Abstract
The aim of this study was to isolate, identify and select, with respect to acidification and proteolytic activities, the autochthonous mesophilic lactic acid bacteria (LAB) present in milk and Caciotta della Garfagnana, a cheese produced either with raw or thermised cow’s milk in small dairies and family plants of Garfagnana (Tuscany), to obtain LAB strains with attributes suitable to be employed as starter cultures in this type of cheese, particularly when thermised milk is used to control spoilage microflora. Samples of raw milk, curd and cheese were collected from three representative farmers of the production area and used to isolate autochthonous LAB. Phenotypic and genotypic (species-specific PCR assay) identification of isolated LAB was done. Twenty-eight strains of LAB isolated from milk, curd and cheese were screened for acidifying and proteolytic activities. LAB strains with the better attributes were used as mesophilic starter cultures in technological trials: experimental cheeses manufactured with the addition of autochthonous LAB and control cheeses were compared for LAB and pH evolution. Experimental cheeses presented a significant increase in the mesophilic lactic acid microflora up to 14 days of ripening and significantly lower pH values up to seven days of ripening. The use of wild selected mesophilic lactic acid bacteria, together with thermisation of milk, for the Caciotta della Garfagnana looks very promising and could help to both standardise the production and improve quality and traditional characteristics of this type of cheese.
Introduction
The general term Caciotta includes a wide variety of soft cheeses produced in various Italian regions, especially of central Italy. The term usually refers to small-/medium-sized cheeses (0.8–2 kg) of cylindrical shape with low height (4–8 cm) and diameter of 8–10 cm up to 16 cm, made with cow’s milk (tipo dolce), ewe’s milk (tipo saporito o laziale) or both, ripened from one to several weeks (CitationSalvadori del Prato, 1998).
Caciotta della Garfagnana is traditionally manufactured in the upper valley of the Serchio river, called Garfagnana. This region is situated in the north-west of Tuscany, one of the central Italian regions, in the Lucca province. The physical morphology of this area and the small size of the farms characterise Garfagnana’s livestock farming. This activity has an important role in development strategies, although its low productive efficiency, assuring enviroment preservation and biodiversity are maintained, which can be transferred to locally made, typical products. In fact, the quality of typical cheeses is closely associated with the territory of production and local traditions: interactions between pedoclimatic characteristics, autochthonous genetic variations and anthropic components create an environment so specific that it would be extremely difficult to reproduce it elsewhere. To safeguard these products means to preserve the uniqueness of their historical and cultural environment.
Caciotta della Garfagnana is manufactured in small dairies and family plants using cow’s raw or thermised milk produced in local, small-scale farms. It is a cylindrical-shaped cheese with flat faces (8–10 cm of diameter) and a straight or slightly convex rim (4 cm), of 800 g to 1 kg weight. The rind is thin, irregular and yellowish-white, while the inner part is whitish and characterised by small eye-like spots and a semi-soft texture. As not all dairies apply the same manufacturing procedure, a wide variety of cheeses can be obtained. Moreover, the hygienic quality of raw milk used for cheese production is often variable. This situation is usually related to inappropriate management of milking and causes defects of microbial origin, owing to proliferation of spoilage microrganisms during ripening (early gas-blowing). The early gas defect is characterised by an incomplete ripening of the cheese and heterogeneous plastic mass, and the occurring holes are irregular in shape; sometimes a multitude of small holes may arise, unfavourably affecting the aspect of the cheese when cut. In addition, the organoleptic characteristics of these cheeses are altered, with an unpleasant taste and sponge texture (CitationSalvadori del Prato, 1998; CitationMucchetti and Neviani, 2006). To reduce cheese defects, many farmers apply milk thermisation. This treatment, which is important for the reduction of spoilage bacteria, particularly coliforms, causes the loss of lactic acid species important for milk fermentation and cheese ripening, even if its mildness can allow survival of some mesophilic lactic acid bacteria (LAB) strains (CitationSalvadori del Prato, 1998). The selection of LAB, to be used to produce cheese with thermised milk, allows the restoration of the wild microflora to maintain the unique character and taste of the cheese. The optimisation of the wild microbial biodiversity plays an important role in characterisation and improvement of dairy products on an artisan and/or industrial scale. Moreover, the activity of starter is crucial for the control of coliforms by decreasing the pH and the amount of lactose in the curd.
In our study, the selection procedure of autochthonous mesophilic LAB present in both raw milk and thermised Caciotta della Garfagnana cheese is described. LAB strains with the better attributes have been used as starter in experimental cheese-making trials carried out with thermised milk.
Materials and methods
Traditional cheese-making and sampling
Cow’s milk and Caciotta della Garfagnana samples, at different ripening times, were collected from three farmers, representative of the production area (Garfagnana), who habitually apply milk thermisation. All dairies adopted the same local traditional manufacturing procedure, even if every dairy farm habitually uses its own cheese-making techniques, which may vary. Cow’s milk was thermised (58–68°C, 15 s), cooled to 37°C and coagulated with a liquid commercial calf rennet (30 mL/q), with a coagulation time of 15–20 min after the rennet addition. The curd was manually cut into hazelnut grains and was transferred into perforated moulds of 10 cm in diameter and 4 cm in depth, pressed to drain the whey and then transferred to an artisanal warm room at 23–26°C for 2–4 h (stufatura). The cheeses were removed, dry salted and ripened for 60 days at about 10°C and 90% relative humidity. The milk samples were collected directly from the vat, before thermisation, and the curd samples were collected after the moulding. Sampling of cheeses was carried out at 2, 7, 14, 21, 28, 35, 45 and 60 days of ripening using the standard methods (CitationIDF, 1995). The samples were chilled to 4°C and analysed within 24 h.
Isolation, phenotypic and genotypic identification of lactic acid bacteria
Milk (10 mL), curd and cheese (10 g) samples were diluted and homogenised in 90 mL of sterile solutions (peptone saline solution for milk samples and 2% sodium citrate solution for curd and cheeses) in a Stomacher apparatus (400 Circulator, PBI International, Milan, Italy). The homogenates were serially diluted in sterile solutions and plated on the specific media required for the different microbial groups: M17 agar (Oxoid, Basingstoke, UK) incubated at 30°C for 24 h and MRS agar (Oxoid) incubated anaerobically (Gas Generating Kit BR0038, Oxoid) at 37°C for 72 h were used for lactococci and mesophilic lactobacilli, respectively. Plates with 30 to 300 colonies were selected for LAB isolation. For each sampling point, one to five colonies were randomly picked from the countable M17 and MRS agar plates and streaked out three times on the same media used for the isolation to check for purity.
Phenotypic strains identification was carried out according to the CitationBergey’s Manual of Systematic Bacteriology (1986); carbohydrate fermentation patterns were determined using the API 50 CHL test kit (API System bioMérieux, Marcy l’Etoile, France). Biochemical identification was validated by species-specific PCR using primers described by CitationCorroler et al. (1998) for Lactococcus lactis subsp. lactis/cremoris and CitationTilsala-Timisjiarvi and Alatossava (1997) for Lactobacillus rhamnosus and Lactobacillus paracasei. For the DNA extraction, bacterial colonies were suspended in sterile water and harvested by centrifuging at 7500 rpm for 10 min. DNA was extracted by the DNeasy tissue kit (Qiagen GmbH, Hilden, Germany) according to the supplier’s procedures. Amplifications were performed in a thermocycler (GeneAmp PCR System 2700, Perkin-Elmer, Norwalk, CT, USA) in a final volume of 25 µL. Aliquots (5 µL) of PCR products were routinely checked on 1.5% agarose gels.
Strain characterisation
After genotypic identification, single strains were evaluated for acidifying and proteolytic activities. All the analyses were carried out in triplicate.
Acidifying activity
The selected strains were first inoculated in M17 and MRS broths (Oxoid) and then activated twice in sterile 10% reconstituted skimmed milk (Skim Milk Powder, Oxoid). Finally, sterile reconstituted skimmed milk (100 mL) was inoculated with 1% of an 18–20 h activated culture, and pH was measured by using a GLP 21 pH meter (Crison Instruments S.A., Barcelona, Spain) during incubation at 30°C after 6, 18 and 24 h. The values were expressed as a pH decrease (ΔpH), calculated as a difference between the pH value of skimmed milk not inoculated (control) and the values registered at each determination.
Proteolytic activity
Proteolytic activity of the different strains was detected by the o-phthaldialdehyde (OPA) spectrophotometric method (CitationChurch et al., 1983). The increase in optical density at 340 nm (OD340) relative to the control was determined using a spectrophotometer (Ultraspec 2000, Pharmacia Biotech, Uppsala, Sweden). The OPA solution contained: 2 mL of 20% (w/v) sodium dodecyl sulphate (SDS), 25 mL of 100 mM sodium tetraborate (Sigma Chemical Co., St Louis, MO, USA), 40 mg of OPA (Sigma Chemical Co.), previously dissolved in 1 mL of methanol, 100 µL of 2-mercaptoethanol (Merck, Darmstadt, Germany) and distilled water up to a 50 mL final volume.
LAB cultures were inoculated in 10% reconstituted skimmed milk and incubated at 30°C for 18–20 h. An aliquot of 2.5 mL from each tested strain culture was mixed with 5 mL of 0.72 N trichloroacetic acid, then the mixture was filtered using Watman no.1 paper. The filtrate (100 µL) was added to 2 mL of OPA reagent and, after 2 min at room temperature (20°C), absorbance of the solution was measured by a spectrophotometer at 340 nm. The proteolytic activity of these bacterial cultures was expressed as µg glycine released/mL using a standard curve of glycine (BDH Chemicals Ltd., Poole, UK).
Ability to grow in association
After the strain characterisation, three selected strains of lactococci and four strains of lactobacilli, chosen on the basis of their acidifying and proteolytic activities, were examined for their ability to grow on sterilised whole cow’s milk, both as single strains and in association (every lactococcus with every lactobacillus strain, for a total of 12 combinations). The selected strains were first inoculated in M17 and MRS broths (Oxoid), respectively, and then activated twice in sterilised whole cow’s milk. Finally, sterilised whole cow’s milk was inoculated with 1% of a 18–20 h activated culture of every single strain to test its ability to grow alone. At the same time, every selected couple of strains was tested for the ability to grow in association. After an incubation at 30°C of 18–20 h, counts of lactococci and lactobacilli (single strains and associations) were performed, after preparing serial dilutions on peptone saline solution, on M17 agar (Oxoid) incubated at 30°C for 24 h for lactococci, and on MRS agar (Oxoid) incubated anaerobically at 37°C for 72 h for lactobacilli. Results were expressed as the mean of the values obtained from triplicate trials of each single strain and association.
Preparation of experimental starter
Autochthonous starter was prepared in the laboratory by mixing three strains of LAB (one strain of lactococcus and two strains of lactobacilli), selected on the basis of the results of the strain characterisation and test of ability to grow in association. This chosen association was preliminarily tested, as indicated previously, also for simultaneous growth. For starter preparation, the three strains, opportunely activated, were separately inoculated (1%) in sterile reconstituted skimmed milk, incubated at 30°C for 24 h and then used to finally inoculate the same medium, in the final ratio of 3:1:1, with the same incubation. Before the cheese-making trial, the acidification profile of experimental starter was evaluated by measuring the pH decrease versus the control (sterile reconstituted skimmed milk) after 6, 18 and 24 h of incubation.
Cheese-making trials
Cheese-making trials were performed by three dairies by preparing experimental cheeses with milk inoculated with autochthonous lactic acid starter and starter-free control cheeses. For both control and experimental batches of each farm, the same lot of milk was employed. Milk was thermised at 58–68°C for 15 s. In the experimental batches, thermised milk was inoculated with selected autochthonous starter prepared in the laboratory. Starter was added at a level of 1% of the total milk, obtaining in milk a pH decrease after starter inoculation of not higher than 0.20. Manufacturing trials were carried out in accordance with the traditional procedure. For each vat, nine cheeses were produced. The evolution of pH and lactic acid microflora was monitored in both types of cheese (curd, 0, 2, 7, 14, 21, 28, 35, 45, 60 d of ripening). The curd samples were collected before the moulding and the samples of cheese at time 0 were collected after the stufatura. The pH was measured on a single cheese at the time of every analysis using the Crison GLP21 pH meter previously cited, provided with a Hanna FC 200B penetration electrode (Hanna Instruments, Padova, Italy). The LAB microrganisms were enumerated on a single cheese for every analysis time, on M17 and MRS agar (Oxoid) for lactococci and lactobacilli, respectively, as previously described.
Statistical analysis
Results from microbial counts were previously converted into log cfu/mL or g. For the test of the ability of the LAB strains to grow in association, the Student t-test was performed to determine if significant differences between counts from every single strain and every association occurred. One-way analysis of variance (ANOVA) was used for the test of starter acidification curve to estimate possible differences between mean values of ΔpH derived from the association of strains selected for starter preparation and the single strains at 6, 18 and 24 h.
In the same way, parameters from microbiological analyses (lactococci and lactobacilli counts at 10 analysis times) and pH values (at the same analysis times) were evaluated by one-way ANOVA to estimate possible differences between means (experimental vs control cheeses). Single pairs of means were evaluated by the Tukey test. Differences were considered significant at P<0.05.
Results and discussion
Isolation, phenotypic and genotypic identification of lactic acid bacteria
Seventy-three bacterial strains, presumptively identified as LAB (48 lactococci and 25 lactobacilli) by physiological tests (morphology, Gram staining, catalase reaction and growth with 2% and 4% NaCl for lactococci; morphology, Gram staining, catalase reaction, growth at 15°C for lactobacilli) were isolated from raw milk, curd and Caciotta della Garfagnana cheese. The genera Lactococcus and Lactobacillus were identified with the abbreviations Lc and Lb, respectively. Fermentation profiles of carbohydrates indicated that 34 isolates (20 lactococci and 14 lactobacilli) were identified with a good degree of reliability (identification %, ID>85%). These strains were identified as Lactococcus lactis subsp. lactis (20 strains), Lactobacillus paracasei (12 strains) and Lactobacillus rhamnosus (two strains).
Genotypic identification by species-specific PCR confirmed the results of biochemical identification for 15 Lactococcus lactis subsp. lactis, 12 Lactobacillus paracasei and one Lactobacillus rhamnosus, listed in , together with their source of isolation. These LAB species have often been isolated from other artisanal raw cow’s milk cheeses (CitationWouters et al., 2002; CitationPoznansky et al., 2004; CitationDi Cagno et al., 2007; CitationTerzic-Vidojevic et al., 2007). In this type of dairy products, non-starter lactic acid bacteria (NSLAB), such as Lactobacillus paracasei and Lactobacillus rhamnosus, support the cheese-making process and play a relevant role, increasing the flavour development of cheese, as previously described (CitationCorroler et al., 1998; CitationGrappin and Beuvier, 1998).
Table 1 Lactic acid bacteria strains from raw milk and Caciotta della Garfagnana cheese submitted to strain characterisation.
Strain characterisation
The results of acidifying and proteolytic activities of the 28 isolates of LAB, genotypically confirmed and ascribed to species Lactococcus lactis subsp. lactis, Lactobacillus paracasei and Lactobacillus rhamnosus, generally recognised as safe and potentially useful as starters, are reported in .
Table 2 Strain characterisation of lactic acid bacteria isolated from raw milk and Caciotta della Garfagnana cheese.
As concerns the acidifying activity, after 6 h of incubation in milk (ΔpH6), none of the examined strains was identified as a fast acid producer because they did not decrease the pH below 5.0 in 6 h (CitationHuggins and Sandine, 1984; CitationDurlu-Ozkaya et al., 2001); at the same time, three strains, one Lactococcus lactis subsp. lactis (Lc45) and two Lactobacillus paracasei (Lb27 and Lb101), showed good acidifying activity (ΔpH>1). At 18 h (ΔpH18), three strains (Lc45, Lb76 and Lb101) were identified as high acid producers (ΔpH>2). Two of these LAB strains (Lc45 and Lb101) showed a high acidifying activity after 24 h as well.
In general, lactobacilli strains showed lower proteolytic ability compared with Lactococcus lactis, as previously reported (CitationBottazzi, 1993; CitationDurlu-Ozkaya et al., 2001; CitationDagdemir and Ozdemir, 2008). Proteolytic activity of lactobacilli strains varied from 23.49 to 69.78 µg glycine/mL; the highest proteolytic activities (45.76 and 69.78 µg glycine/mL) were shown by Lb101 and Lb91. Proteolytic activity of lactococci strains ranged between 89.83 and 230.52 µg glycine/mL; the highest values of proteolytic activities (>200 µg glycine/mL) were found in three strains (Lc45, Lc72, Lc104).
Ability to grow in association
The associations Lc45/Lb91, Lc45/Lb101 and Lc104/Lb91 gave the best results in the test of ability to grow in association (). No significant decrease resulted from the counts on selective media of their pure cultures and associations.
Table 3 Ability to grow in association of selected lactic acid bacteria isolated from Caciotta della Garfagnana cheese.
Preparation of experimental starter
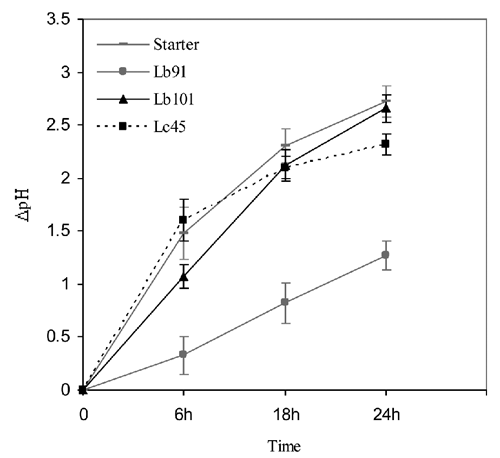
Finally, Lc45, Lb91 an Lb101 were chosen for the preparation of starter. Lc45 showed good acidifying ability, with a ΔpH24 value of 2.34, a suitable proteolytic activity (230.52 µg glycine/mL) and good capacity to grow in association with Lb91 and Lb101. Analogously, the two strains of lactobacilli were chosen because they presented the best performances within the isolates of the same species. The three strains together revealed good growth, with no significant difference compared with the growth values of the single strains (Lc45, 8.68 log cfu/mL; Lb91, 7.60 log cfu/mL; Lb101, 7.78 log cfu/mL). Autochthonous starter was prepared by mixing the three chosen strains of selected LAB, as previously described. The acidification curve () of this mixed culture showed that this association presented good acidifying activity, with significantly lower pH values than those shown by the single lactobacilli strains (with the only exception of Lb91 at 24 h, with no significant difference). In comparison with the Lc45 strain, selected starter gave better results at 18 and 24 h, but not at 6 h, with significant differences at each analysis time.
Cheese-making trials
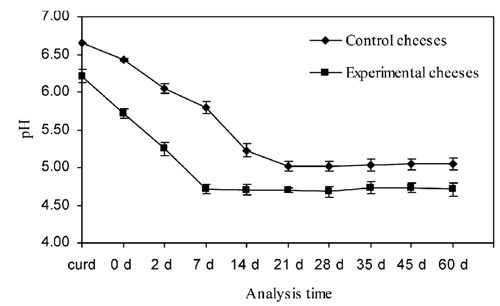
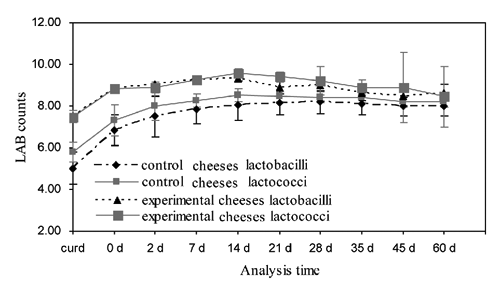
The evolution of pH and lactic acid microflora was monitored both in Caciotta della Garfagnana cheeses obtained using experimental starter and in control cheeses. The evolution of pH of both control and experimental cheeses is shown in . After stufatura (0 days), pH values were below 5.6 in experimental cheeses, while control cheeses at the same time presented a higher value of pH (6.4). Significant differences between pH value means (experimental vs control) were found in curd and cheese at 0, 2 and 7 days. The addition of the autochthonous selected strains resulted in an increase in the mesophilic lactic acid microflora in experimental cheeses compared with the controls, with significant differences in curd and cheese at 0, 2, 7 and 14 days for both lactobacilli and lactococci, while at 21 days differences were significant only for lactococci. As shown in , both lactococci and lactobacilli quickly developed in experimental cheeses, reaching levels higher than 8 log cfu/g beginning from 0 days. Both lactic acid categories reached the highest levels after 14 days of ripening (9.56 and 9.33 log cfu/g, respectively, for lactococci and lactobacilli) and remained at levels higher than 8 log cfu/g up to the end of the ripening period, even with loads not significantly higher than in control cheeses.
Conclusions
Caciotta della Garfagnana is a cow’s milk cheese traditionally manufactured in small dairy plants of this Tuscany area. The qualitative variability of the raw milk and the differences in manufacturing procedures do not allow the standardisation of this cheese, which often has defects of microbiological origin. To reduce these problems, the use of autochthonous cultures of LAB together with a mild heat treatment of milk were verified.
The choice not to use starter exclusively composed of lactococci, well known as strong acidifiers, but instead an important amount of mesophilic lactobacilli was determined by the fact that it was fundamental to preserve the original characteristics of the cheese, without heavily interfering with the natural balance between lactococci and lactobacilli. The starter appeared to be able to determine an increase of lactic acid microflora and a lowering of the pH values of the cheese, both significant in the curd and in the initial phase of ripening. Moreover, the organoleptic evaluation of the experimental and control cheeses after 21 days of ripening, carried out by expert staff of the cheese factories, detected the best characteristics in cheeses made with autochthonous cultures. Particularly, texture, flavour and taste of the experimental cheese were described as agreeable and typical.
In conclusion, the use of wild mesophilic lactic acid bacteria as starter or adjunct cultures for the Caciotta della Garfagnana looks very promising and could help to produce a standardised and more stable product, preserving the traditional characteristics of this type of cheese.
References
- Bergey's Manual of Systematic Bacteriology 1986 Vol. 2 Williams & Wilkins Baltimore, MD, USA
- BottazziV. 1993 Microbiologia lattiero-casearia Edagricole, Bologna Italy
- ChurchF.C. SwaisgoodH.E. PorterD.H. CatignaniG.L. 1983 Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins J. Dairy Sci 66 1219 1227
- CorrolerD. ManginI. DesmasuresN. GueguenM. 1998 An ecological study of lactococci isolated from raw milk in the Camembert cheese registered designation of origin area Appl. Environ. Microbiol 64 4729 4735
- DagdemirE. OzdemirS. 2008 Technological characterization of the natural lactic acid bacteria of artisanal Turkish white pickled cheese Int. J. Dairy Technol 61 133 140
- Di CagnoR. BuchinS. De CandiaS. De AngelisM. FoxP.F. GobbettiM. 2007 Characterization of Italian cheeses ripened under non-conventional conditions J. Dairy Sci 90 2689 2704
- Durlu-OzkayaF. XanthopoulosV. TunailN. Litopoulou-TzenatakiE. 2001 Technologically important properties of lactic acid bacteria isolates from Beyaz cheese made from raw ewes milk J. Appl. Microbiol 91 861 870
- GrappinR. BeuvierE. 1998 Possible implications of milk pasteurization on the manufacture and sensory quality of ripened cheese Int. Dairy J 7 751 761
- HugginsA.R. SandineW.E. 1984 Differentiation of fast and slow milk-coagulating isolates in strains of lactic streptococci J. Dairy Sci 67 1674 1679
- IDF 1995 Milk and milk products. Guidance on methods of sampling. Standard 50C:1995 International Dairy Federation ed Brussels, Belgium
- MucchettiG. NevianiE. 2006 Microbiologia e tecnologia lattiero-casearia. Qualità e sicurezza Tecniche Nuove Milano, Italy
- PoznanskiE. CavazzaA. CappaF. CocconcelliP.S. 2004 Indigenous raw milk microbiota influences the bacterial development in traditional cheese from an alpine natural park Int. J. Food Microbiol 92 141 151
- Salvadori del PratoO. 1998 Trattato di tecnologia casearia Edagricole, Bologna Italy
- Terzic-VidojevicA. VukasinovicM. VeljovicK. OstojicM. TopisirovicL. 2007 Characterization of microflora in homemade semi-hard white Zlatar cheese Int. J. Food Microbiol 114 36 42
- Tilsala-TimisjiarviA. AlatossavaT. 1997 Development of oligonucleotide primers from the 16S–23S rRNA intergenic sequences for identifying different dairy and probiotic lactic acid bacteria by PCR Int. J. Food Microbiol 35 49 56
- WoutersJ.T.M. AyadE.H.E. HugenholtzJ. SmitG. 2002 Microbes from raw milk for fermented dairy products Int. Dairy J 12 91 109