Abstract
In this study, we analyzed the developmental expression of estrogen receptors (ESR1 and ESR2), prolactin receptors (PRLR) and casein genes (CSN1S1, CSN1S2, CSN2 and CSN3) in the ewe mammary glands from prepubertal stage to involution. Using Real-time PCR we showed that the activation of casein genes transcription was up regulated during lactation and significantly down regulated before lambing and at involution in comparison to the expression measured in the prepubertal group. The highest expression of ESR1 and ESR2 genes occurred in prepubertal group compared to adult group. The PRLR expression of the short and long forms was up regulated before lambing and down regulated during lactation and involution.
Thus, the mRNA expression data for ESRs and PRLR show clear regulatory changes suggesting involvement of these receptors in sheep mammary glands during development to involution. Casein genes transcription could be primed through PRLR signal, but other factors may be necessary for milk protein longterm expression during lactation.
Introduction
In dairy sheep, genetic selection has caused deep morphological changes in the udder and physiological changes in the whole body of the animal. The former is identified by the higher mammary cistern volume, the latter by neurohormonal changes. These changes allow the alveoli to have a longer life span and maintain a metabolic status that favours the switch of energy and nutrients to the mammary gland instead of body reserves (CitationPulina et al., 2007). The pattern of the lactation curve is influenced by the number of secretory cells in the mammary gland at each day in milking (DIM) and by the synthetic activity of each secretory cell. Growth and differentiation of the glandular epithelium during puberty and pregnancy are important determinants of the total area of secretory epithelium and consequently of milk yield (CitationPulina et al., 2009).
These physiological changes are orchestrated by systemic and local factors, which control synthetic and secretory mechanisms by transcriptional control of key mammary genes (CitationStefanon et al., 2002).
In dairy species, it is generally believed that there is normally little mammary growth during established lactation (CitationAkers 2002); however, in human breast the onset of secretory activity is accompanied by glandular-alveolar growth and expansion of acini (CitationBattersby and Anderson, 1988). Moreover, in many tissues it exists a dividing transit population of cells in which signs of proliferation, phenotypic differentiation and functional differentiation are displayed simultaneously (CitationPotten and Loeffler, 1990). In a recent paper, we reported for the first time the turnover of mammary cells and the interaction of their signals during the complete lactation cycle in sheep (CitationColitti and Farinacci, 2009); we concluded that mammary glands of dairy ewes seem to operate in a much more dynamic state than other lactating animals and this is particularly important in the construction of mechanistic models of lactation. In general, these models are based on the assumption that milk production at each time of lactation depends on the number of active cells and on the secretory activity (CitationDimauro et al., 2007). Therefore, mechanistic models of lactation could represent a useful tool to evaluate possible effect of selection for increasing lactation persistency in different breed and production scenarios. In a complementary paper (CitationPulina et al., 2009), based on the same experimental units, we concluded that the milk production around lactation peak (30 L) is sustained by the higher epithelium volume and higher milk secretion rate per secretory tissue unit.
The estrogen receptor-α (ESR1) is a critical transcription factor that regulates epithelial cell proliferation and ductal morphogenesis during postnatal mammary gland development (CitationFeng et al., 2007). Between the two isoforms α and β (ESR1 and ESR2, respectively), ESR1 is considered the primary receptor for mammary gland development and function; it induces proliferation of the mammary tissue, but the mechanism is not clear, since the proliferating mammary cells do not contain this receptor (CitationClarke et al., 1997). Moreover, ESR2 modulates ESR1 action in tissues where they are co-expressed (CitationHall and McDonnell, 1999).
The role of prolactin in milk synthesis is probably related to the fact that it inhibits mammary apoptosis by suppressing the actions of IGF binding protein (IGFBP-5), which antagonizes the effects of IGF-I on the survival of mammary epithelial cells (CitationTonner et al., 2000). Proliferation and differentiation of secretory mammary epithelium are also dependent on the presence of the prolactin receptor (CitationOrmandy et al., 1997) and the downstream Jak2-Stat5 pathway (CitationLiu et al., 1998). The prolactin receptor (PRLR) belongs to the superfamily of cytokine receptors (CitationKelly et al., 1991) and exists in different isoforms, generated by alternative splicing, that are identical in their extracellular ligand-binding domain, but differ in the length and sequence of their intracellular domain (CitationBole-Feysot et al., 1998). cDNAs encoding a long and a short form of PRLR have been isolated from different species (CitationBignon et al., 1997; CitationShirota et al., 1990) and are differentially expressed in different tissues, suggesting that they can activate distinct signalling pathways (CitationSchuler et al., 1997). The long form of PRLR actives Jak2, a cytoplasmic protein tyrosine kinase, which in turn can serve as docking sites for the SH2 domains in STAT5 (CitationGroner, 2002). Activated STAT5 binds to DNA sites in the nucleus known as GAS elements and modulates the activity of target genes, as the β-casein gene (CitationKazansky et al., 1995; CitationJohn et al., 1999). However, the lack of correspondence of STAT5a gene expression and β-casein gene expression suggests that STAT5 activation may facilitate the interaction of other factors binding within composite response elements identified recently in the milk protein gene promoters. Responsive elements are responsible for the stable expression of milk protein genes in terminally differentiated mammary epithelial cells (CitationKazansky et al., 1995). In lactating animals, STAT5a induces expression of milk protein genes, largely in response to prolactin (CitationNevalainen et al., 2002) together with laminin-1, which is a major basement membrane component required for milk protein expression (CitationStreuli et al., 1995; CitationXu et al., 2009).
Moreover, in a recent paper it has been demonstrated that in ruminants, the increased milking frequency enhances the expression of the long and short isoform of prolactin receptors and β casein on the mammary epithelial cells and reduces cell apoptosis by modulating hormone sensivity (CitationBernier-Donner et al., 2010).
Caseins comprise a group of four proteins (αs1, αs2, β, and κ) resulting from the expression of four structural genes (CSN1S1, CSN1S2, CSN2 and CSN3, respectively) (CitationBevilacqua et al., 2006). These proteins represent on average 82% of sheep milk Total Nitrogen (N × 6.38; CitationPulina and Nudda, 2004), but there is a large variability from one species to another (CitationMiranda et al., 2004).
Since a surprisingly very high proliferation index, measured by Ki-67 immunostaining, was observed during lactation in mammary glands of Sardinian sheep (CitationColitti and Farinacci 2009), the goal of this study was to investigate in the same mammary tissues the expression of CSN1S1, CSN1S2, CSN2 and CSN3, markers of functional differentiation. The expression patterns of ESR1 and ESR2, and PRLR long and short forms, which play an active role in morphogenesis, growths and functional differentiation, were also studied in mammary glands of sheep prior to lambing to involution.
Materials and methods
Animals
Tissue was collected from mammary glands of thirty Sardinian sheep that were slaughtered at different developmental stages: prepubertal (30±5 days, group P), 10 days before lambing (group LateP), 30, 60, 150 DIM (groups 30L, 60L, 150L, respectively) and 8 days after the end of lactation (group 8IN). At each sampling periods, five animals were randomly selected from a flock of grazing sheep and a clinical examination was conducted in vivo to ascertain animal health and to exclude mastitis. Sardinian sheep are a breed primarily used to produce milk; the typical breeding system implies one lambing per year, with the mating season starting in late spring for mature ewes and in early autumn for maiden ewes and with lactation starting in autumn and in late winter, respectively. Dry-off occurs simultaneously in mid summer for yearlings and mature ewes (lactation length 150 DIM and 240 DIM, approximately) when nutritional value of pastures collapses due to lack of rain in this season. In this study milk yield ranged from 1600 g/d to 900 g/d at 150 DIM. Ewes at 30 DIM were allowed to suckle their lamb; the other groups (60L, 150L) were mechanically milked twice daily and manually ten minutes before slaughtering, therefore just before tissue collection.
Samples of tissue were collected in TRIzol® (Invitrogen, Milano, Italy), frozen in liquid nitrogen and kept one week at −80°C till RNA extraction. The experiment was carried out in accordance with state and local laws and ethical regulations (CitationItalian Regulation, 1992).
RNA extraction and primer design
Total RNA was extracted from about 30 mg of mammary tissues using TRIzol® Plus RNA Purification System (Invitrogen, Milano, Italy), following the manufacturer’s instructions. The concentration of the extracted total RNA was quantified using a spectrophotometer (NanoDrop 1000 Spectrophotometer, Thermo-Scientific, Wilmington, DE, USA) and the assessment of the purity of RNA samples ranged between 1.8–1.9 The RNA integrity was evaluated through the observation of 18S and 28S ribosomal bands after electrophoresis on 1% agarose gel, in the presence of ethidium bromide. In sample analysis, the β-actin (U39357) expression was used as an internal control, confirming thorough integrity of the RNA.
A Primer3 Input software (CitationRozen and Skaletsky, 2000) was used to design the primer sequences encoding for: CSN1S1 (X03237), CSN1S2A (X03238), CSN2 (X79703), CSN3 (AY237637), PRLR long form (AF041257), PRLR short form (AF041977), ESR1 (AY033393), ESR2 (AF177936) and 18S rRNA (AY753190). Primers and product lengths for each gene are listed in according to the HUGO Gene Nomenclature Committee.
Table 1 Oligonucleotide primer sequences and reaction conditions for SybrGreen qRT-PCR.
Reverse transcription
Reverse transcriptions were performed with 2 µg of extracted total RNA by using Improm-II Reverse Transcriptase (Promega, Milano, Italy) as following described. Two micrograms of total RNA with 1 µL oligo(dT)18 primers (0.5 µg/µL MBI Fermentas, Italy) and nuclease free water to a final volume of 20 µL, were incubated at 70°C for 5 min in a PTC-100 thermocycler (MJ Research Inc., Waltham, MA, USA). Then, a mix was prepared with 4 µL of Improm-II Reverse Transcriptase buffer (5× Promega, Milano, Italy), 1.2 µL MgCl2 (50 mM), 1 µL of Improm-II Reverse Transcriptase and 1 µL of dNTP (10 mM) was added to the reaction and incubated at 37°C for 90 min and finally at 94°C for 5 min. The final concentration of cDNA was assumed as 100 ng/µL.
Standard curves analyses
For each gene, an aliquot of cDNA samples was pooled and standard curves with serial dilution of pool were used to optimize PCR conditions and to calculate the efficiency, fluorescence baseline and threshold. The expression of target genes was normalized using the 18S rRNA gene, which is known to be constitutively expressed (CitationRobinson et al., 2007) and was retro transcribed also with 1 µL random hexamers (100 µM, MBI Fermentas, Milano Italy).
Real time PCR quantitation
Realtime PCRs were performed in triplicate form using Platinum® SYBR® Green qPCR SuperMix-UDG (Invitrogen, Milano, Italy). For these reactions, a master mix with the following components was prepared to the indicated end concentration: 1 µL of cDNA, 9.5 µL water, 1 µL of each primer and 12.5 µL of 2× Platinum SYBR Green qPCR SuperMix-UDG for a total volume of 25 µL. cDNA concentrations and primers molarities were different for each gene and determined with standard curves analyses performed before Real time PCR reactions. cDNA and primers concentrations are showed in .
PCR amplifications were conducted applying 45 cycles (1 sec at 95°C, 30 sec at the specific annealing temperature, 30 sec at 72°C) in a 96-well spectrofluorometric thermal cycler (DNA Engine Opticon 2; MJ Research, Inc., Waltham, MA USA). The melting curve analysis of amplification products was performed at the end of each PCR reaction to confirm that a single PCR product was detected.
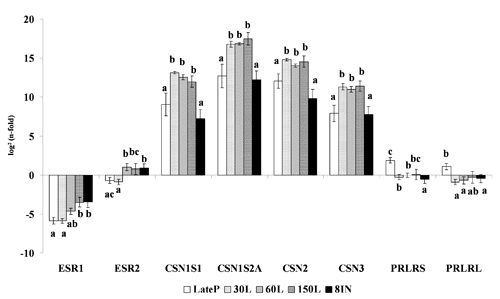
The expression level of a given target gene in each experimental group was analyzed by the 2−▵▵Ct method (CitationBustin, 2000; CitationPfaffl, 2001) where 2−▵▵Ct represents the difference of a given target gene between each group before lambing and during lactation (groups from LateP to 8IN) vs. the group P. More precisely, individual ▵▵Ct was calculated for each sample of group (LateP to 8IN) as ▵▵CT=▵CT (sample group) - mean ▵Ct (group P). The n- fold expression of a given target gene was calculated as log2(2−▵▵CT) ().
Statistical analysis
All the recorded variables were submitted to analysis of variance using the ANOVA model to assess significant differences between groups; Duncan’s least significant difference test was used to compare the means (CitationSPSS Inc., 1997).
Results
18S rRNA expression was quantified in all samples and resulted in constant expression levels. No significant differences between the groups could be shown in the investigated ovine mammary tissues. Expression of ERS1, ERS2, PRLRs and caseins mRNA were normalised according to the relative 18S rRNA expression of each sample.
The n-fold values, reported in as log2(n-fold), indicate the relative abundance of each target gene in comparison with the P group (prepubertal).
The relative expression of CSN1S1, CSN1S2A, CSN2 and CSN3 genes in the sheep mammary glands indicated the same significant pattern of difference (P<0.05) among groups. These genes were down-regulated at LateP and 8IN and up-regulated during lactation (30L, 60L, 150L).
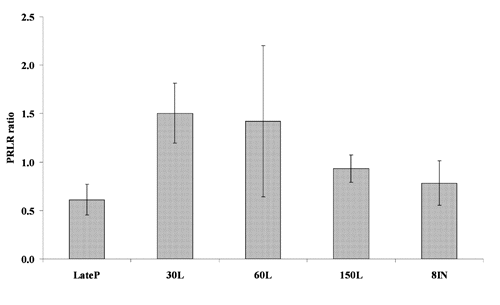
The relative transcription of ESR1 significantly increased (P<0.05) from LateP to 8IN; the same parameter for ESR2 significantly increased at 60L and remained constant and up regulated (even if not significantly for 150L) until 8IN (P<0.05). The PRLR expression of the short and long isoforms showed similar trend, being significantly up regulated at LateP and down regulated during lactation and involution. Statistical analysis of the ratio between the long and the short form of PRLR did not significantly differ among groups, but showed a trend in which the mRNA encoding the long form of the ovine PRLR predominated on the short one, in every group of sheep ().
Discussion
In a previous paper (CitationColitti and Farinacci, 2009), where cell turnover and gene activity in mammary gland of Sardinian sheep was evaluated, we suggested that sheep mammary glands seem to operate in a much more dynamic state than those of other domestic ruminants. In particular, to explain the high apoptosis to proliferation ratio we suggested that, as reported by CitationPotten and Loeffler (1990), there was a dividing transit population of cells in which signs of proliferation, phenotyopic differentiation and functional differentiation were displayed simultaneously. This was in agreement with the results reported by CitationSuzuki et al. (2000) that also found cells, in the breast tissue of pregnant women, positive to marker of proliferation, Ki-67 and to markers of mammary functional differentiation, β-casein and κ-casein. We evaluated markers of functional differentiation like CSN1S1, CSN1S2A, CSN2, CSN3 by Real time PCR analyses.
In this study, we showed that activation of casein genes transcription, relatively to the prepubertal group (P), is up regulated during lactation and significantly down regulated before lambing and at involution. This is concomitant to the enhancement, although not significantly, of the ratio between the long and short form of prolactin receptors (). This is in agreement with the data found by CitationCassy et al. (1998) in which they suggested that the short form of the ovine PRLR may have a dominant negative action in the activation of milk protein gene transcription. In fact, the authors reported that the activation of caseins gene transcription was concomitant with the enhancement of the ratio of the long to the short form of the ovine PRLR, which may play a key role in the shift between growth and differentiation of the mammary gland.
Compared to P group, the long form of PRLR is up regulated before lambing and this is in agreement with the trend of STAT5a expression that was lower during lactation and higher after the end of lactation (CitationColitti and Farinacci, 2009). As already reported, STAT5a expression resulted negatively related to that of lactalbumin, a major milk protein gene for ruminants, which significantly increased from lambing to lactation and it is also related to the expression of caseins. Therefore, in agreement with CitationKazansky et al. (1995) the activation of STAT5a, induced by PRLR, may prime milk gene expression, but other factors are necessary for milk protein long-term expression during lactation.
Our mRNA expression results demonstrated a high expression of ESR1 and ESR2 genes in prepubertal group. In fact, at the beginning of lactation period and during lactation (lactogenesis and galactopoiesis) the n-folds were significantly lower for ERS1 gene and also for ESR2 gene (). These is in agreement with CitationSchams et al. (2003), who found the presence of high ERS1 and ERS2 levels before the start of lobulo-alveolar development and ESR1 significantly lower expression during pregnancy and lactogenesis. This is due to distinct regulatory mechanisms that involved the receptors, being the ESR1 regulated at post-transcriptional level and ESR2 at transcriptional level (CitationChang et al., 2005). In fact, the receptors present opposite expressions in presence of estradiol (CitationSchams et al., 2003). This is in agreement with our results that showed a higher espression of ESR2 during lactation. Moreover, ESR2 is expressed, in mammary glands, not only in the luminal cells but also in myoepithelial and stromal cells, suggesting different roles for this gene within the glands (CitationSpeirs et al., 2002). In fact, the colocalization of ESR1 and ESR2 expressions with that of proliferation marker (Ki-67) could be remarkable to clarify the nature of cells in which they are expressed and the pathways by which hormones modulate proliferation.
These observations suggest a possible and important role of these receptors for the initiation of alveolar development, maybe in cooperation with proliferative growth factors.
Conclusions
This study, designed to investigate in mammary tissues the expression of markers of functional differentiation as αs1, αs2, κ, β caseins, the expression patterns of α and β receptors for estrogen and prolactin receptors long and short forms, showed that: i) activation of casein genes transcription, relatively to the P group, is upregulated during lactation and significantly downregulated before lambing and at involution; ii) a high expression of of α and β receptors for estrogen genes occurs in P group compared with adult group; iii) the prolactin receptor expression of the short and long isoform are upregulated at LateP in comparison with P group, and down-regulated during lactation and involution.
References
- AkersR.M. 2002 Mammary development, anatomy and physiology Iowa State Press (eds.) Lactation and the mammary gland Blackwell Publishing Co Ames, IA, USA 45 65
- BattersbyS. AndersonT.J. 1988 Proliferative and secretory activity in the pregnant and lactating human breast Virchows Arch. A 413 189 196
- Bernier-DodierP. DelbecchiL. WagnerG.F. TalbotB.G. LacasseP. 2010 Effect of milking frequency on lactation persistency and mammary gland remodeling in mid-lactation cows J. Dairy Sci 93 555 564
- BevilacquaC. HelblingJ.C. MirandaG. MartinP. 2006 Translational efficiency of casein transcripts in the mammary tissue of lactating ruminants Reprod. Nutr. Dev 5 567 578
- BignonC. BinardN. OrmandyC. SchulerL.A. KellyP.A. DjianeJ. 1997 Long and short forms of the ovine prolactin receptor: cDNA cloning and genomic analysis reveal that the two forms arise by different alternative splicing mechanisms in ruminants and in rodents J. Mol. Endocrinol 19 109 120
- Bole-FeysotC. GoffinV. EderyM. BinartN. KellyP.A. 1998 Prolactin (PRL) and its receptor: actions signal transduction pathways and phenotypes observed in PRL receptor knockout mice Endocr. Rev 19 225 268
- BustinS.A. 2000 Absolute quantification of m-RNA using real-time reverse transcription polymerase chain reaction assays J. Mol. Endocrinol 25 169 193
- CassyS. CharlierM. BelairL. GuillomotM. CharronG. BlochB. DjianeJ. 1998 Developmental expression and localization of the prolactin receptor (PRL-R) gene in ewe mammary gland during pregnancy and lactation: estimation of the ratio of the two forms of PRL-R messenger ribonucleic acid Biol. Reprod 58 1290 1296
- ChangG. LiY. OmotoY. WangY. BergT. NordM. VihkoP. WarnerM. PiaoY.S. GustafssonJ.A. 2005 Differential regulation of estrogen receptor (ER)α and ERβ in primate mammary gland J. Clin. Endocr. Metab 90 435 444
- ClarkeR.B. HowellA. PottenC.S. AndersonE. 1997 Dissociation between steroid receptor expression and cell proliferation in the human breast Cancer Res 57 4987 4991
- ColittiM. FarinacciM. 2009 Cell turnover and gene activities in sheep mammary glands prior to lambing to involution Tissue Cell 41 326 333
- DimauroC. Cappio-BorlinoA. MacciottaN.P.P. PulinaG. 2007 Use of a computer-aided design to develop a stress simulation model for lactating dairy sheep Livest. Sci 106 200 209
- FengY. MankaD. WagnerK-U. KhanS.A. 2007 Estrogen receptor-α expression in the mammary epithelium is required for ductal and alveolar morphogenesis in mice P. Natl. Acad. Sci. USA 104 14718 14723
- GronerB. 2002 Transcription factor regulation in mammary epithelial cells Domest. Anim. Endocrin 23 25 32
- HallJ. M. McDonnellD. P. 1999 The estrogen receptor beta-isoform (ERbeta) of the human estrogen receptor modulates ERalpha transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens Endocrinology 140 5566 5578
- Italian Regulation 1992 Application of the Council Directive (EEC) No. 86/609 regarding the protection of animals used for experimental and other scientific purposes. LD 116/1992 Official Journal No. 294 15 12 1992 5 24
- JohnS. VinkemeierU. SoldainiE. DarnellJ.E. LeonardW.J. 1999 The significance of tetramerization in promoter recruitment by Stat5 Mol. Cell. Biol 19 1910 1918
- KazanskyA.V. RaughtB. LindseyS.M. WangY.F. RosenJ.M. 1995 Regulation of mammary gland factor/Stat5a during mammary gland development Mol. Endocrinol 9 1598 1609
- KellyP.A. DjianeJ Postel-VinayM.C. EderyM. 1991 The prolactin/growth hormone receptor family Endocr. Rev 12 235 251
- LiuX. GallegoM.I. SmithG.H. RobinsonG.W. HennighausenL. 1998 Functional rescue of Stat5a-null mammary tissue through the activation of compensating signals including Stat5b Cell Growth Differ 9 795 803
- MirandaG. MahéM.F. LerouxC. MartinP. 2004 Proteomic tools to characterize the protein fraction of Equidae milk Proteomics 4 2496 2509
- NevalainenM.T. XieJ. BubendorfL. WagnerK.U. RuiH. 2002 Basal activation of transcription factor signal transducer and activator of transcription (Stat5) in nonpregnant mouse and human breast epithelium Mol. Endocrinol 16 1108 1124
- OrmandyC.J. CamusA. BarraJ. DamotteJ.D. LucasB. ButeauH. EderyM. BrousseN. BabinetC. BinartN. KellyP.A. 1997 Null mutation of the prolactin recetor gene producesmultiple reproductive defects in the mouse Genes Dev 11 167 178
- PfafflM.W. 2001 A new mathematical model for relative quantification in real-time RTPCR Nucleic Acids Res 29 e45
- PottenC.S. LoefflerM. 1990 Stem cells: attributes, cycles, spirals, pitfalls and uncertainties: lesson for and from the crypt Development 110 1001 1020
- PulinaG. ColittiM. FarinacciM. MazzetteA. CanuG. Castanares CastroN. NuddaA. 2009 The evolution of mammary glands at different stages in Sarda dairy ewes: preliminary results Ital. J. Anim. Sci 8(suppl. 2) 652 654
- PulinaG. NuddaA. 2004 Milk production PulinaG. Dairy sheep nutrition CAB International Cambridge, MA, USA 1 12
- PulinaG. NuddaA. MacciottaN.P.P. BattaconeG. RassuS.P.G. CannasA. 2007 Non-nutritional factors affecting lactation persistency in dairy ewes: a review Ital. J. Anim. Sci 6 115 141
- RobinsonT.L. SutherlandL.A. SutherlandJ. 2007 Validation of candidate bovine reference genes for use with real-time PCR Vet. Immunol. Immunopathol 115 160 165
- RozenS. SkaletskyH.J. 2000 Primer3 on the WWW for general users and for biologist programmers KrawetzS. MisenerS. Bioinformatics Methods and Protocols: Methods in Molecular Biology Humana Press Totowa, NJ, USA 365 386
- SchamsD. KohlenbergS. AmselgruberW. BerishaB. PfafflM.W. SinowatzF. 2003 Expression and localisation of oestrogen and progesterone receptors in the bovine mammary gland during development, function and involution J. Endocr 177 305 317
- SchulerL.A. NagelR.J. GaoJ. HorsemanN.D. KesslerM.A. 1997 Prolactin receptor heterogeneity in bovine fetal and maternal tissues Endocrinology 138 3187 3194
- ShirotaM. BanvilleD. AliS. JolicoeurC. BoutinJ.M. EderyM. DjianeJ. KellyP.A. 1990 Expression of two forms of prolactin receptor in rat ovary and liver Mol. Endocrinol 4 1136 1143
- SpeirsV. SklirisG.P. BurdallS.E. CarderP.J. 2002 Distinct expression patterns of ERα and ERβ in normal human mammary gland J. Clin. Pathol 55 371 374
- SPSS 1997 Advanced Statistics 7.5 SPSS Inc Chicago, IL, USA
- StefanonB. ColittiM. GabaiG. KnightC.H. WildeC.J. 2002 Mammary apoptosis and lactation persistency in dairy animals J. Dairy Res 69 37 52
- StreuliC.H. SchmidhauserC. BaileyN. YurchencoP. SkubitzA.P.N. RoskelleyC. BisselM.J. 1995 Laminin mediates tissue-specific gene expression in mammary epithelia J. Cell Biol 129 591 603
- SuzukiR. AthertonA.J. O’HareM.J. EntwistleA. LakhaniS.R. ClarkeC. 2000 Proliferation and differentiation in the human breast during pregnancy Differentiation 66 106 115
- TonnerE. AllanG.J. FlintD.J. 2000 Hormonal control of plasmin and tissue-type plasminogen activator activity in rat milk during involution of the mammary gland J. Endocrinol 167 265 273
- XuR. NelsonC.M. MuschlerJ.L. VeisehM. VonderhaarB.K. BissellM.J. 2009 Sustained activation of STAT5 is essential for chromatin remodeling and maintenance of mammary-specific function J. Cell Biol 184 57 66