Abstract
The biodiversity safeguard is an important goal of poultry production in every developed country. Nowadays, the high chicken meat demand from the world market has been leading to a large spread of strongly producing commercial chicken lines. The creation of these standard types is causing a progressive loss of genetic variability. Ancona and Livorno are two Italian autochthonous chicken breeds which represent a great resource in terms of specific genetic richness. Aim of this study is to investigate the genetic diversity of these breeds as potential valuable genetic variability source. In fact, in spite of their endangered status, these chicken breeds are very appreciated for their ability to adapt themselves to extensive organic rearing systems. Blood samples from 131 individuals were collected and genotyped through a thirty microsatellitesbased analysis. All the observed descriptive statistical indexes suggested a heterozygosity deficiency and an inbreeding level (mean observed heterozygosity = 0.46, mean expected heterozygosity = 0.53, Fis in Ancona and Livorno = 0.251 and 0.086). The tree from inter-individual DAS distance using Neighbour-Joining algorithm and the FCA analysis showed a higher internal variability in Livorno than in Ancona. STRUCTURE analysis showed the genetic uniqueness of the breeds and the presence of sub-groups in Ancona originating from a possible genetic isolation. This research could be a suitable starting point to set up improved selection schemes and a potential preliminary genotypic test for all the cocks to be used in the selection.
Introduction
The poultry biodiversity safeguard is a strong objective in every developed country (CitationZanetti et al., 2007). The breed genetic variability gives the chance to select the individuals more able to be adapted to climatic changes, diseases and market variations. Because of the several different environments, up to decades ago Italy showed a considerable biodiversity in livestock breeds and populations. Within the last one-hundred years, the number of the endangered autochthonous breeds is dramatically increased (CitationZanon and Sabbioni, 2001), leading to an irreversible loss of genetic resources. The reasons of this negative trend are mainly the use of a few breeds selected to maximise the yields and the creation of specialised crossbreeds for the several productions. As a consequence of this loss of genetic diversity, many chicken local breeds reared in Italy until some decades ago are now disappeared (CitationGandini and Villa, 2003). The autochthonous extant breeds, which have been excluded from intensive rearing systems for a long time, represent an important source of variability. Their disappearance might lead to the loss of a potentially useful genetic patrimony. Ancona and Livorno (Leghorn Italian type), are two of these autochthonous chicken breeds (CitationFAO, 2010). The Ancona produces white or sometimes tinted eggs and is also considered an excellent layer because of its good all-year-round egg laying capacity. The Livorno is worldwide spread with different livery colors; this breed is an excellent white egg layer. The mean production can reach two-hundred and eighty eggs per year; the feed-to-egg conversion rate is excellent.
The production systems standardisation takes advantage of commercial strains which have been selected for improved performance and intensive rearing system; such cosmopolitan types are affected by a progressive reduction of genetic variability, which on the other hand is still present in the local traditional breeds (CitationSpalona et al., 2007), particularly suitable for extensive rearing systems.
Microsatellites markers are one of the most common and powerful tool to investigate genetic variability. Such molecular markers have been widely used in several studies regarding genetic diversity of domestic animals such as pig (CitationVicente et al., 2008), sheep (CitationLasagna et al., 2011), cattle (CitationLi et al., 2009), goat (CitationMahmoudi et al., 2009), horse (CitationGiacomoni et al., 2008) and chicken (CitationHillel et al., 2003).
Aim of this study is to investigate the genetic diversity of the autochthonous Ancona and Livorno breeds. In fact, these local breeds are under threat of extinction, as demonstrated by their drastic decline in number and their low consistency (CitationMugnai et al., 2009). In spite of their endangered status, these chicken breeds are very appreciated for their ability to adapt themselves to extensive organic rearing systems. Besides that, they were proposed as egg layers models for an en plain air rearing system (CitationCastellini et al., 2006; CitationMugnai et al., 2009; CitationDal Bosco et al., 2011). The molecular results on these breeds will be useful to set up improved selection schemes and to conserve strategies to avoid inbreeding.
Materials and methods
Animal sampling and microsatellite analysis
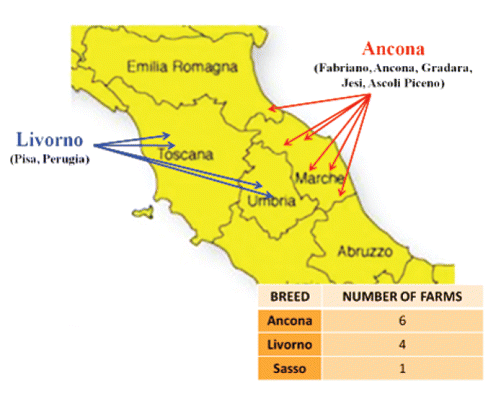
Blood samples were randomly collected from different animals belonging to Ancona (50), White Livorno (51) and Sasso (30) breeds. Animals from the French breed Sasso were included to have an out-group. These animals were chosen, as far as we were able to manage, in different farms in order to avoid closely related individuals and to have a representative sample of the breeds. shows geographical areas and number of farms which the individuals belonging to different breeds were sampled from. The most important area where Ancona is reared includes the Italian regions Marche and part of Emilia-Romagna. Genomic DNA was extracted from blood using the GenElute Blood Genomic DNA kit (Sigma Aldrich, St. Louis, MO, USA). Thirty loci microsatellites () were chosen on the basis of their position in the chicken genome. Twenty-nine of them had already been used in the AVIANDIV project (CitationAviandiv, 2011) and the microsatellite marker LEI0192 (CitationGroenen et al., 2000) was also added. The markers were subjected to a standard multiplex PCR amplification using a Biometra TGradient 96 at the following conditions: initial denaturation step of 5 min at 95°C, 35 cycles of 30 sec at 90°C, 45 sec at the annealing T° of each multiplex PCR, 30 sec at 72°C and a final extension of 15 min at 72°C. The multiplex PCR products were pooled in order to analyze many microsatellites in each electrophoresis. Analyses of fragments were performed using an automated DNA sequencer (ABI PRISM 3130xl, Applied Biosystems, Foster City, CA, USA) and a computer software (GeneMapper version 4.0, Applied Biosystems). Allele calling was adjusted to AVIANDIV project nomenclature (CitationAviandiv, 2011) including nine standard DNA reference samples.
Table 1 Microsatellite markers, chromosomes involved, alleles detected, size range and mean polymorphism information content per locus.
Statistical analysis
The 30 microsatellites PIC values calculated according to CitationBotstein et al. (1980) and observed and expected heterozygosity in the analyzed breeds were estimated by the EXCEL MICROSATELLITE TOOLKIT 3.1.1 (CitationPark, 2001). The number of alleles observed in each locus and mean number of alleles per breed were counted using POPGENE 3.2 software package (CitationYeh et al., 1999). The number of private alleles was calculated through direct count on allelic frequencies calculated by the software CONVERT (CitationGlaubitz, 2004). The Hardy-Weinberg equilibrium was tested by the software GENEPOP 4.0 (CitationRaymond and Rousset, 1995). A Markov Chain Monte Carlo method (20 batches, 5000 iterations per batch, and a dememorisation number of 10,000) was applied to perform exact probability tests, according to the algorithm described by CitationGuo and Thompson (1992). To assess the population genetic structure of the chicken breeds, Wright’s F-statistic was estimated. Fixation indices per locus (Fis, Fit and Fst) were calculated according to CitationWeir and Cockerham (1984) using the software GENETIX 4.05 (CitationBelkhir et al., 1996–Citation2004), which was also employed to obtain the Fis per population calculated with 1000 bootstraps. The significance of the fixation indices was tested using the software ARLEQUIN 3.11 (CitationSchneider et al., 1997), according to the analysis of molecular variance (AMOVA). The DAS genetic distance (CitationChakraborty and Jin, 1993) among the individuals was calculated using the software POPULATIONS 1.2.28 (CitationLangella, 2002). The Neighbour-Joining methodology was applied and a tree was built from the inter-individual distances by using the MEGA 4 package (CitationTamura et al., 2007). Factorial correspondence analysis (FCA) (CitationBenzécri, 1982), assessed by the employment of GENETIX 4.05, was used in order to investigate further the differentiation of the individuals within each population. STRUCTURE version 2.2 (CitationPritchard et al., 2000) was employed to confirm the genetic pattern of each individual belonging to the different breeds and to reveal possible clustering substructures. The Bayesian assignment of individuals to populations considered an ancestry model with admixture and correlated allele frequencies. Ten independent runs with 1,000,000 MCMC (Markov Chain Monte Carlo) iterations and a burn-in of 300,000 were carried out for 2≤K≤6 (K, number of clusters) to estimate the most likely number of clusters present in the data set. This numerical value was then established by calculating ΔK, as in CitationEvanno et al. (2005). The clustering pattern was visualised using the software DISTRUCT 1.1 (CitationRosenberg, 2004).
Results and discussion
In spite of the presence of some loci microsatellites showing a low level of polymorphism, the used panel turned out to be good and reliable for genetic diversity analysis. CitationHillel et al. (2003) got comparable results in a study involving more chicken populations. The total number of alleles found in the thirty microsatellite markers was 177. In Spanish chicken breeds, CitationDávila et al. (2009) detected a lower number of total alleles across all the population. LEI0234 showed the highest number of alleles observed in each locus (14), whereas MCW0248 and MCW0103 the lowest (2). With regard to PIC per locus, about half markers showed slightly high values (>0.50), while the others revealed lower values (<0.50) (). These results were not much different from those which CitationTadano et al. (2007) pointed out in a study involving Japanese chicken breeds. However, the informativeness of this microsatellites panel was lower if compared with the results obtained by CitationQu et al. (2006) and CitationBeigi-Nassiri et al. (2007). The mean number of alleles per breed () ranged from 3.50 for Ancona to 4.03 for Sasso. CitationRosenberg et al. (2001) found higher values in a study which took twenty European chicken breeds into account. The same findings arose from the analysis of the genetic diversity of Chinese indigenous chicken breeds (CitationQu et al., 2006), which were characterised by a more substantial number of alleles. An explanation of the lower number of alleles found in Ancona and Livorno could be due to the fact that the genetic variability parameters are generally lacking in small autochthonous chicken breeds, compared with larger and more differentiated populations. The Ancona breed showed 17 private alleles whereas Livorno 26 (). On the other hand Sasso was characterised by the highest number of breed-specific alleles (32) and this is consistent with his cosmopolitan status and with the fact that this breed was genetically influenced by other breeds not included in this work. The mean values of observed heterozygosity (0.49) and expected heterozygosity (0.52) in the total analysed population (data not showed) are not very high, suggesting a low genetic variability. In more details, Sasso displayed the highest value of observed heterozygosity (0.68) while Ancona the lowest (0.35) (). The mean expected heterozygosity ranged from a maximum of 0.60 in Sasso to a minimum of 0.47 in Ancona. With regard to Ancona breed, the numerical deviation of the observed heterozygosity compared to the expected heterozygosity is consistent with the values found by CitationDalvit et al. (2009). In their analyses, CitationQu et al. (2006) obtained higher values, probably due to the presence of more populations in the study. However, the results found in this work are comparable with those observed by CitationDalvit et al. (2009) in other two Italian autochthonous chicken breeds (Robusta Maculata and Ermellinata di Rovigo). It might therefore be logic speculating the presence of a general low level of genetic variability within the Italian autochthonous chicken breeds. The Fis calculated in each breed () were significantly different from zero (P<0.05) in Ancona (0.251) and Livorno (0.086), indicating heterozygosity deficiency in these breeds. The positive and significantly different from zero Fis values might arise from the presence of inbreeding or the presence of sub-populations within the breeds. It is reasonable to speculate that both the hypotheses are possible for the studied breeds, especially for Ancona. Ancona is a small breed and exchange of genetic material among breeders rearing it is not very common. Sasso showed a negative Fis value (−0.142), revealing a heterozygosity excess. This situation is clearly confirmed and actually is the consequence of the observed heterozygosity value which is higher than the expected heterozygosity. Negative Fis values are generally present in populations showing geneflow due to the introduction of individuals belonging to other breeds for the reproduction. Twenty-six loci, out of thirty, deviated (P<0.05) from the Hardy-Weinberg equilibrium in the whole population composed by the pooled samples (Table S1, Appendix). This high percentage of deviation from the equilibrium ideal condition is due to a non-random mating which led to a homozygote excess and it is indeed confirmed by the markers Fis values. Deviations from the Hardy-Weinberg equilibrium are expected if individual populations are sub-structured into flocks within populations that are isolated from each other (CitationGranevitze et al., 2007). CitationDalvit et al. (2009) highlighted a very highly significant deviation from the Hardy-Weinberg equilibrium in two Italian local chicken breeds before they started out an in situ marker assisted conservation scheme. In Table S1 (Appendix), Wright fixation indices per locus in the whole population are shown. The mean Fis value was significantly different from zero (0.082) (P<0.05) confirming again the presence of heterozygosity deficiency and not completely random matings in the studied sample. As expected, the mean Fit index was 0.307 (P<0.05), highlighting the presence of some factors which influenced the normal gene flow among the animals resulting in a strong heterozygote deficiency in the total population. The value of the last mean fixation index, Fst (0.245) (P<0.05), displayed the existence of a significant segmentation and a very great genetic differentiation among the different breeds. CitationArcos-Burgos and Muenke (2002) stated that Fst could be significantly greater than zero when a population establishes a pattern of subdivision from other ones because of some kind of genetic isolation, which eventually lead to a condition of homozygote excess.
Table 2 Studied breeds, sample size of each breed, mean number of observed alleles, private alleles, mean observed and expected heterozygosity and Fis per breed.
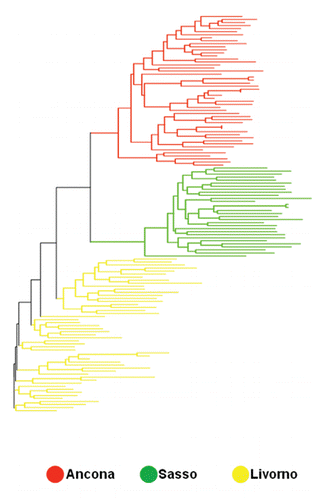
In this study, Livorno and especially Ancona could reasonably be in this situation. The tree from inter-individual DAS distance using Neighbour-Joining algorithm () displayed a very defined cluster for all the investigated breeds. The spatial representation of the genetic inter-individual distances highlights that Ancona and Livorno are characterised by homogeneous genetic patterns. The animals belonging to the different breeds were placed in three well defined areas; however, very curious is the situation occurring in Livorno.
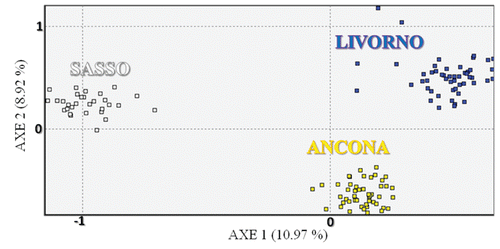
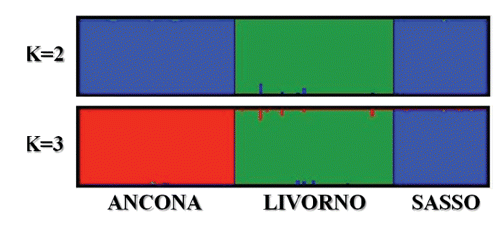
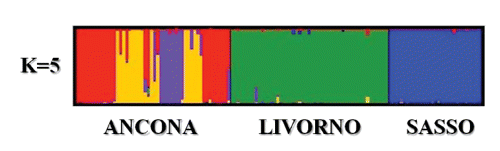
It is worth noting that this breed differed somewhat from the other two breeds, for his taking place at various nodes, and that is in accordance with a greater within-breed inter-individual distance reflecting more internal variability. It is well known that in chicken, where no pedigree information is available and no breeding plans are usually organised, every animals nucleus is a sub-group of the whole population and it is characterised by more genetic variability than the entirety of the total animals sample (CitationRosenberg et al., 2001). The differentiation of the individuals within each breed was further assessed with the FCA by the construction of a two-dimen-sional plot in which the different animals took place (). This analysis gives the chance to show the results through a graphic model with a considerable descriptive value (CitationGuinand, 1996). The first axis explained the 10.97% of the total variation and separates the different breeds from each other, whereas the second axis explained the 8.92%. Other authors, such as CitationFerreira et al. (2006) and CitationWheeldon and White (2009) took advantage of this methodological approach for genetic analysis on animal populations obtaining comparable statistical results. In the present study Livorno and Ancona animals formed two separated and well-defined groups. The Livorno showed only some animals which moved themselves away from the ideal grouping area, whereas within the Ancona all the animals took part in a very homogeneous area. This is consistent with the presence of more internal variability in Livorno. Anyway Livorno and Ancona are the closest breeds in the graphical representation. STRUCTURE-based analysis was carried out to estimate the most likely number of clusters present in the data set, to detect the underlying genetic structure among a set of individuals genotyped at multiple markers and to possibly reveal the potential presence of substructures within the breeds. Following CitationEvanno et al. (2005), the most likely number of cluster turned to be 3, since the highest ΔK value was obtained for K=3 (Figure S1, Appendix). This result was expected, since the most likely number of clusters was the same as the number of the studied breeds, and this genetic frame reflects what we found with the interindividual genetic distance tree and FCA-based analyses. Taking advantage of various methodological approaches, all these analyses in different ways confirmed the genetic uniqueness of the studied breeds. Analysis of the percentage of correctly assigned individuals (q>0.90) for K= 3 () showed the highest values for Ancona and Sasso (100%), with all the animals correctly assigned. With respect to Livorno, fifty animals out of fifty-one were correctly assigned (98%). The proportion of membership in the different clusters is totally comparable among the breeds, even if Ancona exhibited the highest value (0.994) (). All the breeds displayed a very high percentage of assignment (0.994, 0.993 and 0.985 for Ancona, Sasso and Livorno, respectively). These data numerically confirmed the results showed by the FCA analysis and the spatial representation of the genetic inter-individual distances. shows the clustering pattern arising from the STRUCTURE analysis. At K=2 Sasso and Ancona surprisingly clustered together, whereas Livorno clustered separately. This first subdivision was not expected since Sasso is the non Italian breed and was taken as an out-group in this study. An explanation could be that genetic similarities exist more between Ancona and Sasso than between Ancona and Livorno, even though they come from the same country. At K=3, which is the most likely number of partitions, the three breeds perfectly clustered in three really definite clusters. All the animals were correctly assigned to their clusters, with just extremely small amounts of shared genetic components. As already stated, the studied breeds, particularly Livorno and Ancona, represent specific and unique genetic extents, and therefore they should be considered genetic resources to be preserved. Even though we found the highest ΔK for K=3 following Evanno’s method, which perfectly and easily describes the genetic structure of the studied breeds, it is worth showing and discuss the picture we get if we consider the clustering for K=5 (). At K=5 Sasso and Livorno did not change their clustering pattern, whereas Ancona resulted sub-structured. Ancona was characterised by a sub-clustering frame: it was therefore possible to distinguish three different genetic contributions for this breed, which could reflect a geographic partition, as confirmed by the highest Fis value (0.251) detected just in Ancona. The animals forming the Ancona cluster resulted segmented according to the different farms where they were sampled from. In fact, the STRUCTURE analysis for K=5, even though with some exceptions, did not show an admixture pattern within the single individuals, but it mainly showed an admixture pattern among the individuals, which generally reflects a farming subdivision. It is worth saying that, even though 3 reasonably was the correct estimation of the most likely number of partitions, the genetic pattern showed at K=5 was very interesting and noteworthy. On one hand, K=3 clearly showed that the three breeds were consistently and perfectly separated from each other and did not share any significant common genetic pattern; on the other hand K=5 showed that the genetic features of Ancona perfectly follow what the local breeders practically do in the reality. The Ancona is a small autochthonous breed, mainly spread across Marche and part of Emilia Romagna. The different breeders have been permanently working at his defence, protection and development in order to safeguard and preserve his existence and his typical peculiarities. Every farm could be considered a conservation temple, where Ancona is maintained at his original genetic standard without any possible contamination from outside. This situation leads to two main consequences. On one hand Ancona keeps his phenotypic and genotypic characteristics unchanged, and that is important for the safeguard of this breed, on the other hand every farm experiences a kind of genetic isolation because of the lack of Ancona males. Every nucleus includes several hens and a few cocks, resulting in matings always based on the same fertilising males. This eventually leads to inbreeding and to a situation called breeding effect, which is the same as genetic drift. This situation is so marked that it could be possible to speculate the presence of potential sub-populations within the same main breed.
Table 3 Percentage of correctly assigned animals with q>0.90 and proportion of membership of the three chicken populations for K=3.
Conclusions
To sum up, this study highlights the general lack of genetic variability in the Italian local studied breeds, Ancona and Livorno. After all, the autochthonous breeds are thought to progressively lose their genetic variability because of the wider and wider spreading of commercial breeds; this negative trend was confirmed in Ancona and Livorno through the employment of molecular tools such microsatellites. Microsatellites also resulted a powerful tool to study the genetic diversity and the evolution of domestic animals such the local chicken breeds Ancona and Livorno.
Interestingly, microsatellites gave the chance to demonstrate the genetic uniqueness of the considered breeds and the presence of potential sub-populations within the Ancona breed due to genetic isolation. It would be therefore desirable to set up improved selection schemes in order to save the genetic diversity, to avoid inbreeding and to overcome the presence of population sub-structures.This study confirmed the possibility to discriminate with molecular markers among different breeds by using statistical assignment analysis. These results also might give a suitable starting point to set up a potential preliminary genotypic test for all the cocks to be used in the fertilisation plans, in order to genetically characterise individuals having specific and valuable genetic features and belonging to specific breeds, and to avoid therefore the employment of undefined animals.
Acknowledgements:
the authors would like to thank Steffen Weigend and Michele Tixier-Boichard for providing the AVIANDIV standard samples.
They would also thank the two anonymous referees for their valuable comments to the manuscript and their constructive suggestions.
A further acknowledgment to all the breeders and to Prof. Isabella Romboli (University of Pisa) who supplied biological samples.
This research was financially supported by Regione Marche and the Ministry of Education, University and Research (MIUR, Italy), Project No. PRIN 2008 and 2008FN93B3_002.
References
- Arcos-BurgosM MuenkeM 2002 Genetics of Population Isolates Clin. Genet 61 233 247
- AVIANDIV Project 2011 Development of Strategy and Application of Molecular Tools to Assess Biodiversity in Chicken Genetic Resources Available from: http://aviandiv.tzv.fal.de/
- Beigi-NassiriM.T HamidiZ TavakoliS 2007 The investigation of genetic variation at microsatellite loci in Mazandaran native chickens Int. J. Poultry Sci 9 675 678
- BelkhirK BorsaP ChikhiL RaufasteN BonhommeF 1996–2004 GENETIX 4.05. logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome. Populations Interactions: CNRS UMR 5000 Université de Montpellier II Montpellier, France Available from: http://www.genetix.univ-montp2.fr/genetix/genetix.htm
- BenzécriJ.P 1982 L'Analyse des Données, 2. L'Analyse des Correspondances Dunod Ed. Paris, France
- BotsteinD WhiteR.L SkolnickM DavisR.W 1980 Construction of a genetic linkage map in man using restriction fragment length polymorphisms Am. J. Hum. Genet 32 314 331
- CastelliniC MugnaiC PedrazzoliM Dal BoscoA 2006 Productive performance and carcass traits of Leghorn chickens and their crosses reared according to organic farming system Proc. 12th Eur. Poultry Conf. Verona, Italy Available from: http://www.equizoobio.it/materiale_divulgativo.html
- ChakrabortyR JinL 1993 A unified approach to study hypervariable polymorphisms: statistical considerations of determining relatedness and population distances ChakrabortyJ EpplenT JeffreysA.J DNA Fingerprinting: State of the Science Birkhauser Verlag Basel, Switzerland
- Dal BoscoA MugnaiC CastelliniC 2011 Performance and meat quality of pure and crossed Ancona chickens organically reared Arch. Geflugelk 75 7 12
- DalvitC ZanettiE CassandroM 2009 Estimation of genetic diversity over time in an in-situ marker assisted conservation scheme of local chicken breeds Ital. J. Anim. Sci 8 (Suppl.2) 63 65
- DávilaS.G GilM.G Resino-TalavánP CampoJ.L 2009 Evaluation of diversity between different Spanish chicken breeds, a tester line, and a White Leghorn population based on microsatellite markers Poultry Sci 88 2518 2525
- EvannoG RegnautS GoudetJ 2005 Detecting the number of cluster of individuals using the software STRUCTURE: a simulation study Mol. Ecol 14 2611 2620
- FAO 2010 Domestic Animal Diversity Information System Available from: http://dad.fao.org/
- FerreiraE SoutoL SoaresA.M.V.M FonsecaC 2006 Genetic structure of the wild boar (Sus scrofa L.) population in Portugal Wildl. Biol. Pract 2 17 25
- GandiniG.C VillaE 2003 Analysis of the cultural value of local livestock breeds: a methodology J. Anim. Breed. Genet 120 111 111
- GiacomoniE.H Fernández-StolzG.P FreitasT.R.O 2008 Genetic diversity in the Pantaneiro horse breed assessed using microsatellite DNA markers Genet. Mol. Res 7 261 270
- GlaubitzJ.C 2004 CONVERT: A user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages Mol. Ecol. Notes 4 309 310
- GranevitzeZ HillelJ ChenG.H CucN.T.K FeldmanM EdingH WeigendS 2007 Genetic diversity within chicken populations from different continents and management histories Anim. Genet 38 576 583
- GroenenM.A.M ChengH.H BumsteadN BenkelB.F Elwood-BrilesW BurkeT BurtD.W CrittendenL.B DodgsonJ HillelJ LamontS Ponce de LeonA SollerM TakahashiH VignalA 2000 A Consensus Linkage Map of the Chicken Genome Genome Res 10 137 147
- GuinandB 1996 Use of a multivariate model using allele frequency distributions to analyse patterns of genetic differentiation among populations Biol. J. Linn. Soc 58 173 195
- GuoS.W ThompsonE.A 1992 A Monte Carlo method for combined segregation and linkage analysis Am. J. Hum. Genet 51 1111 1126
- HillelJ GroenenM.A.M Tixier-BoichardM KorolA.B DavidL KirzhnerV.M BurkeT Barre-DirieA CrooijmansR.P.M.A EloK FeldmanM.W FreidlinP.J Mäki-TanilaA OortwijnM ThomsoneP VignalA WimmersK WeigendS 2003 Biodiversity of 52 chicken populations assessed by microsatellite typing of DNA pools Genet. Sel. Evol 35 533 557
- LangellaO 2002 Population 1.2.28. Logiciel de génétique des populations Laboratoire populations, génétique et évolution CNRS UPR 9034 Ed. Gif-sur-Yvette, France
- LasagnaE BianchiM CeccobelliS LandiV Martínez MartínezA Vega PlaJ.L PanellaF Delgado BermejoJ.V SartiF.M 2011 Genetic relationships and population structure in three Italian Merinoderived sheep breeds Small Ruminant Res 96 111 119
- LiM.H KantanenJ 2009 Genetic structure of Eurasian cattle (Bos taurus) based on microsatellites: clarification for their breed classification Anim. Genet 41 150 158
- MahmoudiB BabayevM.S 2009 The investigation of genetic variation in Taleshi goat using microsatellite markers Res. J. Biol. Sci 4 644 646
- MugnaiC Dal BoscoA, Castellini, C 2009 Effect of rearing system and season on the performance and egg characteristics of Ancona laying hens Ital. J. Anim. Sci 8 175 188
- ParkS.D.E 2001 Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection Degree Diss University of Dublin Ireland
- PritchardJ.K StephensM DonnellyP 2000 Inference of population Structure using multilocus genotype data Genetics 155 945 959
- QuL LiX XuG ChenK YangH ZhangL WuG HouZ XuG YangN 2006 Evaluation of genetic diversity in Chinese indigenous chicken breeds using microsatellite markers Sci. China Ser. C 49 332 341
- RaymondM RoussetF 1995 An exact test for population differentiation Evolution 49 1280 1283
- RosenbergN.A 2004 Distruct: a program for the graphical display of population structure Mol. Ecol. Notes 4 137 138
- RosenbergN.A BurkeT EloK FeldmanM.W FreidlinP.J GroenenM.A.M HillelJ Mäki-TanilaA Tixier-BoichardM VignalA WimmersK WeigendS 2001 Empirical Evaluation of Genetic Clustering Methods Using Multilocus Genotypes From 20 Chicken Breeds Genetics 159 699 713
- SchneiderS KuefferJ.M RoessliD ExcoffierL 1997 ARLEQUIN: a software for population genetic data analysis. version 1.1 Genetics and Biometry Lab Department of Anthropology, University of Geneva Ed. Geneva, Switzerland
- SpalonaA RanvigH Cywa-BenkoK ZanonA SabbioniA SzalayI BenkovaJ BaumgartnerJ SzwaczkowskiT 2007 Population size in conservation of local chicken breeds in chosen European countries Arch. Geflügelk 71 49 55
- TadanoR SekinoM NishiboriM TsudzukiM 2007 Microsatellite marker analysis for the genetic relationships among Japanese long-tailed chicken breeds Poultry Sci 86 460 469
- TamuraK DudleyJ NeiM KumarS 2007 MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 Mol. Biol. Evol 24 1596 1599
- VicenteA.A CarolinoM.I SousaM.C.O GinjaC SilvaF.S MartinezA.M Vega-PlaJ.L CarolinoN GamaL.T 2008 Genetic diversity in native and commercial breeds of pigs in Portugal assessed by microsatellites J. Anim. Sci 86 2496 2507
- WeirB.S CockerhamC.C 1984 Estimating F-statistics for the analysis of population structure Evolution 38 1358 1370
- WheeldonT WhiteB.N 2009 Genetic analysis of historic western Great Lakes region wolf samples reveals early Canis lupus/ lycaon hybridization Biol. Lett 5 101 104
- YehF.C YangR.C BoyleT 1999 Popgene version 1.31. Microsoft window-based freeware for population genetic analysis Department of Renewable Resources University of Alberta Ed. Edmonton, Canada
- ZanettiE DalvitC De MarchiM Dal ZottoR CassandroM 2007 Genetic characterisation of Italian chicken breeds using a panel of twenty microsatellite markers Agriculture 13 197 200
- ZanonA SabbioniA 2001 Identificazione e salvaguardia genetica delle razze avicole italiane Annali della Facoltà di Medicina Veterinaria di Parma 21 117 134