Abstract
Fusisporium solani was described as the causal agent of a dry rot of potato in Germany in the mid 19th century. As Fusarium solani, the species became known as a plurivorous plant pathogen, endophyte, decomposer, and opportunistic pathogen of humans and nutritional symbiont of insects. In parallel, it became evident that the morphologically defined species F. solani represents a phylogenetically and biologically complex group of often morphologically cryptic species that has come to be known in part as the F. solani species complex (FSSC), accommodating several formae speciales and mating populations/biological species. The FSSC currently includes more than 60 phylogenetic species. Several of these have been named, but the majority remains unnamed and the identity of F. solani sensu stricto is unclear. To promote further taxonomic developments in the FSSC, lectoand epitypification is proposed for Fusisporium solani. Although no type material for F. solani is known to exist, the species was abundantly illustrated in the protologue. Thus, a relevant illustration provided by von Martius is selected as the lectotype. The epitype selected here originates from a rotting potato collected in a field in Slovenia. This strain causes a dry rot of artificially inoculated potatoes. It groups in the heretofore unnamed phylogenetic species 5, which is nested within clade 3 of the FSSC (FSSC 5). Members of this phylogenetic species have a wide geographic distribution and include soil saprotrophs and plant and opportunistic human pathogens. This typification is consistent with the original description of Fusisporium solani and the concept of F. solani as a widely distributed soil inhabitant and pathogen.
Introduction
Fusarium solani (Mart.) Sacc. is a name under broad usage in the literature of plant pathology, applied to an extremely diverse assemblage of fungi with respect to host/substrate, pathogenicity, geographic distribution, morphological characteristics (including presence or absence of microconidia and macroconidia, macroconidial characteristics, ascospore size) and homo- and heterothallic sexual stages when known.
CitationWollenweber and Reinking (1935) delimited F. solani within section Martiella, with eight subspecific varieties and forms. CitationSnyder and Hansen (1941) combined all species, varieties and forms from Wollenweber and Reinking’s sections Martiella and Ventricosum into F. solani. This treatment has been the most widely used (e.g. CitationNelson et al. 1983, CitationLeslie and Summerell 2006). CitationBooth (1971) also combined sections Ventricosum and Martiella, retaining four species within section Martiella, including F. solani. CitationGerlach and Nirenberg (1982) synonymized four of Wollenweber and Reinking’s varieties under F. solani var. solani, and retained four additional species within section Martiella. While an association between section Martiella and F. solani sensu lato has been supported by phylogenetics, section Ventricosum represents a clade distantly removed from F. solani (CitationGräfenhan et al. 2011, CitationO’Donnell et al. 2013, CitationLombard et al. 2015). Based on sexual compatibility, CitationMatuo and Snyder (1973) described several mating populations within F. solani that became known as Nectria haematococca mating populations I–VII. Each of these mating groups represented one of the formae speciales that corresponded to pathogenicity on specific plant host species and cultivars. Phylogenetic analyses have consistently demonstrated that F. solani represents a complex of mostly morphologically cryptic species, now known as the F. solani species complex (FSSC; CitationO’Donnell 2000, CitationZhang et al. 2006, CitationO’Donnell et al. 2008, CitationNalim et al. 2011, CitationShort et al. 2013). These analyses suggest that the FSSC consists of three major clades encompassing over 60 phylogenetic species. Representatives of a vast majority of these morphologically homogeneous species had previously been identified as F. solani, but this name has never been firmly applied to a single phylogenetic species, supported by typification. To facilitate better communication, a haplotype nomenclatural system was introduced in which each phylogenetic species received a numerical identifier, with unique sequence types within species designated by lower case letters (CitationChang et al. 2006, CitationO’Donnell et al. 2008). Some of these species have been named (e.g. CitationO’Donnell 2000, CitationCovert et al. 2007, CitationNalim et al. 2011, CitationShort et al. 2013) or connected to previously known species (CitationO’Donnell 2000, CitationSummerbell and Schroers 2002), but most remain unnamed. One barrier to developing a taxonomy for the FSSC is a lack of understanding of the identity of F. solani sensu stricto.
The fungus that we know as F. solani was first described as Fusisporium solani by von Martius in 1842 as the cause of a potato tuber rot. The original illustrations show relatively long phialides and sparsely branched or unbranched conidiophores (); these conidiophores have become diagnostic for fusaria belonging to section Martiella (approximating the FSSC) in several Fusarium revisions (CitationWollenweber and Reinking 1935, CitationBooth 1971, CitationGerlach and Nirenberg 1982, CitationNelson et al. 1983). The objective of the present research was to collect a Fusarium from potato in Central Europe that agrees with the original morphological and ecological concept of Fusisporium solani and to use molecular phylogenetic tools to associate it with an appropriate phylogenetic species in the FSSC. With the identity of F. solani stabilized, the taxonomic logjam that is the FSSC can be broken.
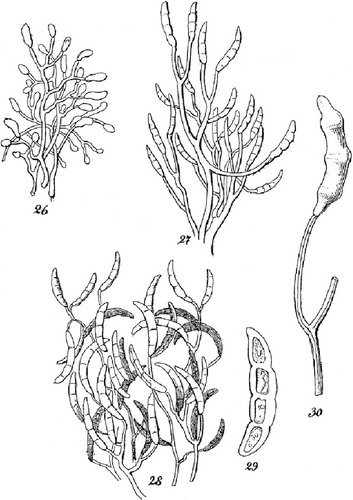
Fig. 1 Reproduction of selected elements of the original drawings by Citationvon Martius (1842) illustrating Fusisporium solani, the causal agent of potato dry rot. Original figure numbers were kept. 26–28. Repeatedly branched conidiophores, comparable to sporodochial conidiophores illustrated in , bearing multi-septate, Fusarium-like macroconidia. 29. Fusarium-like macroconidium with three pairs of lateral constrictions indicating the location of 3 putative septa. 30. Sparsely branched conidiophore with a still attached Fusarium-like macroconidium.
Materials and methods
Collection and isolation of strains
Isolations were made from potato tubers left lying in fields after harvest in Germany and Slovenia in 2009. Typically, these potatoes were partly covered by soil. Potatoes were collected if they were still in a reasonably good condition but showing rot and fungal colonization in an early stage. Potatoes were placed singly in paper bags, transported to the laboratory, stored overnight in a refrigerator at ca. 4 C and then processed the next day. Using a dissecting microscope, sporulating structures (mycelium with conidiophores or sporodochial pustules) were removed from the surface of the tubers with a needle and suspended into a drop of sterile water on a glass microscope slide. Suspensions were checked for Fusarium macroconidia microscopically at magnifications up to 320 × and drawn by capillary action into fine sterile glass capillary tubes, the tips of which were then moved over the surface of diluted potato dextrose agar enriched with antibiotics (dPDA+; 1 L: 14 g potato dextrose [Biolife, Italy], 10 g technical agar [Biolife, Italy], 12 mg penicillin G potassium salt [Fluka, Buchs, Switzerland], 54 mg streptomycin sulfate salt [Sigma-Aldrich]) for the distribution of macroconidia. Macroconidia that were well separated from each other were identified microscopically at low magnification, allowed to germinate overnight and isolated on synthetic nutrient-poor agar (SNA) (CitationNirenberg 1976). Nine to 12 macroconidia were isolated from each suspension and kept in slanted SNA stock tubes. Representatives of these were deposited at the ARS culture collection (NRRL, USDA National Center for Agricultural Utilization Research, Peoria, IL USA), the Fusarium Research Center (FRC, Penn State University, Department of Plant Pathology and Environmental Microbiology, University Park, PA, U.S.A.) and the Centraalbureau voor Schimmelcultures (CBS, Fungal Biodiversity Centre, Utrecht, the Netherlands). G. J. S. cultures are preserved in the culture collection of the Systematic Mycology and Microbiology Laboratory, U.S. Department of Agriculture, Beltsville, MD U.S.A. The locations that were sampled in August–October 2009 and the strains taken from each are listed in supplementary table I.
Molecular methods
DNA sequences were obtained from three gene regions, using the primers and PCR methods for the FSSC multilocus sequence typing scheme utilized by CitationO’Donnell et al. (2008): (i) an intron-rich portion of the translation elongation factor 1-α gene (TEF1, CitationGeiser et al. 2004), (ii) a region of the rRNA gene repeat including nuc rDNA ITS1-5.8S-ITS2 (ITS) and the 28S D1-D2 region (28S), and (iii) the gene for the second largest subunit of RNA polymerase II (RPB2). Bidirectional DNA sequences for each region were generated at the Penn State Genome Core Facility (University Park, Pennsylvania) on an ABI 3730XL automated DNA sequencer. Edited sequences were added to existing alignments with representatives of the known phylogenetic species within Clade 3 of the FSSC, with NRRL 22316 (F. staphyleae [= Geejayessia atrofusca] [CitationSchroers et al. 2011]) assigned as outgroup. Alignments were deposited at www.treebase.org (accession number S18168). Maximum likelihood (ML) analyses performed using raxmlGUI 1.3.1 (CitationSilvestro and Michalak 2012, CitationStamatakis 2014) applied the general time-reversible (GTR) substitution model with gamma model of rate heterogeneity in 100 inferences on the original alignment and ML bootstrap analyses using the convergence criteria autoMRE. Heuristic searches for shortest trees in maximum parsimony analyses (MP) were generated with PAUP 4.0b10 (CitationSwofford 2003) as described (CitationSchroers et al. 2011) with a maxtrees setting of 1000 and starting trees obtained with 1000 (heuristic searches) or 10 (bootstrap analyses) stepwise, random addition sequences. Bayesian analyses were performed using MrBayes 3.2.1 (CitationRonquist et al. 2012) and jModeltest 2.1.4 (CitationDarriba et al. 2012) as described (CitationSchroers et al. 2011). Bayesian and bootstrap analyses (MP, ML) were performed with and without FSSC 5-f (NRRL 22783) and FSSC 5-jj (NRRL 36208) for inferring FSSC 5 species boundary. Bayesian and bootstrap analyses (ML) were run on each of the partitioned datasets (rDNA, RPB2, TEF1) for testing gene tree concordance.
Pathogenicity tests
Organically produced potato tubers of cv. ‘Agria’ were purchased from a farmer in Ljubljana (Slovenia) in April 2011. The tubers were washed first with tap water to remove soil and debris, then rinsed three times in sterile distilled water (dH2O), surface-sterilized for 15 min in a 2% sodium hypochlorite solution (Kemika, Zagreb, Croatia) and rinsed again in dH2O. Whole potatoes were placed between pieces of sterile filter paper and air dried overnight. Two wound cavities per tuber (5 mm diam, 5–8 mm deep) were obtained with a sterile cork borer. Cavities of groups of 12 potatoes were inoculated with NRRL 66303, 66304, or, respectively, FRC S-2368. These strains were newly isolated from rotten potato in Slovenia and nested within FSSC 5 according to TEF1 sequences. The inoculum consisted of an agar plug colonized by sporulating mycelium that was transferred into one of the two wound cavities. In the other wound cavity, 100 μL of a spore suspension was pipetted using a medical syringe. Agar plugs were excised from the margin of actively growing, 14-d-old SNA cultures. Conidia for the spore suspensions were collected from 14-d-old cultures (SNA plus filter paper) and adjusted to 106 spores/mL using a Bürker-Türk hemocytometer (Brand, Germany). As a negative control, potatoes were inoculated with a sterile agar plug (SNA) or with 100 μL dH2O, respectively. Ten–12 potatoes were treated in control experiments or inoculated with each of the selected strains and placed in plastic containers on top of sterile filter paper. The plastic containers were placed in loosely closed plastic bags for allowing air exchange and incubated at 20 C in the dark. After 4 wk incubation at 20 C, a few arbitrarily selected potatoes were examined. After 8 wk, longitudinal sections were made through the center of the wound cavities and documented photographically. To retrieve rot fungi, either small pieces of material at the interface between healthy potato flesh and rot was excised with a sterile scalpel, or mycelium was removed from the developing rots with sterile glass needles and plated on dPDA+. Fungi were identified using morphological criteria. When a fungus fitting the broad morphological concept of F. solani was encountered, it was identified using TEF1 sequences (results not shown).
Morphology
Morphological characters were documented from colonies grown on carnation leaf agar (CLA) (CitationFisher et al. 1982) in 9 cm diam plastic Petri plates for approximately 10 d at 25 C under cool white fluorescent light (12 h) alternating with 12 h darkness. Microscopic observations were made from water mounts. Measurements were taken, where possible, from at least 30 conidia from pionnote sporodochia and erect/aerial conidiophores. The largest and smallest observations are presented in parentheses, and ranges based on the mean ± one standard deviation were calculated. For photo-documentation, structures were stained in 1% (aq.) phloxine, as needed. Colony characters were documented from cultures grown on PDA (Difco) and SNA grown in 9-cm-diam plastic Petri plates at 25 C under light conditions described above for 10 d. Colony colors were assigned following CitationKornerup and Wanscher (1978).
Results
Ten collections from potato fields in Slovenia yielded fusaria that conformed to the original description of Fusisporium solani () and were nested phylogenetically within FSSC 5 (). Selected representatives of these 10 strains caused a dry rot of potato tubers in pathogenicity tests () resembling the symptoms described by Citationvon Martius (1842).
Fig. 2 Majority rule consensus phylogram from MrBayes analyses of the F. solani species complex (FSSC) inferred from rDNA (104/986 parsimony informative positions), TEF1 (117/709) and RPB2 (303/1738) sequences. Alphanumeric designations next to strain numbers indicate the three-locus haplotypes and species as described in CitationO’Donnell et al. (2008). The phylogenetic species FSSC 5 is inferred as F. solani sensu stricto. Arrows indicate divergent haplotypes 5-f and 5-jj and boldface ‘ET’ the ex-epitype isolate NRRL 66304 (= FRC S-2364, CBS 140079, G.J.S. 09-1466). Values shown at nodes indicate ML and MP bootstrap support > 60%; asterisks above branches represent node support of 100% BS. Bold branches refer to nodes with a PP value of 0.95–1.00. Gray highlight is used to identify isolates from potato. The novel phylogenetic species FSSC 44 isolated from potato in Slovenia is represented by NRRL 34617 and G.J.S. 09-1459.
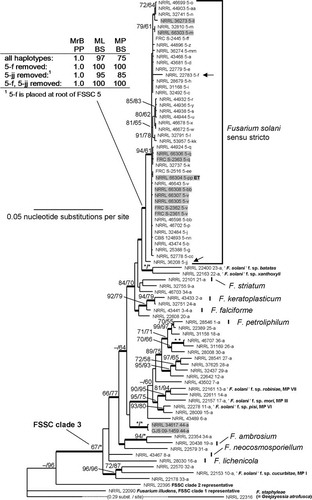
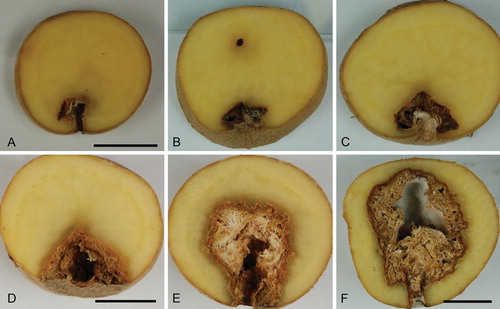
Fig. 3 A–F. Dry rot symptoms of potato tubers artificially infected with Fusarium solani. Tubers in A and B infected with strain NRRL 66303 (FRC S-2561, CBS 140075, G.J.S. 09-1464); C, D, E with ex-epitype strain NRRL 66304 (FRC S-2364, CBS 140079, G.J.S. 09-1466); F with FRC S-2368 (G.J.S. 09-1471). Scale bars: A, D, F = 2 cm. A applies for B, C; D applies for E.
Phylogenetic support for FSSC phylogenetic species 5
Trees with identical overall topologies and resolving a monophyletic FSSC 5 as shown in the MrBayes consensus tree () were encountered in all of the 100 ML inferences and in the MP and ML bootstrap consensus trees. Analyses of all three-locus haplotypes of FSSC 5 resolved a moderately or highly supported monophyletic group (maximum likelihood bootstrap (ML BS) = 97%; maximum parsimony bootstrap (MP BS) = 75%; Bayesian posterior probability (PP) = 1.00), consistent with findings in previous studies focused on the FSSC (CitationZhang et al. 2006, CitationO’Donnell et al. 2008, CitationShort et al. 2011). The FSSC 5 clade received highest possible support when multilocus haplotype FSSC 5-f, represented by strain NRRL 22783 (= IMI 252021, ex gray seal skin lesion, Washington, DC), was excluded from the analyses (ML BS = 100%, MP BS = 100% and PP = 1.00). Scrutiny of individual gene genealogies showed that NRRL 22783 nested within FSSC 5 according to TEF1 and rDNA sequences, but the RPB2 allele grouped it with the undescribed phylogenetic species 9 (NRRL 32755), with weak to moderate statistical support (ML BS = 73%; PP = 1.00; results not shown), thus clearly outside the statistically supported FSSC 5 species clade (ML BS = 82%; PP = 0.99; results not shown). The FSSC 5 multilocus sequence type 5-jj, represented by strain NRRL 36208, fell distinctly basal within the FSSC 5 clade. Removal of this haplotype from the analysis altered the support for the FSSC 5 clade slightly (ML BS = 95%, MP BS = 85%, PP = 1.00), and removal of haplotypes 5-f and 5-jj increased support to 100/100/1.00. Strains NRRL 66303–66308 and FRC S-2361–2363 isolated from potato collected in Slovenia were nested within FSSC 5 in all analyses. All strains from Slovenia matched previously known haplotypes (5-m, 5-q, 5-v, 5-bb) except for the exepitype NRRL 66304, which was assigned to novel haplotype FSSC 5-pp. A monophyletic FSSC 5 was also resolved in partitioned analyses of rDNA, TEF1 and, following removal of FSSC 5-f, RPB2 sequences. rDNA sequences resolved FSSC 22-a and 23-a as closest sister taxa of FSSC 5, however, the relationship was not statistically supported. RPB2 and TEF1 did not suggest a clear sistergroup relationship of FSSC 5 to any other species or haplotype. Two additional potato isolates from Slovenia, NRRL 34617 and G.J.S. 09-1459, represent a putatively novel phylogenetic species in Clade 3 of the FSSC. These isolates, assigned as FSSC 44-a, had identical three-locus haplotypes.
Pathogenicity tests
After 4 wk incubation few arbitrarily selected potatoes that had been inoculated with FSSC 5 members NRRL 66303 and 66304 and FRC S-2368 showed visible rots. After 8 wk, ca. 50% of tubers (i.e. ca. 5 from each trial) revealed a small rot in longitudinal sections () while extensive areas of the potato flesh became rotten in 20–50% of tubers (i.e. 2–5 from each trial) (). In some inoculated potatoes no rot was detected; none of the potatoes used as negative controls showed rot symptoms. PCR experiments based on strains isolated from the interface between rotten and healthy potato flesh detected TEF1 sequences that were identical with sequences obtained from strains used for inoculations (results not shown). Typically, a mottled pattern of yellowish brown or sometimes dark, brownish to blackish colors was observed (). Cavities filled with white or grayish-white mycelium were also observed in sections through dry rotten potatoes artificially infected with the selected strains.
Taxonomy
Fusarium solani (Mart.) Sacc., Michelia 2(7): 296. 1881.
≡ Fusisporium solani Mart., Die Kartoffel-Epidemie der letzten Jahre oder die Stockfäule und Räude der Kartoffeln: 20. 1842.
Typification: MBT203351: [icon]: Citationvon Martius (1842), taf. III, Fig. 29. GERMANY. Macroconidium illustrated from dry-rotten potato (lectotype of Fusarium (Fusisporium) solani, designated here). MBT203352: SLOVENIA. Doljenska District: Radohova vas (Dinaric valley systems and corrosion plains), dried culture on SNA with carnation leaf pieces, isol. ex decaying tuber of Solanum tuberosum in field following harvest, 18 Aug 2009, H.-J. Schroers 1434): (epitype of Fusarium (Fusisporium) solani, designated here, CBS H-22335). Ex-epitype culture: NRRL 66304 (= FRC S-2364, CBS 140079, G.J.S. 09-1466). GenBank accession numbers for DNA sequences derived from type: ITS/28S = KT313633, TEF = KT313611, RPB2 = KT313623.
Characteristics of F. solani on field-collected potatoes ()
Sections of rotting potatoes highly convoluted with cushion-shaped protuberances, partly covered with cottony to arachnoid white mycelium, accompanied by a buff-colored slime, sometimes appearing as tuberculate conidial masses. Unbranched conidiophores typically arising from the aerial mycelium, 60–250 μm long, 7–10 μm at the base, with few septa, the terminal cell comprising a phialide; conidia 0–3-septate; 0–1-septate conidia ellipsoidal to slightly clavate, 3-septate conidia clavate to falcate, basal cell indistinct. Conidia forming in a slime comprising a palisade of short conidiophores consisting of single phialides or tight whorls of few phialides; phialides cylindrical, 15–24(−40) μm long, 4.0–5.5(−7.0) μm wide, collarette thickened, not flared; conidia arising from these conidiophores falcate with a prominent, pedicellate basal cell; primarily 3-septate, septa often obscure.
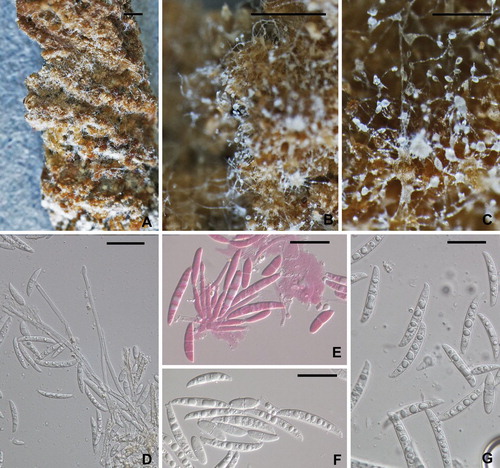
Fig. 4 Fusarium solani from potato collected in the field. A–C. Macroscopic view of mycelium, conidiophores and conidia. Note the long conidiophores and slimy heads of conidia in B and C. D–G. Conidiophores and conidia. E stained in 1% (aq.) phloxine. All from NRRL 66304. Scale bars: A = 1 mm, B = 0.5 mm, C = 100 μm, D–G = 20 μm.
0-septate (n = 39): (6.5−)9.0–18.0(−30.0) × (2.5−)3.5–6.0(−7.5) μm
1-septate (n = 60): (10.0−)20.5–22.5(−33.0)×(3.5−)4.0–6.0(−7.5) μm
2-to-3-septate (n = 76): (20.0−)26.0–37.0(−47.0) × (3.0−)5.0–7.0 μm
Chlamydospores not observed.
Characteristics obtained from artificial media (, )
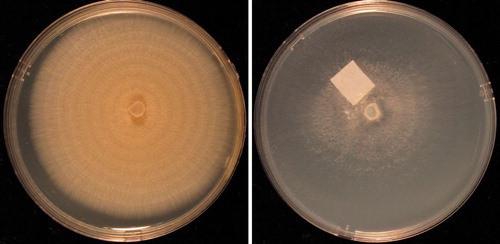
Colonies grown 10 d at 25 C under cool white fluorescent light 12 h/darkness: On PDA (69–)70–80(−82) mm diam at 25 C, averaging 7.5 (6.5–8.5) mm/d at 25 C and 9.5 (7.0–10.5) mm/d at 30 C over first three days of growth (CitationShort et al. 2013); colony pigmentation from above pale yellow to pale salmon; colony reverse pigmentation in a gradient from K&W 4A6 (Maize Yellow) in the middle to 4A2 (yellowish white) at the margin; conidia forming from the surface of the colony and in the aerial mycelium; conidia arising from the surface of the colony embedded in yellowish beige slime; conidia forming from the aerial mycelium in translucent drops of clear liquid at the tips of long, unbranched conidiophores; aerial mycelium cottony, forming more or less distinct concentric rings. On SNA 80–90 mm diam after 10 d at 25 C, aerial mycelium cottony, white, dense over the center of the colony, nearly invisible elsewhere; conidia forming abundantly from erect, typically unbranched conidiophores from the surface of the agar and in aerial mycelium; blue pigment not observed. On CLA little or no aerial mycelium; cream colored sporodochia formed abundantly on carnation leaf pieces, pionnotes less frequently developed on agar surface; long, unbranched conidiophores arising in abundance from the agar surface; conidia held in hyaline drops of liquid. Conidiophores (27–)67–123(−230) μm long, (2.0–)3.5–5.0 (−7.0) μm wide at the base, unbranched or branched up to three times, straight, thin-walled, smooth; each branch terminating in a single phialide, tapering uniformly from base to tip, collarette of phialides thickened, not conspicuously flared. Conidia typically arising from unbranched conidiophores 0–3(4–5)-septate; 0-septate conidia ellipsoidal; 1, 2-septate conidia ellipsoidal to falcate; 3-septate conidia more or less clavate, broad at the base tapering uniformly to a subacute tip (‘flame-shaped’); 4 and 5-septate rare from unbranched conidiophores, falcate, slightly curved with a pedicellate basal cell.
Fig. 5 Fusarium solani on carnation leaf agar. A. White to cream colored, slimy conidial masses formed by pionnote sporodochia on pieces of carnation leaf. B, C. Unbranched or sparsely branched conidiophores arising from the surface of the agar. D, E. 0-to-3 septate conidia produced from unbranched or sparsely branched conidiophores. F. Conidiophores from pionnote sporodochium producing up to 5-septate conidia. G–K. Conidia arising from pionnotes. L. Chlamydospores. A, C, E, J from FRC S-2362 (G.J.S. 09-1462); B, D, K, L from FRC S-2361 (G.J.S. 09-1461); F from NRRL 66307 (FRC S-2367, CBS 140077, G.J.S. 09-1469); G from FRC S-2363 (G.J.S. 09-1463); H, I from ex-epitype strain NRRL 66304 (FRC S-2364, CBS 140079, G.J.S. 09-1466). Scale bars: A = 1 mm; B–E, I, K = 20 μm; F–H, J = 30 μm; L = 10 μm.
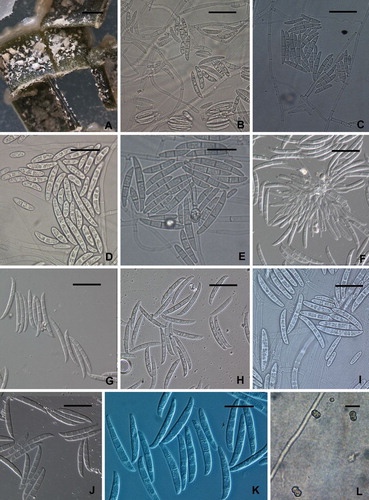
Fig. 6 Fusarium solani cultures on PDA (left) and SNA (right) after 10 days at 25 C under alternating cool white fluorescent light (12 h) and darkness (12 h) in 9-cm-diam Petri plates. From ex-epitype strain NRRL 66304 (FRC S-2364, CBS 140079, G.J.S. 09-1466).
Conidia from aerial conidiophores:
0-septate (n = 240): (5.5−)8.5–13.5(−16.5) × (2.0−)2.5–4.5(−6.0) μm
1-septate (n = 240): (12.0−)17.0–24.0(−28.5) × (2.0−)3.5–5.5(−8.0) μm
2-septate (n = 240): (14.0−)22.0–28.5(−32.0) × (2.5−)4.0–6.0(−6.5) μm
3-septate (n = 240): (23.0−)29.0–39.0(−45.0) × (3.0−)4.5–6.5(−7.5) μm
4-septate (n = 17): (42.0−)43.0–50.0(−53) × (3.5−)4.5–6.5(−7.5) μm
5-septate (n = 3): 48.0–53.0 × 6–7 μm
Pionnotal conidiophores comprising fascicles of phialides; phialides cylindrical or slightly enlarged in the middle, 14–17 × 2.5–3.5 μm, straight or slightly curved, collarette slightly thickened, not flared. Conidia arising from pionnote conidiophores/sporodochia, mostly 3–4 (0–5)-septate, falcate slightly curved and often more strongly curved at the tip, base broadly pedicellate.
Pionnotal conidia:
3-septate (n = 241): (24.0−)36.0–44.0(−48) × (2.0−)4.5–6.0(−8.0) μm
4-septate (n = 226): (31.0−)42.0–48.0(−52.0) × (3.0−)4.5–6.0(−7.5) μm
5-septate (n = 129): (41.0−)45.0–51.0(−56.0) × (2.5−)4.5–6.0(−8.0) μm
Chlamydospores abundant on CLA, one- or two-celled, each globose, 6.5–8.5 μm diam, pale brown, roughened.
Other material examined: NRRL 66305 (= FRC S-2365, CBS 140074, G.J.S. 09-1467), NRRL 66303 (= FRC S-2561, CBS 140075, G.J.S. 09-1464), NRRL 66306 (= FRC S-2366, CBS 140076, G.J.S. 09-1468), NRRL 66307 (= FRC S-2367, CBS 140077, G.J.S. 09-1469), NRRL 66308 (= FRC S-2562, CBS 140078, G.J.S. 09-1470), FRC S-2361 (= G.J.S. 09-1461), FRC S-2362 (= G.J.S. 09-1462), FRC S-2363 (= G.J.S. 09-1463), and FRC S-2368 (= G.J.S. 09-1471) represented as strains isolated from potato collected in Slovenia. Additional worldwide collections of F. solani from a variety of other sources, representing all known multilocus sequence types for the species, are listed in Supplementary table I.
Variation within FSSC 5
Seven strains of phylogenetic species FSSC 5 that were isolated from human subjects were included in this study, each representing a different haplotype (Supplementary table I, Fig. 2; CitationO’Donnell et al. 2008). Colony characters of these strains were the same as described above for the strains isolated from potato (, ). As is the case for many FSSC species, strains of FSSC 5, including NRRL 32810 from human eye, sometimes formed a blue pigment in the agar of nutritionally rich media such as PDA or a gray-blue pigment in pionnotal conidial masses. Most produced macro- and microconidia and long, unbranched conidiophores in the aerial mycelium and pionnotal conidiophores producing white conidial masses. 0–3-septate conidia predominated from aerial conidiophores; 3-septate conidia tended to be clavate to ‘flame-shaped’, as described above. Pionnotal conidia were primarily 3–4(−5)-septate, straight but with a more strongly curved tip cell and a more or less distinctly pedicellate basal cell.
The potato strains and most of the medical strains were uniform in producing predominantly 3-septate conidia and no aseptate conidia from pionnotes; FSSC 5 isolates varied in the production of 4-to-5 septate conidia. Some clinically relevant strains produced 0, 3–5-septate conidia (NRRL 32810). Although potato strain FRC S-2361 produced 3-septate pionnotal conidia that were smaller than the 3-septate conidia in all other collections (n = 30, (23.0–)25.0–35.0(−40.0) × (4.0–)4.5–5.0(−6.0) μm), otherwise this strain could not be distinguished from the other FSSC 5 strains isolated from potato.
Discussion
Inferences from the von Martius text
In the original description (Citationvon Martius 1842) Fusisporium solani is described as the cause of a dry potato rot. The monograph by Citationvon Martius (1842) reviews several potato diseases known at that time and describes apparently for the first time a dry rot of tubers. This disease emerged as an epidemic problem in several parts of Germany, specifically regions of the current Hesse and Rhineland-Palatinate provinces, in the ten or so years before von Martius published his monograph. Today, the consensus opinion is that Citationvon Martius (1842) did not describe potato late blight caused by Phytophthora infestans, which was putatively introduced into Europe at about the same time (CitationBourke 1991, CitationAndrivon 1996), and that he described what we now know to be a ‘Fusarium’ species as the causal agent of the dry rot (CitationGerlach and Nirenberg 1982).
According to von Martius, potato dry rot can cause considerable yield losses during harvest. We therefore have interpreted this to be primarily a pre-harvest disease, thus motivating us to sample field-collected potatoes in the search for F. solani. However, he also described it as a disease of stored potatoes causing devastating losses of the stocks within the year of harvest. Von Martius emphasized that the disease was mainly observed on varieties producing white and yellow tubers specifically used for human consumption and in regions where pieces of potatoes rather than whole tubers were used as planting material. In the first stage of the disease, von Martius described a weak rot and mainly minor disturbances of the potato skin. The early second stage of the disease is characterized by blackish discolorations of the potato flesh below the skin, accompanied by dry shriveling, blistering and breakage of the potato skin, followed by the emergence of a white mycelium on the surface of the potatoes. In the late second stage, the potatoes dry out progressively and the rot extends into the flesh. The appearance of the rot is then described as a mottled pattern with grayish yellow, maroon-brown or black colors and the formation of cavities within the rotten potato flesh that can be filled with white mycelium (von Martius plate II, Fig. 8).
Microscopically, von Martius described the fungal mycelium that he removed from white spots in rotten parts below the potato surface as a felt of branched filaments with blind ends, whose tips can produce either ‘small,’ unicellular, round or elliptical, entirely or later, lancet- or sickle-shaped spores. The first formed conidial state was described as ‘varietas β: Sporotrichoides’. The sickle-shaped Fusarium-like macroconidia, described under Fusisporium solani, were 3-to-4 septate and possessed vacuolate and dense cytoplasm (, von Martius plate III, figs. 27–30). These macroconidia were slightly crescent-shaped and they had an attenuated tip that was slightly more strongly curved than the rest of the conidium, and a rounded basal cell. Von Martius concluded that the two conidial forms were produced by a single fungus that causes the rot. Von Martius gave the length of the sickle-shaped, 3–4 septate conidia, described as ‘ausgewachsene(n) Sporen’ (mature spores) as ‘0,075’ mm, or 75 μm. Von Martius also described tuberculate masses and sporodochia (as ‘Höckerchen’ [small knobs]) that can break through the epidermis and develop on the surface of progressively desiccated potatoes (von Martius plate III, Fig. 22, 25).
Comparison of strains isolated from potato in Slovenia to the original description of Fusisporium solani
Assuming that the drawings provided by von Martius are accurate and the 3-septate conidium illustrated in his Fig. 29 (plate III) is 75 μm long, as specified, then it would have to be 14 μm wide. This seems unlikely. From the original illustration, there is no doubt that von Martius’ Fusisporium solani is a Fusarium. However, in this genus it is unlikely that a conidium would measure as much as 75 μm in length and still only possess 3 or 4 septa because as conidia lengthen, they develop additional septa. Thus in Fusarium, a conidium that is 75 μm long would have at least 5 and most likely 7 septa. Moreover, if the 3-septate conidium illustrated by von Martius was actually 14 μm wide it would be very unusual in Fusarium. On the other hand, if the 3-septate conidium of Fig. 29 were 30 μm long, it would be 5 μm wide, which is exactly what we have observed for 3-septate conidia of the Fusarium that associated potato dry rot. It is therefore likely that the 75 μm length specification is based on an erroneous measurement by von Martius. The convoluted and tuberculate surface of affected potatoes and our observation that conidia of FSSC 5 strains can be formed in aerial mycelium and aggregate into slime agree with what was described by von Martius for Fusisporium solani.
The description of the potato disease provided by von Martius is consistent with the experimental results we obtained in pathogenicity tests. After 4 wk incubation, few of the 30–36 potatoes that had been infected with the three isolates showed a visible rot; after 8 wk, most potatoes showed either small () or extensive areas of dry rots (). The observed mottled pattern of yellowish brown or sometimes dark, brownish to blackish colors () and the white or grayish-white mycelium filling cavities of the rotten areas could be described as ‘truffle-like’, a term used several times by von Martius, presumably referring to the convoluted, geode-like gleba of many hypogeous fungi.
Typification of Fusisporium solani
The known collections of von Martius are kept at BR, G, M, MEL and TO (Index Herbariorum, http://sweetgum.nybg.org/ih/; abbreviations following same source). His ‘private collections’, which include the majority of the specimens that Martius gathered after his trip to Brazil in 1817–1820 are preserved in BR (CitationFörther 1994). None of these herbaria contain specimens of Fusisporium solani that can be linked to von Martius (curators of herbaria pers comm). Thus a new type for Fusisporium solani is required. Von Martius’ figures and description clearly illustrate a species of Fusarium. The illustrations are quite literal: this is a species having microconidia and 3 or 4-septate, slightly curved macroconidia measuring (upon reinterpretation) approximately 30 × 5 μm and loosely branched conidiophores with mostly terminal phialides. This suggests the F. solani species complex. Thus, we interpret Citationvon Martius (1842) plate III, Fig. 29 () as being part of the original description of Fusisporium solani and, accordingly, designate it as lectotype for Fusisporium solani.
The selected epitype belongs to the FSSC phylogenetic species 5 that was isolated from dry-rotten potato also in Denmark (NRRL 36273, Ulf Thrane pers comm), Iran (CitationChehri et al. 2014) and Poland (CitationStefańczyk et al. 2016, according to TEF1 sequences) and from various other substrata and regions of the world (CitationZhang et al. 2006, CitationBogale et al. 2009, CitationBalmas et al. 2010, CitationShort et al. 2011, Citation2013). The consensus morphology associated with FSSC 5 corresponds well with F. solani circumscriptions in modern taxonomic treatments (CitationWollenweber and Reinking 1935, CitationBooth 1971, CitationGerlach and Nirenberg 1982, CitationNelson et al. 1983). Therefore, the suggested epitypification conforms well to the original description of Fusisporium solani, preserves the currently understood concept of F. solani as a geographically widely distributed species, and minimizes the disruption to the current use of this name.
It is not clear, however, to which extent von Martius examined each of his samples microscopically or whether he observed the same microscopic characters in studied samples. DNA barcode-based results have indicated that dry potato rot can be caused also by members of the F. oxysporum species complex (CitationGarcia Bayona et al. 2011), and Fusarium species typically known from cereals and grasses such as F. graminearum, F. sambucinum, F. avenaceum and F. culmorum (CitationPeters JC et al. 2008, CitationPeters RD et al. 2008, CitationEstrada et al. 2010, CitationSagar et al. 2011). Also, other members of the FSSC were isolated from dry-rotten potatoes in our recent inventories, including putatively novel phylogenetic species FSSC 44 (). Because von Martius did not illustrate short phialides seated laterally on hyphae, we purport that the lectotype does not include elements of F. oxysporum. Likewise, characteristics of macroconidia and microconidia von Martius described are inconsistent with F. avenaceum, F. culmorum, F. graminearum or F. sambucinum.
Nectria haematococca and Fusarium solani
‘Nectria haematococca’ is not the teleomorph of F. solani. Although the anamorphically typified name F. solani is commonly linked to ‘Nectria haematococca’ (= Haematonectria haematococca, Neocosmospora haematococca [Berk. & Br.] Nalim et al. [CitationNalim et al. 2011]), the teleomorph name is now linked to the anamorphic name Fusarium haematococcum. This species belongs to FSSC clade 2 sensu CitationO’Donnell (2000) and is possibly endemic to Sri Lanka (CitationNalim et al. 2011). Fusarium solani, as we have defined it here, corresponds to phylogenetic species 5 within FSSC clade 3, which is known to accommodate opportunistic human or animal pathogens (CitationO’Donnell et al. 2008). Although both mating-type idiomorphs have been identified within FSSC 5 (N. Zhang unpubl), no sexual stage is known. Clade 3 is particularly species rich, accommodating up to 20 medically relevant phylogenetic species, the biological species or mating populations defined by CitationMatuo and Snyder (1973) and species having putatively temperate as well as tropical endemics. Several species in this clade have been named (CitationSummerbell and Schroers 2002, CitationNalim et al. 2011, CitationShort et al. 2013). Four of the most common species in the F. solani species complex (F. petroliphilum = FSSC 1, F. keratoplasticum = FSSC 2, F. falciforme = FSSC 3+4 and F. solani = FSSC 5) all show a good deal of intraspecific morphological variation, and overlapping morphological traits (CitationMehl and Epstein 2007, CitationShort et al. 2013). Thus, the molecular methods utilized in this paper and previous studies should be used for the identification of these taxa. The broad morphological concept of F. solani sensu lato, however, is generally identifiable in a majority of species across the FSSC. Where this morphology is encountered without molecular identification to phylogenetic species, we strongly encourage the use of ‘F. solani species complex’ rather than ‘F. solani’ to avoid confusion.
Most haplotypes of F. solani sensu stricto resolved as a strongly supported monophyletic group. However, two haplotypes (5-f and 5-jj) possess divergent alleles that reduce the statistical support for the F. solani clade in a combined multilocus analysis. Haplotype 5-f possesses TEF1 and rDNA alleles that are clearly nested within FSSC 5, but its RPB2 allele does not; the RPB2 allele tends to resolve with the undescribed species FSSC 9. The divergent RPB2 allele in 5-f could be explained either by introgression from FSSC 9 or an unknown related species, or the retention of ancestral polymorphism from a common ancestor of both species. In F. keratoplasticum (= FSSC 2), two isolates were genotyped that had rDNA alleles that were identical to those in FSSC 9 (CitationShort et al. 2011, Citation2013). In this case, hybridization was invoked as the likely cause, because of the 100% identity with extant alleles in a known taxon. In the current case, however, the RPB2 allele in F. solani haplotype 5-f is quite divergent from all known RPB2 alleles, so if it was acquired via hybridization, then it was likely from an unknown or extinct species. An alternative hypothesis, that the allelic divergence reflects the retention of ancestral polymorphism, is likely a better explanation, particularly when considered in the light of another divergent F. solani haplotype, 5-jj. In this case, 5-jj possesses TEF1 and RPB2 alleles that resolve basally within the F. solani/FSSC 5 clade, suggesting that there may be a great deal of ancestrally derived diversity in this group. Phylogenomic studies on this taxon may reveal insights that allow rigorous testing of these alternative hypotheses. Despite the possibility of interspecific hybridization and/or incomplete lineage sorting, we conclude that FSSC 5 satisfies the tenets of genealogical concordance phylogenetic species recognition (CitationTaylor et al. 2000).
With this epitypification, it is important to note that only seven of the ca. 45 phylogenetic species in Clade 3 of the FSSC are clearly connected to names. Named species include the most commonly encountered human pathogenic species, F. keratoplasticum, F. petroliphilum (also known as Nectria haematococca mating population (MP) V, ≈ F. solani f. sp. cucurbitae), and F. falciforme, F. lichenicola, F. pseudensiforme, F. neocosmosporiellum and the ambrosia beetle symbionts F. ambrosium and F. euwallaceae (CitationFreeman et al. 2013). Unnamed species include the formae speciales pisi (MPVI), mori (MPIII), robiniae (MPVII), xanthoxyli (MPII), and batatas (MPIV). Fusarium solani f. sp. batatas and F. solani f. sp. xanthoxyli are agents of foot rot of sweet-potato sprouts (CitationMcClure 1951) or, respectively, trunk-blight of Zanthoxylum piperitum (CitationSakurai & Matuo 1961). Fusarium striatum was recently characterized as an agent of a disease of greenhouse grown tomato (CitationMoine et al. 2014) although it was originally described from potato (as F. solani var. striatum) and used by CitationWollenweber and Reinking (1935) to identify dry rot agents on potato. The ability to act as pathogens of immunosuppressed patients and as causal keratitis agents in healthy people seems to be widely distributed in members of clade 3 of the FSSC. It is possible that their ability to produce immunosuppressive secondary metabolites such as cyclosporine A and C, detected in F. falciforme, F. keratoplasticum and what we now define as F. solani sensu stricto (CitationSugiura et al. 1999, CitationShort et al. 2013) may facilitate the infection process by these fungi.
The importance of F. solani is highlighted by the search query ‘Fusarium solani’ receiving more Google Scholar result hits than the binomials of eight of the ten most important plant pathogens defined by CitationDean et al. (2012), and the species name more often cited in titles of academic literature than five of those ten (ISI Web of Knowledge, July 2015). Since most members of the FSSC continue to be referred to as F. solani, taxonomic advancement is facilitated by identifying FSSC 5 as the specific object of comparison in recognizing new species.
Acknowledgments
We gratefully acknowledge the assistance of the curators and staff of the herbaria BR (National Botanic Garden of Belgium, Meise), G (Conservatoire et Jardin botaniques de la Ville de Genève, Switzerland), M (Botanische Staatssammlung München, Germany), MEL (Royal Botanic Gardens, Australia, Victoria, Melbourne), TO (University of Turin, Italy) and W (Naturhistorisches Museum Wien, Austria). We thank Mrs. M. Kralj Kunčič for collecting potatoes from Begunje and Stacy Sink at the United States Department of Agriculture National Center for Agricultural Utilization Research Mycotoxin Prevention and Applied Microbiology Research Unit for the species and haplotype assignments. This work was supported by grants from the National Science Foundation, NSF PEET grant 07-31510 to Priscila Chaverri, Amy Rossman and GJS, and NSF DEB 0089474 to DMG and GJS. This work was supported in part by the National Institute of Food and Agriculture Project PEN 04527 at the Pennsylvania State Agricultural Experiment Station. DPGS was supported by NIFA grant 2010-65110-20488, Education in Genomics-Based Microbial Forensics. The Slovenian Research Agency (ARRS) supported the study through financing the research program Agrobiodiversity P4-0072.
Literature cited
- AndrivonD. 1996. The origin of Phytophthora infestans populations present in Europe in the 1840s: A critical review of historical and scientific evidence. Plant Pathol 45:1027–1035, doi:10.1046/j.1365-3059.1996.d01-196.x
- BalmasVMigheliQSchermBGarauPO’DonnellKCeccherelliGKangSGeiserDM. 2010. Multilocus phylogenetics show high levels of endemic fusaria inhabiting Sardinian soils (Tyrrhenian Islands). Mycologia 102: 803–812, doi:10.3852/09-201
- BogaleMSteenkampETWingfieldMJWingfieldBD. 2009. Diverse Fusarium solani isolates colonise agricultural environments in Ethiopia. Eur J Plant Pathol 124:369–378, doi:10.1007/s10658-008-9422-y
- BoothC. 1971. The genus Fusarium. Kew, England: Commonwealth Mycological Institute. 237 p.
- BourkeA. 1991. Potato blight in Europe in 1845: the scientific controversy. In: LucasJAShattockRCShawDSCookeLR, eds. Phytophthora. Cambridge: Cambridge University Press. p 12–24.
- ChangDCGrantGBO’DonnellKWannemuehlerKANoble-WangJRaoCYJacobsonLMCrowellCSSneedRSLewisFMTSchaffzinJKKainerMAGeneseCAAlfonsoECJonesDBSrinivasanAFridkinSKParkBJ. 2006. A multistate outbreak of Fusarium keratitis associated with use of a new contact lens solution. J Am Med Assn 296:953–963, doi:10.1001/jama.296.8.953
- ChehriKGhasempourHRKarimiN. 2014. Molecular phylogenetic and pathogenetic characterization of Fusarium solani species complex (FSSC), the cause of dry rot on potato in Iran. Microb Pathog 67–68:14–19, doi:10.1016/j.micpath.2014.01.002
- CovertSFAokiTO’DonnellKStarkeyDHollidayAGeiserDMCheungFTownCStromAJubaJScandianiMYangXB. 2007. Sexual reproduction in the soybean sudden death syndrome pathogen Fusarium tucumaniae. Fungal Genet Biol 44:799–807, doi:10.1016/j.fgb.2006.12.009
- DarribaDTaboadaGLDoalloRPosadaD. 2012. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods 9:772, doi:10.1038/nmeth.2109
- DeanRVan KanJAPretoriusZAHammond-KosackKEDi PietroASpanuPDRuddJJDickmanMKahmannREllisJFosterGD. 2012. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13: 414–430, doi:10.1111/j.1364-3703.2011.00783.x
- EstradaRGudmestadNCRiveraVVSecorGA. 2010. Fusarium graminearum as a dry rot pathogen of potato in the USA: Prevalence, comparison of host isolate aggressiveness and factors affecting aetiology. Plant Pathol 59:1114–1120, doi:10.1111/j.1365-3059.2010.02343.x
- FisherNLBurgessLWToussounTANelsonPE. 1982. Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 72:151–153, doi:10.1094/Phyto-72-151
- FörtherH. 1994. Die Geschichte des Martius-Herbariums: seine Brasilienkollektion und Empfehlungen zur Typenwahl. Sendtnera 2:5–24.
- FreemanSSharonMMaymonMMendelZProtasovAAokiTEskalenAO’DonnellK. 2013. Fusarium euwallaceae sp. nov.—a symbiotic fungus of Euwallacea sp., an invasive ambrosia beetle in Israel and California. Mycologia 105:1595–1606, doi:10.3852/13-066
- Garcia BayonaLGrajalesACardenasMESierraRCepero de GarciaMCBernalAJimenezPRestrepoS. 2011. First report of Fusarium oxysporum causing potato dry rot in Solanum tuberosum in Colombia. New Dis. Rep 24:14.
- GeiserDMJiménez-GascoMKangSMakalowskaIVeeraraghavanNWardTJZhangNKuldauGAO’DonnellK. 2004. FUSARIUM-ID 1.0: A DNA sequence database for identifying Fusarium. Europ J Plant Pathol 110:473–479, doi:10.1023/B:EJPP.0000032386.75915.a0
- GerlachWNirenbergHI. 1982. The genus Fusarium—a pictorial atlas. Mitt Biol Bundesanst Land- Forstw Berlin-Dahlem 209:1–406.
- GräfenhanTSchroersHJNirenbergHISeifertKA. 2011. An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Stud Mycol 68:79–113, doi:10.3114/sim.2011.68.04
- KornerupAWanscherJH. 1978. Methuen handbook of colour. London: Methuen.
- LeslieJFSummerellBA. 2006. The Fusarium laboratory manual. Blackwell Publishing, Ames, IA, USA. 388 p.
- LombardLvan der MerweNAGroenewaldJZCrousPW. 2015. Generic concepts in Nectriaceae. Stud Mycol 80:189–245, doi:10.1016/j.simyco.2014.12.002
- MatuoTSnyderWC. 1973. Use of morphology and mating populations in the identification of formae speciales in Fusarium solani. Phytopathology 63:562–565, doi:10.1094/Phyto-63-562
- McClureTT. 1951. Fusarium foot rot of sweet-potato sprouts. Phytopathology 41:72–77.
- MehlHLEpsteinL. 2007. Fusarium solani species complex isolates conspecific with Fusarium solani f. sp. cucurbitae race 2 from naturally infected human and plant tissue and environmental sources are equally virulent on plants, grow at 37°C and are interfertile. Environ Microbiol 9:2189–2199, doi:10.1111/j.1462-2920.2007.01333.x
- MoineLMLabbéCLouis-SeizeGSeifertKABélangerRR. 2014. Identification and detection of Fusarium striatum as a new record of pathogen to greenhouse tomato in Northeastern America. Plant Dis 98:292–298, doi:10.1094/PDIS-08-13-0844-RE
- NalimFASamuelsGJWijesunderaRLGeiserDM. 2011. New species from the Fusarium solani species complex derived from perithecia and soil in the Old World tropics. Mycologia 103:1302–1330, doi:10.3852/10-307
- NelsonPEToussounTAMarasasWFO. 1983. Fusarium species: An illustrated manual for identification. The Pennsylvania State University Press, University Park and London. 193 p.
- NirenbergHI. 1976. Studies on the morphologic and biologic differentiation in Fusarium section Liseola. Mitt Biol Bundesanst Land- Forstw Berlin-Dahlem 169:1–117.
- O’DonnellK. 2000. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 92:919–938, doi:10.2307/3761588
- O’DonnellKRooneyAPProctorRHBrownDWMcCormickSPWardTJFrandsenRJNLysøeERehnerSAAokiTRobertVARGCrousPWGroenewaldJZKangSGeiserDM. 2013. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet Biol 52:20–31, doi:10.1016/j.fgb.2012.12.004
- O’DonnellKSarverBABrandtMChangDCNoble-WangJParkBJSuttonDABenjaminLLindsleyMPadhyeAGeiserDMWardTJ. 2007. Phylogenetic diversity and microsphere array-based genotyping of human pathogenic Fusaria, including isolates from the multistate contact lens-associated U.S. keratitis outbreaks of 2005 and 2006. J Clin Microbiol 45:2235–2248, doi:10.1128/JCM.00533-07
- O’DonnellKSuttonDAFothergillAMcCarthyDRinaldiMGBrandtMEZhangNGeiserDM. 2008. Molecular phylogenetic diversity; multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J Clin Microbiol 46:2477–2490, doi:10.1128/JCM.02371-07
- O’DonnellKSuttonDARinaldiMGSarverBABalajeeSASchroersHJSummerbellRCRobertVACrousPWZhangNAokiTJungKParkJLeeYHKangSParkBGeiserDM. 2010. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J Clin Microbiol 48:3708–3718, doi:10.1128/JCM.00989-10
- PetersJCLeesAKCullenDWSullivanLStroudGPCunningtonAC. 2008. Characterization of Fusarium spp. responsible for causing dry rot of potato in Great Britain. Plant Pathol 57:262–271, doi:10.1111/j.1365-3059.2007.01777.x
- PetersRDMacLeodCSeifertKAMartinRAHaleLRGrauCRMacInnisS. 2008. Pathogenicity to potato tubers of Fusarium spp. isolated from potato, cereal and forage crops. Am J Potato Res 85:367–374, doi:10.1007/s12230-008-9037-z
- RonquistFTeslenkoMvan der MarkPAyresDLDarlingAHöhnaSLargetBLiuLSuchardMAHuelsenbeckJP. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542, doi:10.1093/sysbio/sys029
- SagarVSharmaSJeevalathaAChakrabartiSKSinghBP. 2011. First report of Fusarium sambucinum causing dry rot of potato in India. New Dis. Rep 24:5.
- SakuraiYMatuoT. 1961. Taxonomy of the causal fungus of trunk-blight of Xanthoxylum piperitum and heterothallism in this fungus. Ann Phytopathol Soc Jap 26:112–117, doi:10.3186/jjphytopath.26.112
- SchroersH-JGräfenhanTNirenbergHISeifertKA. 2011. A revision of Cyanonectria and Geejayessia gen. nov., and related species with Fusarium-like anamorphs. Stud Mycol 68:115–138, doi:10.3114/sim.2011.68.05
- ShortDPGO’DonnellKThraneUNielsenKFZhangNJubaJHGeiserDM. 2013. Phylogenetic relationships among members of the Fusarium solani species complex in human infections and the descriptions of F. keratoplasticum sp. nov. and F. petroliphilum stat. nov. Fungal Genet Biol 53:59–70, doi:10.1016/j.fgb.2013.01.004
- ShortDPGO’DonnellKZhangNJubaJHGeiserDM. 2011. Widespread occurrence of human pathogenic types of the fungus Fusarium detected in plumbing drains. J Clin Microbiol 49:4264–4272, doi:10.1128/JCM.05468-11
- SilvestroDMichalakI. 2012. RaxmlGUI: A graphical front-end for RAxML. Org Divers Evol 12:335–337, doi:10.1007/s13127-011-0056-0
- SnyderWCHansenHN. 1941. The species concept in Fusarium with reference to section Martiella. Ame J Bot 28:738–742, doi:10.2307/2436658
- StamatakisA. 2014. RAxML 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313, doi:10.1093/bioinformatics/btu033
- StefańczykESobkowiakSBrylińskaMŚliwkaJ. 2016. Diversity of Fusarium spp. associated with dry rot of potato tubers in Poland. Eur J Plant Pathol ( in press), doi:10.1007/s10658-016-0875-0
- SugiuraYBarrJRBarrDBBrockJWElieCMUenoYPattersonDGJrPotterMEReissE. 1999. Physiological characteristics and mycotoxins of human clinical isolates of Fusarium species. Mycol Res 103:1462–1468, doi:10.1017/S095375629900862X
- SummerbellRSchroersH-J. 2002. Analysis of phylogenetic relationship of Cylindrocarpon lichenicola and Acremonium falciforme to the Fusarium solani species complex and a review of similarities in the spectrum of opportunistic infections caused by these fungi. J Clin Microbiol 40:2866–2875, doi:10.1128/JCM.40.8.2866-2875.2002
- SwoffordDL. 2003. PAUP* 4. Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, Massachusetts.
- TaylorJWJacobsonDJKrokenSKasugaTGeiserDMHibbettDSFisherMC. 2000. Phylogenetic species recognition and species concepts in Fungi. Fungal Genet Biol 31:21–32, doi:10.1006/fgbi.2000.1228
- von MartiusCFP. 1842. Die Kartoffel Epidemie der letzten Jahre oder die Stockfäule und Räude der Kartoffel, geschildert und in ihren ursachlichen Verhältnissen erörtert. München: Verlag der königlich bayerischen Akademie der Wissenschaften. 70 p.
- WollenweberHWReinkingOA. 1935. Die Fusarien, ihre Beschreibung, Schadwirkung und Bekämpfung. Berlin: Paul Parey. 355 p.
- ZhangNO’DonnellKSuttonDANalimFASummerbellRCPadhyeAAGeiserDM. 2006. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J Clin Microbiol 44:2186–2190, doi:10.1128/JCM.00120-06
