conidiophores on aerial hyphae abundant, with bush-like branching, tending to be unilateral, terminating in a single apical conidiogenous cell, erect or repent, up to 100 μm long. Conidiogenous hyphae sparingly branched or laterally branched, 3.0–6.0 μm long, 1.0–1.5 μm wide at base, thick-walled, septa produced basipetally to form arthroconidia. Terminal and single lateral conidia 0-septate, borne on the main fertile hypha or on short lateral branches, subglobose to clavate or broadly ellipsoidal with a truncate base, straight, (1.4–) 1.7–2.5 (–2.7) × (1.2–) 1.4–1.8 (–2.3) μm, thick-walled. Arthroconidia 0(–1)-septate, cylindrical, cuboid, or doliiform, straight, (2.1–)2.7–5.3(–8.5) × (1.0–)1.2–1.8(–2.8) μm, thick-walled. Chlamydospores not observed.
Cardinal temperatures: optimum 25–30 C, minimum 5 C, maximum 35 C.
Notes: Conidiophore branching patterns of Sp. frutex and Sp. pyramidalis are similar, but the former has less branched conidiophores that lack a pyramidal aspect. Other characters such as conidial size and growth rates are different (–). Based on our 18S + 28S and ITS phylogenetic analyses, this species is closely related to Sp. pyramidalis (, ) but clearly genetically distinct.
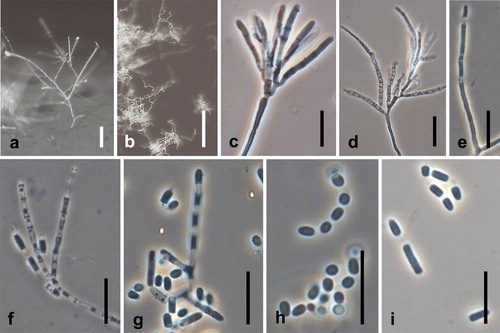
Spiromastix kosraensis Hirooka, Tanney & Seifert, sp. nov. ,
Fig. 10. Spiromastix kosraensis. Ex-type culture on MEA. A, B. Aerial conidiophores. C–G. Conidiophores and conidia. H, I. Terminal and single lateral conidia and arthroconidia. Bars: A = 50 μm, B = 100 μm, C–I = 10 μm.
MycoBank MB811858
Typification: FEDERATED STATES OF MICRONESIA: Kosrae: Lelu, N05.35748 E163.01065, dried culture from settled dust in a private residence, 1 Apr 2009, W. Law (holotype DAOM 571578). Ex-type culture DAOMC 250079 = CBS 138267.
Etymology: kosraensis, referring to the geographical origin of the ex-type strain.
Colony diameter (mm) at 7 d, 25 C: 20SMEA 5; MEA 4; OA 5; WA 3; MLA 3; SGA 5; CZA 2–3; CREA 3; 14 d: 20SMEA 10–11; MEA 10; OA 14; WA 10–11; MLA 8; SGA 11–12; CZA 9–10; CREA 7–8. Colonies after 14 d at 25 C: on 20SMEA flat, center white to pale yellow (3A3) with a yellow (3A7) concentric ring toward margin, aerial mycelia velvet-like, margin diffuse, margin, reverse yellowish brown (5D8); on MEA slightly convex, white to pastel yellow (3A4), aerial mycelia woolly, margin diffuse, white, reverse golden brown (5D7); on OA flat to slightly convex, deep yellow (4A8) with abundant light brown (6D5) soluble pigment in agar, aerial mycelia velvet-like, margin slightly diffuse, white margin. Arthroconidia abundant on all media.
Somatic hyphae on MEA septate, branched, thin-walled, 1.0–2.0 μm diam. Conidiophores on aerial hyphae abundant, verticillate, branching three or four times, many branches terminating in a single apical conidiogenous cell, erect or repent, up to 300 μm long. Conidiogenous hyphae sparingly branched or laterally branched, 14.5–50.5 μm long, 1.0–2.0 μm wide at base, thick-walled, septa produced basipetally to form arthroconidia. Terminal and single lateral conidia 0-septate, borne on the main fertile hypha or on short lateral branches, subglobose to broadly ellipsoidal with a truncate base, straight, (2.1–)2.5–3.5(–4.2) × (1.3–)1.6–2.0 (–2.4) μm, thick-walled. Arthroconidia 0(–1)-septate, cylindrical, cuboid, or doliiform, straight, (2.8–)3.8–6.8 (–9.5) × (1.4–)1.6–2.2(–2.7) μm, thick-walled. Chlamydospores not produced.
Cardinal temperatures: optimum 25–30 C, minimum 20 C, maximum 35 C.
Notes: Spiromastix kosraensis has erect conidiophores up to 300 μm long and relatively long conidiogenous hyphae (). This conidiophore morphology is unique in Spiromastix and its relatives. Our ITS phylogenetic analysis placed Sp. kosraensis close to Sp. princeps, which lacks a known asexual state (). In our study no ascomata of Sp. kosraensis were observed even after 1 y incubation on nine different media. Spiromastix minimus Hirooka, Tanney & Seifert, sp. nov ,
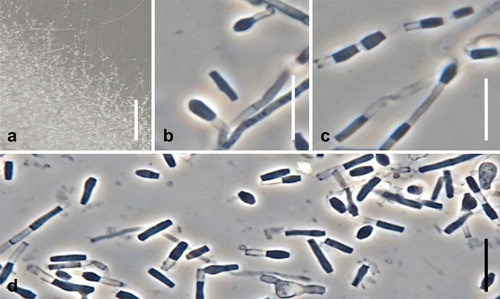
Fig. 11. Spiromastix minimus. Ex-type culture on WA. A. Submerged or aerial conidiophores. B. Conidiophores and conidia. C, D. Terminal and single lateral conidia and arthroconidia. Bars: A = 500 μm, B–D = 10 μm.
MycoBank MB811859
Typification: NEW ZEALAND: Otago; Dunedin, 284 Coast Road, Warrington, dried culture isolated from house dust, 1 May 2009, T. Atkinson (holotype DAOM 571579). Ex-type culture DAOMC 250080 = CBS 138268.
Etymology: minimus (Latin), referring to the little differentiated morphology of the fungus.
Colony diameters (mm) 7 d, 25 C: 20SMEA 2; MEA 1–2; OA 2; WA 1; MLA 1; SGA 1–2; CZA 2; CREA 1–3; 14 d: 20SMEA 8–9; MEA 4–5; OA 7–8; WA 4–5; MLA 6; SGA 6–7; CZA 6–8; CREA 6–9. Colonies after 14 d: On 20SMEA convex, white, aerial mycelia cottony, margin diffuse, colorless, reverse white to yellowish white (4A2); on MEA convex, white, aerial mycelia cottony and more abundant than on 20SMEA, margin diffuse, margin, reverse yellowish white (4A2); on OA flat, moist, colorless to white, with centrally tufted aerial mycelia, margin diffuse, colorless, reverse colorless to white; on WA flat, surface and reverse colorless to white, aerial mycelia sparse, margin diffuse, colorless. Arthroconidia sparse on WA, not observed on other media.
Somatic hyphae on MEA septate, branched, thin-walled, 1.0–1.5 μm diam. Conidiophores on submerged or aerial hyphae, sparse, unbranched, slightly curved, up to 15 μm long. Conidiogenous hyphae sparingly branched or laterally branched, 7.0–13.0 μm long, 1.5–2.0 μm wide at base, thick-walled, septa produced basipetally to form arthroconidia. Terminal and single lateral conidia 0-septate, borne on the main fertile hypha or on short lateral branches, clavate or broadly ellipsoidal with a truncate base, straight, (2.7–)3.0–3.8(–4.0) × (1.6–)1.7–1.9(–2.1) μm, thick-walled. Arthroconidia 0(–1)-septate, cylindrical, cuboid, or doliiform, straight, (4.4–) 4.8–7.2(–9.1) × (0.8–)1.1–1.5(–1.8) μm, thick-walled. Chlamydospores terminal or intercalary, solitary, 0-septate, subglobose to oblong, 3.0–5.5 μm, thick-walled.
Cardinal temperatures: optimum 20–25 C, minimum 10 C, maximum 30 C.
Notes: Conidia of Sp. minimus were produced only when the fungus was cultivated on WA, and even on this medium the production of conidia and conidiophores was sparse (). Whether the greatly reduced morphology is actually diagnostic or indicates culture degeneration, the optimum growth temperature of 20 C after 7 d () distinguishes Sp. minimus from other species.
Spiromastix princeps Udagawa & Uchiy., Mycoscience 40:300. 1999.
MycoBank MB459773
≡ Acanthogymnomyces princeps (Udagawa & Uchiy.) Udagawa & Uchiy., Mycotaxon 76:417. 2000.
Notes: Spiromastix princeps was isolated from soil in Kenya by CitationUdagawa and Uchiyama (1999) and is the only species of the genus producing ascomata with straight or slightly undulate appendages and oblate ascospores with an equatorial rim and sulcate ornamentation. In our ITS phylogeny it belongs to Spiromastix (), but appendage shape and ascospore ornamentation differ from other Spiromastix species and are similar to Acanthogymnomyces terrestris, the type of that genus. Following our molecular evidence, we conclude that A. princeps was originally correctly classified as Sp. princeps.
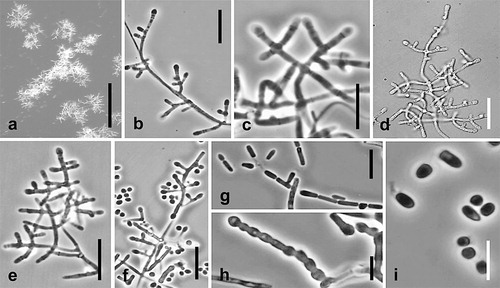
Spiromastix pyramidalis Hirooka, Tanney & Seifert, sp. nov. ,
Fig. 12. Spiromastix pyramidalis. Ex-type culture on MEA. A. Aerial conidiophores. B–H. Conidiophores and conidia. I. Terminal and single lateral conidia and arthroconidia. Bars: A = 500 μm, B–G = 10 μm, H, I = 5 μm.
MycoBank MB811860
Typification: AUSTRALIA: Tasmania; Hobart, Sandy Bay, dried culture from house dust, 10 Feb 2009, B. Horton (holotype DAOM 571580). Ex-type culture DAOMC 250081 = CBS 138269.
Etymology: pyramidalis (Latin), referring to the pyramidal aspect of the conidiophores.
Colony diameter (mm) at 7 d, 25 C: 20SMEA 8–9; MEA 4–5; OA 6; WA 6; MLA 5–6; SGA 6–7; CZA 5; CREA 5–6; 14 d: 20SMEA 19–20; MEA 6; OA 8–9; WA 9; MLA 11–12; SGA 13–14; CZA 9–10; CREA 6–7. Colonies after 14 d at 25 C: On 20SMEA flat, white, aerial mycelia cottony, occurring in radial patches, margin entire, colorless, reverse white to yellowish gray (4B2); on MEA slightly convex, white with greenish yellow (1A6) tinge in the center, aerial mycelia velvet-like, margin entire, white, reverse grayish yellow (4B5); on OA slightly convex, white with grayish brown (6D3), center honeycomb or lattice-like because of exudate droplets, aerial mycelia cottony, margin entire, colorless, reverse grayish yellow (4B5), with abundant colorless exudate droplets on colony surface. Arthroconidia produced abundantly on all media.
Somatic hyphae on MEA septate, branched, thin-walled, 1.0–2.0 μm diam. Conidiophores on aerial hyphae common, with pyramidal branching, tending to be unilateral, terminating in a single apical conidiogenous cell, erect or repent, up to 150 μm long. Conidiogenous hyphae sparingly branched or with a mixture of unilateral and opposite branching, 3.0–22.0 μm long, 1.0–1.5 μm wide at base, thick-walled, septa produced basipetally to form arthroconidia. Terminal and single lateral conidia 0-septate, borne on the main fertile hypha or on short lateral branches, subglobose to clavate or broadly ellipsoidal with a truncate base, straight, (2.1–)2.4–3.2(–3.9) × (1.2–)1.4–2.0(–2.3) μm, thick-walled. Arthroconidia 0(–1)-septate, cylindrical, cuboid, or doliiform, straight, (3.8–) 4.5–7.1(–8.7) × 1.2–1.8(–2.3) μm, thick-walled. Chlamydospores not produced.
Cardinal temperatures: optimum 20–30 C, minimum 10 C, maximum 30 C.
Notes: The branching pattern of the conidiophores of Sp. pyramidalis is somewhat similar to that of Sp. frutex, but the conidiogenous hyphae of Sp. pyramidalis are longer (, , , ). The ITS phylogeny supports the genetic distinctiveness of these two species (, ). Because of the presence of some opposite branching in the conidiophores, Sp. pyramidalis is also reminiscent of the two Sigleria species.
Spiromastix sugiyamae Hirooka, Tanney & Seifert, sp. nov. ,
Fig. 13. Spiromastix sugiyamae. Ex-type culture on WA. A, B. Ascomata on WA. C, D. Ascomata appendages. E, F. Asci. G. Ascospores. H. Scanning electron micrograph (SEM) of ascomata. I. SEM of ascomata appendages. J. SEM of ascospores. Bars: A = 500 μm, B = 100 μm, C–E, H = 10 μm, F, G, I = 5 μm, J = 1 μm.
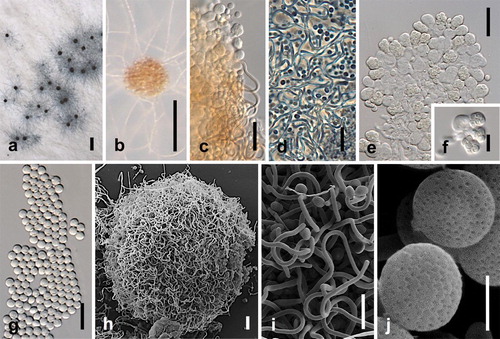
MycoBank MB811861
Typification: USA: FL, dried culture isolated from soil, 1 Sep 1998, M. Sugiyama (holotype DAOM 571581). Ex-type culture JCM 11276 (= CBS 140478).
Etymology: sugiyamae, honoring Dr Masato Sugiyama, Yokohama Research Center, Kanagawa, Japan, in recognition of his many contributions to the molecular phylogeny of onygenalean fungi.
Colony diameter (mm) at 7 d, 25 C: 20SMEA 10–12; MEA 7–8; OA 11–12; WA 8; MLA 3; SGA 9–11; CZA 8–11; CREA 1–3; 14 d: 20SMEA 24–30; MEA 18–19; OA 19–20; WA 19–20; MLA 11–16; SGA 20–22; CZA 19–21; CREA 15–16. Colonies after 14 d at 25 C: on 20SMEA flat to slightly convex, white, aerial mycelia woolly, margin entire, colorless, reverse white to light yellow (4A5); on MEA flat to slightly raised, white, aerial mycelia woolly, occurring in irregular patches, margin diffuse, colorless, reverse yellowish brown (5D6); on OA flat to slightly convex, white, aerial mycelia dense, velvet-like, margin entire, white, reverse white to brownish orange in center (5C5); on WA flat, white, aerial mycelia sparse, margin diffuse, colorless, reverse white. Arthroconidia not observed on any media tested.
Somatic hyphae septate, branched, thin-walled, 1.0–1.5 μm diam. Ascomata on WA superficial, discrete or confluent in small clusters, globose to subglobose, sometimes irregularly shaped, 42–190 μm diam, with appendages, at first yellowish white, becoming brownish red to dark brown, maturing within 1 mo. Peridial hyphae, irregularly branched, undulate to helical, forming a loose network 0.5–1.0 μm diam, forming a loose network, ending in slender appendages; appendages arising from the peridial hyphae as free end of branches, pale brown, with unbranched or sparingly branched, sinuate, aseptate, thick-walled, (11.2–) 16.0–30.0(–40.3) × (0.6–)0.7–1.1(–1.2) μm, slightly swollen at the rounded apex. Asci singly borne, eight-spored, ovoid or pyriform, short-stipitate, (4.8–)5.6–7.2(–8.4) × (3.0–)3.9–4.7(–5.5) μm, hyaline to pale yellowish brown, evanescent. Ascospores hyaline, yellowish orange, oblate, globose in face view, (1.1–)1.4–1.8(–2.0) × (1.7–)2.2–2.6(–2.9) μm, thick-walled, regularly punctate. Asexual state not observed.
Cardinal temperatures: optimum 25 C, minimum 15 C, maximum 30 C.
Notes: Spiromastix sugiyamae initially was identified as Spiromastix sp. by CitationSugiyama and Mikawa (2001) based on morphology and molecular phylogenetic analysis. Our analyses show that Sp. sugiyamae is distinct from other species of Spiromastix, with Sp. warcupii its closest relative, as CitationSugiyama and Mikawa (2001) indicated (, ). This close genetic relationship also is supported by similar morphological characters. There are four known Spiromastix species with sexual states: Sp. saturnispora, Sp. sphaerospora, Sp. princeps and Sp. warcupii. Based on our observations Sp. sugiyamae is almost identical to Sp. warcupii but the ascospores of Sp. sugiyamae are shorter. SEM also reveals that ascospores of Sp. sugiyamae are regularly punctate but lack the few fine grooves in the polar region reported in the otherwise punctate ascospores of Sp. warcupii (CitationSugiyama and Mikawa 2001, based on CBS 576.63) (). CitationKuehn and Orr (1962) cultured Sp. warcupii on eight media and found that all except MEA induced the sexual state. We observed ascomata of Sp. sugiyamae on only WA.
Discussion
The isolation of six new species from house dust by dilution to extinction and two new species from other substrates has increased species sampling and provided insights into the taxonomy of Onygenales, the genera of Spiromastigaceae and the morphological characters of Spiromastix. Unfortunately most of our species are represented by single strains, a condition all too frequent for this family. Only Sigleria carmichaelii was isolated in abundance. All our species of Spiromastix and Sigleria were isolated with 20SMEA and also grew fastest on the isolation medium. They should be considered moderately xerotolerant. The combination of dilution to extinction, media with low water activities, combined with baits such as cellulose, may be an effective strategy to increase sampling of this family. However, the fact that we isolated more than 8000 strains with a method designed to isolate slow-growing fungi and recovered only single strains of most species suggests that they are infrequent in the built environment and that their primary habitats are elsewhere.
Until recently Spiromastix was partly defined by a lack of asexual states. The description of Sp. asexualis (CitationRizzo et al. 2014), which produced only immature ascomata, introduced conidial morphs to the generic concept. Our rDNA phylogenetic analyses supported the recognition of additional species of Spiromastix and the proposal of a sister genus, Sigleria, but ascomata were not observed in seven of the eight species we studied. CitationSigler (2002) noted that while onygenalean fungi are often heterothallic Auxarthron G.F. Orr & Kuehn includes homothallic and heterothallic species as well as some species that possess and others that lack an asexual morph, a situation reflecting our revised circumscription of Spiromastix. We grew our strains on a broad range of agar media hoping to observe homothallic ascomata and crossed strains of Si. carmichaelii hoping to induce heterothallic mating. No ascomata were observed despite repeated attempts for more than 2 y. This does not preclude the possible existence of ascomata under other conditions. If the species from house dust are heterothallic and out-crossing is rare, as we surmise, then conidia are responsible for dispersal in the indoor environment. The frequent disturbance of the indoor habitat from human activities may favor the rapid sporulation associated with asexual reproduction or sexually competent fungi such as Chaetomium or Eurotium that tend to be homothallic and can complete their sexual cycle quickly. Perhaps prolific asexual sporulation allowed our “domestic” Spiromastix and Sigleria species to be isolated from house dust even though their natural habitats lie beyond the walls of human dwellings. The probability of both mating types of heterothallic species being introduced during sporadic, random and low level colonization may explain why we have failed so far to observe mating in our cultures.
Asexual species in Spiromastigaceae have a little differentiated morphology that makes identification of the phylogenetically delimited genera challenging but not impossible. In our experience, conidiophore branching patterns generally were reliable to distinguish Spiromastix with unilateral branching from Sigleria species with opposite branching, but the reduced conidiophores of Sp. minima and Sp. asexualis cannot be interpreted from this perspective. Despite this problem in identifying the genus, for each species of Spiromastix and Sigleria, asexual morphologies are diagnostic at the species level. Although the taxonomic history of the order has focused on ascomata, it is essential to accurately characterize the asexual states of all Onygenales because sexual states may never be discovered for a significant number of species.
Dual nomenclature in Onygenales never achieved a satisfactory one to one correlation between teleomorph and anamorph genera. The two best known anamorph genera, Chrysosporium and Malbranchea, are both associated with multiple teleomorph genera and unsurprisingly molecular phylogenies reveal both as polyphyletic (CitationVidal et al. 2000, CitationSigler et al. 2002). Anamorphs of Spiromastigaceae have micromorphology reminiscent of both of these concepts. In the strict sense Chyrsosporium should be limited to onygenalean fungi with keratinolytic activity, which Spiromastix and Sigleria species lack (CitationVidal et al. 2000, CitationPitt et al. 2013). The anamorphs of some Spiromastix species have erect conidiophores that differ from the more repent or reduced conidiophores of Malbranchea (CitationSigler and Carmichael 1976). The highly branched conidiophores of Sp. kosraensis, Sp. pyramidalis, Sp. frutex, Sigleria carmichaelii and Si. amenda also have some similarity to Geomyces Traaen (Pseudoeurotiaceae, order uncertain, CitationMinnis et al. 2013).
As treated here Spiromastigaceae includes Spiromastix, Pseudospiromastix and the newly described Sigleria and is phylogenetically distinct from Onygenaceae, which remains polyphyletic (CitationSugiyama and Mikawa 2001; CitationSugiyama et al. 1999, Citation2002). Based on ML analyses of the D1-D2 domains of the nuc 28S rDNA, CitationRizzo et al. (2014) proposed the new order “Spiromastixales” and the new family “Spiromastigaceae” (both invalid as noted above). Our rDNA analyses, with considerably more taxon sampling, placed Spiromastigaceae firmly within Onygenales, with 1.00 BI-PP and 72% ML-BP support. Thus we do not accept “Spiromastixales” as a distinct order and did not validate the name, acknowledging that a more comprehensive multigene phylogeny will allow a more definitive conclusion.
We were unable to obtain sequences for all known species of Spiromastix and a re-evaluation of the phylogenetic significance of two characters traditionally considered significant in the taxonomy of Onygenales (i.e. ascospore morphology and ascomata appendage form) was not feasible. Because of its ascospores with a distinctive equatorial rim, the phylogenetic position of Sp. saturnispora would be interesting to explore but its ex-type culture is missing. In our ITS phylogenetic analysis, Sp. princeps, with ascospores with a similar equatorial rim, clustered with Sp. kosraensis, so far known only as an asexual species. Spiromastix sphaerospora, which also lacks an ex-type culture, has brown, coarsely roughened ascomata appendages that differ from the usual smooth-walled, helical appendages of other species in the genus, suggesting that its taxonomy needs to be reconsidered. In our ITS analysis Sp. princeps, with similar ascomata appendages to S. sphaerospora, is in the Spiromastix clade but there are only asexual Spiromastix species as near relatives of the fungus. If sequence data of Sp. sphaerospora could be obtained, it would be helpful for understanding the significance of variation in ascomata appendages for the definition of Spiromastix.
Key to genera and species of Spiromastigaceae
umyc_a_1080135_sm0001.docx
Download MS Word (62.9 KB)Acknowledgments
We thank Olga Koppel, Kalima N. Mwange, Ed Whitfield, Nicolas Poliquin, Catherine Brown, Martin Bidartondo, Toni Atkinson, Anthony Amend, Wayne Law, Bryony Horton and Paramee Noonim who collected house dust or isolated or preserved cultures or generated DNA sequences. Lynne Sigler and Connie Gibas kindly provided ex-type cultures or type specimens. Walter Gams and Scott Redhead provided nomenclatural and linguistic assistance. David Malloch offered useful interpretation of some aspects of the biology of the fungi that we described. We thank Keith Hubbard for his support with SEM. Shigeru Uchiyama and Gen Okada kindly provided information on ex-types and other specimens.
This research was supported by grants from the Microbiology of the Built Environment Program of the Alfred P. Sloan Foundation.
Literature Cited
- AmendASSeifertKASamsonRBrunsTD. 2010. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc Natl Acad Sci USA 107:13748–13753, doi:10.1073/pnas.1000454107
- BazinetALZwicklDJCummingsMP. 2014. A gateway for phylogenetic analysis powered by grid computing featuring GARLI 2.0. Syst Biol 63:812–818, doi:10.1093/sysbio/syu031
- BennyGLKimbroughJW. 1980. A synopsis of the orders and families of Plectomycetes with keys to genera. Mycotaxon 12:1–91.
- CarmichaelJW. 1962. Chrysosporium and some other aleurosporic hyphomycetes. Can J Bot 40:1137–1173, doi:10.1139/b62-104
- CockAWAM deLevesqueCA. 2004. New species of Pythium and Phytophthora. Stud Mycol 50:481–487.
- CurrahRS. 1985. Taxonomy of the Onygenales: Arthrodermataceae, Gymnoascaceae, Myxotrichaceae and Onygenaceae. Mycotaxon 24:1–216.
- CurrahRS. 1994. Peridial morphology and evolution in the prototunicate ascomycetes. In: HawksworthDL, ed. Ascomycete systematics: problems and perspectives in the Nineties. New York: Plenum Press. p 281–293.
- CurrahRSLocquin-LinardM. 1988. Spiromastix grisea sp. nov. and its relationship to other Onygenaceae with helical appendages. Can J Bot 66:1135–1137, doi:10.1139/b88-162
- DarribaDTaboadaGLDoalloRPosadaD. 2012. jModel-Test 2: more models, new heuristics and parallel computing. Nat Methods 9:772, doi:10.1038/nmeth.2109
- de HoogGSGuarroJFiguerasMJGenéJ. 2000. Atlas of clinical fungi. 2nd ed. Baarn, The Netherlands and Reus, Spain: Centraalbureau voor Schimmelcultures and Universitat Rovira I Virgili. 1126 p.
- DoweldAB. 2013. Index Fungorum 30:1.
- FrivsadJC. 1985. Creatine sucrose agar, a differential medium for mycotoxin producing terverticillate Penicillium species. Lett Appl Microbiol 1:109–113, doi:10.1111/j.1472-765X.1985.tb01500.x
- GuarroJSummerbellRCSamsonRA, eds. 2002. Onygenales: the dermatophytes, dimorphics and keratin degraders in their evolutionary context. Stud Mycol 47:1–220.
- GouyMGuindonSGascuelO. 2010. SeaView 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224, doi:10.1093/molbev/msp259
- GreifMDCurrahRS. 2003. A functional interpretation of the role of the reticuloperidium in whole-ascoma dispersal by arthropods. Mycol Res 107:77–81, doi:10.1017/S0953756202007104
- GuarroJGenéJde VroeyC. 1993. A new species of Spiromastix from Africa. Mycotaxon 46:307–313.
- HallTA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Res 41:95–98.
- KatohKStandleyDM. 2013. MAFFT 7: multiple sequence alignment software: improvements in performance and usability. Mol Biol Evol 30:772–780, doi:10.1093/molbev/mst010
- KirkPMCannonPFMinterDWStalpersJA. 2008. Dictionary of the Fungi. 10th ed. Wallingford, UK: CAB International. 771 p.
- KornerupAWanscherJH. 1978. Methuen handbook of color. 3rd ed. London, UK: Eyre Methuen. 252 p.
- KuehnHHOrrGF. 1962. A new genus of Gymnoascaceae. Mycologia 54:160–167, doi:10.2307/3756666
- KuehnHHOrrGFGhoshGR. 1961. A new and widely distributed species of Pseudoarachniotus. Mycopath Mycol Appl 14:215–229, doi:10.1007/BF02057328
- LumbschHTHuhndorfSM. 2007. Outline of Ascomycota 2007. Myconet 13:1–58.
- MallochD. 1981. Molds: their isolation, cultivation and identification. Toronto, Ontario: Univ. Toronto Press. 97 p.
- McNeillJBarrieFRBuckWRDemoulinVGreuterWHawksworthDHerendeenPSKnappSMarholdKPraadoJPrud’Homme van ReineWFSmithGFWiersemaJHTurlandNJ. 2012. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code), Adopted by the 18th International Botanical Congress Melbourne, Australia, Jul 2011. Bratislava, Slovakia: International Association for Plant Taxonomy.
- MinnisAMLinderDL. 2013. Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans comb. nov., in bat hibernacula of eastern North America. Fungal Biol 117:638–649, doi:10.1016/j.funbio.2013.07.001
- NguyenHDTNickersonNLSeifertKA. 2013. Basidioascus and Geminibasidium: a new lineage of heat-resistant and xerotolerant basidiomycetes. Mycologia 105:1231–1250, doi:10.3852/12-351
- PerfilievBW. 1929. Spiromastix verrucosus nov. g. n. sp. n. fam. – Vertreter eines neuen Typus der Flagellaten. Mikrobiol Zurn (Leningrad) 9:132–137, 183.
- PittJILantzHPetterssonOVLeongSL. 2013. Xerochrysium gen. nov. andBettsia, genera encompassing xerophilic species of Chrysosporium. IMA Fungus 4:229–241, doi:10.5598/imafungus.2013.04.02.08
- RittenourWRCiaccioCEBarnesCSKashonMLLemonsARBeezholdDHGreenBJ. 2014. Internal transcribed spacer rRNA gene sequencing analysis of fungal diversity in Kansas City indoor environments. Environ Sci Process Impacts 16:33–43, doi:10.1039/c3em00441
- RizzoLSuttonDAWiederholdNPThompsonEHFriedmanRWickesBLCano-LiraJFStchigelAMGuarroJ. 2014. Isolation and characterisation of the fungus Spiromastix asexualis sp. nov. from discospondylitis in a German shepherd dog and review of Spiromastix with the proposal of the new order Spiromastixales (Ascomycota). Mycoses 57:419–428, doi:10.1111/myc.12178
- RonquistFHuelsenbeckJP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574, doi:10.1093/bioinformatics/btg180
- SabouraudR. 1910. Les Teignes. Paris: Masson et Cie. 988 p.
- SamsonRAHoubrakenJThraneUFrisvadJCAndersenB. 2010. Food and indoor fungi. CBS laboratory manual series 2. Utrecht, The Netherlands: CBS-Fungal Biodiveristy Centre. 390 p.
- SamsonRAUntereinerWA. 2004. Determination of keratin degradation by fungi using keratin azure. Med Mycol 42:239–246, doi:10.1080/13693780310001644680
- SeifertKAMorgan-JonesGGamsWKendrickB. 2011. The genera of hyphomycetes. Biodiversity series 9. Utrecht, The Netherlands: CBS-Fungal Biodiveristy Centre. 997 p.
- SiglerL. 1997. Chrysosporium and molds resembling dermatophytes. In: KaneJSummerbellRSiglerLKrajdenSLandG, eds. Laboratory handbook of dermatophytes: a clinical guide and laboratory manual of dermatophytes and other filamentous fungi from skin, hair and nails. USA: Star Publishing Press. p. 261–311.
- SiglerL. 2002. The onygenaceae and other fungi from the order onygenales. In: HowardDH, ed. Pathogenic fungi in humans and animals. New York: Marcel Dekker Inc. p 195–314.
- SiglerLCarmichaelJW. 1976. Taxonomy of Malbranchea and some other hyphomycetes with arthroconidia. Mycotaxon 4:349–388.
- SiglerLHambletonSFlisALParéA. 2002. Auxarthron teleomorphs for Malbranchea filamentosa and Malbranchea albolutea and relationships within Auxarthron. Stud Mycol 47:111–122.
- SugiyamaMOharaAMikawaT. 1999. Molecular phylogeny of onygenalean fungi based on small subunit ribosomal DNA (SSU rDNA) sequences. Mycoscience 40:251–258, doi:10.1007/BF02463962
- SugiyamaMMikawaT. 2001. Phylogenetic analysis of the non-pathogenic genus Spiromastix (Onygenaceae) and related onygenalean taxa based on large subunit ribosomal DNA sequences. Mycoscience 42:413–421, doi:10.1007/bf02464337
- SugiyamaMSummerbellRCMikawaT. 2002. Molecular phylogeny of onygenalean fungi based on small subunit (SSU) and large subunit (LSU) ribosomal DNA sequences. Stud Mycol 47:5–23.
- SummerbellRC. 2000. Form and function in the evolution of dermatophytes. In: KushwahaRKSGuarroJ, eds. Biology of dermatophytes and other keratinophilic fungi. Bilbao, Portugal: Revista Iberoamericana de Micología. p. 30–43.
- SwoffordDL. 2002. PAUP* 4: phylogenetic analysis using parsimony (*and other methods). Sunderland, Massachusetts: Sinauer Associates.
- TamuraKNeiMKumarS. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101:11030–11035.
- TamuraKStecherGPetersonDFilipskiAKumarS. 2013. MEGA 6: molecular evolutionary genetics analysis. Mol Biol Evol 30:2725–2729, doi:10.1093/molbev/mst197
- UchiyamaSKamiyaSUdagawaS. 1995. Spiromastix saturnispora, a new species from Indonesian soil. Mycoscience 36:353–357, doi:10.1007/BF02268612
- UdagawaS. 1997. Taxonomic studies on Plectomycetes (cleistothecial ascomycetes). Nippon Kingakukai Kaiho 38:143–157.
- UdagawaSUchiyamaS. 1999. Taxonomic studies on new or critical fungi of non-pathogenic Onygenales 2. Mycoscience 40:291–305, doi:10.1007/BF02463966
- UdagawaSUchiyamaS. 2000. Acanthogymnomyces, a new genus of gymnothecial ascomycetes with setae and sulcate ascospores. Mycotaxon 76:411–418.
- UntereinerWAScottJANaveauFABachewichJ. 2002. Phylogeny of Ajellomyces, Polytolypa and Spiromastix (Onygenaceae) inferred from rDNA sequence and non-molecular data. Stud Mycol 47:25–35.
- UntereinerWAScottJANaveauFASiglerLBachewichJAngusA. 2004. The Ajellomycetaceae, a new family of vertebrate-associated Onygenales. Mycologia 96:812–821.
- VidalPVinuesaMAPuellesJMSGuarroJ. 2000. Phylogeny of the anamorphic genus Chrysosporium and related taxa based on rDNA internal transcribed spacer sequences. In: KushwahaRKSGuarroJ, eds. Biology of dermatophytes and other keratinophilic fungi. Bilbao, Spain: Revista Iberoamericana de Micrologia. p 22–29.
- VilgalysRHesterM. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246.
- VisagieCMHirookaYTanneyJBWhitfieldEMwangeKMeijerMAmendASSeifertKASamsonRA. 2014. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud Mycol 78:63–139, doi:10.1016/j.simyco.2014.07.002
- von ArxJA. 1987. A re-evaluation of the Eurotiales. Persoonia 13:273–300.
- WarcupJH. 1957. Studies on the occurrence and activity of fungi in a wheat-field soil. Trans Br Mycol Soc 40:237–IN233, doi:10.1016/S0007-1536(57)80010-2
- ZwicklDJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. GARLI 2.0 available online at code.google.com/p/garli/
