Abstract
The Italian Podolian is an autochthonous breed belonging to the grey steppe cattle group. In the past, the Podolian was present throughout all of the Adriatic and the southern part of Italy, but now it is restricted to four regions and displays morphological differences according to the region. It is thought that the Podolian contributed heavily to the genetic make-up of the Chianina and Romagnola breeds, while contrasting opinions exist regarding the relationship between the Podolian and the Piemontese. This paper aimed at characterizing genetic variability and structure by means of microsatellite loci to assess the relationship between the Podolian and the most relevant European breeds, and to detect evidence of selection in the considered breeds by investigating the existence of outlier loci. The DNA of 134 Italian Podolian individuals from 14 different herds in 4 breeding areas were collected, genotyped at 30 microsatellite loci and compared to 13 Italian and European breeds, including the grey type ones and using N’dama as the outgroup. The Podolian showed the highest values of observed heterozygosity and polymorphism content. Moreover in a subset of 50 samples (chosen according to geographic origin), polymorphism data analysis evidenced the absence of a geographically determined genetic structure. The hierarchical population clustering evidenced a Podolian cluster at K=5 and the phylogenetic tree based on Reynold’s distances confirmed the genetic links between the Podolian breeds. Two loci presented significantly low FST values, suggesting that they could be under the effect of balancing selection. It can be concluded that Italian Podolian has no genetic subdivision, contains a notable amount of genetic variability and is closely related to the other Podolian type breeds.
Introduction
The Italian Podolian (ITPOD) is one of the grey cattle breeds reared in Italy (including the Romagnola and Chianina) and is part of a larger European family (i.e. the Hungarian Grey, Istrian).
The presence of Podolian in Italy has been shown since the ancient times (CitationBaker and Manwell, 1980; CitationCiani and Matassino, 2001; Citation2007). In the past, the breed was reared throughout all of the Adriatic and the southern part of the country, but currently the breeding areas are restricted to the regions of Southern Italy. The Italian Podolian originated in the Podolian region of Eastern Europe, as indicated by its name, and it is thought that the breed related to Chianina and Romagnola breeds (CitationAstolfi et al., 1983) and also to Istrian Grey (CitationFAO, 2011). The breed is well adapted for exploiting the Mediterranean scrub and highlands, so it can be considered as a genetic resource for future breeding options aimed at enhancing animal production in marginal areas by using the environment in a sustainable way. In the eighties, a selection scheme aimed at improving meat production was implemented, but the use of artificial insemination is still limited and the selection scheme is not fully applied. For this reason, a genetic structure that differentiates Podolian populations reared in different areas may be hypothesized due to the genetic isolation between population and could justify the slight morphological differences observed between the regions. Over the last few decades, this breed has experienced a drastic reduction in numbers and now about 24,000 animals are registered in the herd book (CitationANABIC, 2011), while the total population is estimated to consist of more than 100,000 heads.
In order to ascertain genetic variability and population structure at the DNA level, different types of markers can be investigated. A fundamental requisite is the absence of selection pressure on the considered loci, since it was demonstrated that the inclusion of a few outliers among neutral loci can greatly affect the estimate of population indexes (CitationLuikart et al., 2003). Outlier loci can be efficiently identified by estimating single loci FST values (CitationBeaumont and Nichols, 2003). Microsatellites have been extensively used to characterize the population genetics of local and cosmopolitan cattle breeds (CitationBrenneman et al., 2007; CitationCanon et al., 2008; CitationDadi et al., 2008; CitationDalvit et al., 2008; CitationNdumu et al., 2008) and to unravel the genetic distance and relationship between breeds (CitationMcHugh et al., 1994; CitationMoazami-Goudarzi et al., 1997; CitationCiampolini et al., 1995; CitationCanon et al., 2001; CitationDel Bo et al., 2001; CitationWiener et al., 2004; CitationEuropean Cattle Genetic Diversity Consortium, 2006; CitationMartin-Burriel et al., 2007; CitationSun et al., 2008). Using the same molecular markers, the genetic variability of other Podolian breeds of Eastern Europe has been investigated CitationGeorgescu et al., 2009; CitationManatrinon et al., 2008), as well as that of the Italian Podolian (CitationMoioli et al., 2004; CitationD’Angelo et al., 2006). However, in the previous studies the existence of genetic structure in the Italian Podolian as well as its relationship with the main European breeds have never been investigated. Moreover, the existence of outlier loci has never been tested. The aim of this paper was to characterize the genetic variability and structure of the Italian Podolian and to ascertain the genetic relationships between this breed and the Italian breeds included in the RESGEN project and the most relevant European breeds. The genomes were analyzed by means of microsatellite markers and the existence of outlier loci and their effect on genetic distance and structure were also considered.
Materials and methods
A total of 134 unrelated individuals belonging to 14 different herds in 4 Italian regions (2 from Molise, 1 from Puglia, 10 from Basilicata and 1 from Campania) were sampled by collecting peripheral blood samples. The DNA was isolated from lymphocytes by classical phenol-chloroform extraction methods (CitationSambrook et al., 1989). The individual genotypes were determined using the panel of 30 microsatellites suggested by FAO (CitationISAG-FAO, 2004) for cattle biodiversity investigations (BM1818, BM1824, BM2113, CSRM60, CSSM66, ETH3, ETH10, ETH152, ETH185, ETH225, HAUT24, HAUT27, HEL1, HEL5, HEL9, HEL13, ILST005, ILST006, INRA005, INRA023, INRA0032, INRA0035, INRA037, INRA063, MM12, SPS115, TGLA53, TGLA122, TGLA126, TGLA227). Amplification was performed as suggested by the ISAG/FAO Standing Committee Recommendations (CitationISAG/FAO, 2004).
Furthermore, a subset of 50 out of the 134 individuals balanced for the region of origin was selected and the obtained genotypes werw standardized using three reference samples on order to make them comparable to the genotypes obtained by the European Cattle Genetic Diversity Consortium in the RESGEN project N. CT98-118 of the European Union 5th Framework Programme. The breeds considered for comparison were the Italian Grey Podolian type breeds: Romagnola (ITROM), Chianina (iTCHI), and Piemontese (ITPIm), other Italian breeds: Rendena (ITREN), Cabannina (ITCAB), Pezzata Rossa Italiana (ITPRI) and Grigio Alpina (ITGRI), a grey coated breed apparently not related to the Podolian breed, and Istrian (CRIST), a Podolian breed from Croatia. Some of the most representative European and cosmopolitan breeds were also considered: Swiss Brown (CHSWB), Simmental (CHSIM), Holstein Friesian (NLFRH), and Limousine (FRLIM). N’Dama (NDA) breed was selected as outgroup. An average of 40 genotyped individuals per breed was analyzed. Genotypes from the Podolian dataset were elaborated with the GENEPOP ver. 3.1d computer package (CitationRaymond and Rousset, 1995) to obtain allelic frequencies and the number of alleles for each locus, and the observed and expected heterozygosities were obtained with the ARLEQUIN ver. 2.000 (CitationSchneider et al., 2000) computer package by applying the method of CitationGuo and Thompson (1992). The computations required to detect outlier loci were performed using the approach proposed by Beaumont & CitationNichols (1996). The detection of outlier loci was performed following the FST outlier approach developed by CitationBeaumont and Nichols (1996) further developed by CitationBeaumont and Balding (2004), and implemented in the FDIST2 software (http://www.rubic.rdg.ac.uk/~mab/software.html). This method evaluates the relationship between FST and He (expected heterozygosity) by calculating the expected distribution of Wright’s coefficient FST vs He under an island model of migration with neutral markers. This distribution is then used to identify outlier loci that have excessively high or low FST values compared to neutral expectations. Such outlier loci are candidates for being under directional selection.
Structure software 2.3 (CitationPritchard et al., 2000) was used to reveal the existence of genetic variability partitioning. In order to analyze the relationships between the European breeds, ten independent runs of each K, testing K ranging from 2 to 15, with 100,000 Markov chain Monte Carlo (MCMC) iterations and a burn-in period of 100,000, were performed using the admixture model considering allele frequencies between populations as independent with no prior information about individual membership. The method by Evanno for testing K (CitationEvanno et al., 2005) was applied to detect the strength of the genetic structure detected by the software. DISTRUCT (CitationRosenberg, 2004) software was used to visualize the estimated membership coefficients. PowerMarker software (CitationLiu and Muse, 2005) was applied in order to compute Reynold’s genetic distances between breed pairs and to construct and display the phylogenetic tree using the neighbour-joining (NJ) algorithm, which was visualized using Treeview software (CitationPage, 1996).
Results and discussion
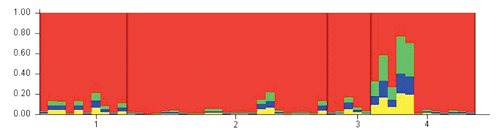
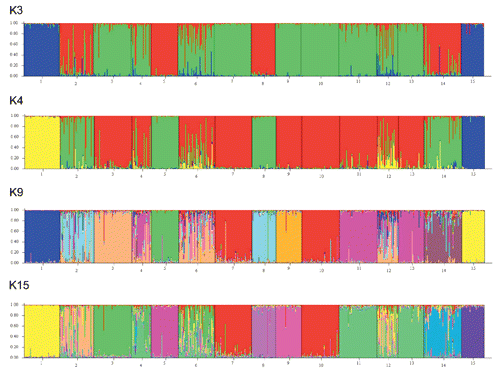
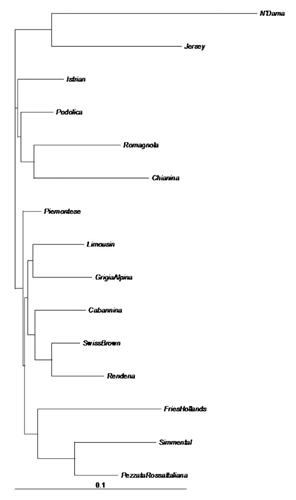
All loci were successfully amplified and found to be polymorphic in the 134 Italian Podolian samples. The average number of alleles per locus was 9.9. A deviation from the Hardy-Weinberg equilibrium (P<0.05) was detected in 11 out of the 30 loci (Table S1, Appendix ). The results of the basic population parameters in all of the examined European breeds using all the 30 loci are shown in . The Italian Podolian breed showed the highest values of expected (0.73) and observed (0.71) heterozygosities, while the Simmental displayed the lowest values (0.59) for both measures. The difference between expected and observed heterozygosity in the Italian Podolian resulted in a slightly positive inbreeding coefficient (0.05). The observed heterozygosities were higher than those reported for the British breeds investigated using the same microsatellite set (CitationWiener et al., 2004), whose values ranged from 0.56 (Highland) to 0.67 (Ayrshire), and for local Spanish, French and Portuguese breeds analyzed at 16 microsatellite loci of the FAO list (values ranging from 0.58 to 0.71 in Salers and Barrosa, respectively) (CitationCanon et al., 2001). The same microsatellites were also investigated in the Hungarian Podolian, where a lower value of observed heterozygosity (0.66) was obtained (CitationManatrinon et al., 2008), whereas in the Romanian Podolian, in which 11 loci were analyzed (CitationGeorgescu et al., 2009), the value was comparable (0.76) to that of the Italian population. Genetic variability in the Italian Podolian breed was investigated in a previous study (CitationD’Angelo et al., 2006), which found a lower value of observed heterozygosity (0.44). This discrepancy can be justified by the fact that the investigation was conducted on different microsatellites and (more relevantly) in a restricted geographic area. On the other hand, in agreement with our findings, an investigation into the STAT5A locus evidenced a larger genetic variability in the Italian Podolian compared to the cosmopolitan breeds (CitationDario et al., 2009). A higher variability in Podolian breeds compared to Italian meat breeds was also recently reported using SNP analysis (CitationPariset et al., 2010). This high variability could be explained considering that selection scheme is not fully adopted and the population is more consistent than other local breeds. The search for outliers evidenced significant values for two microsatellites (BM1818 and HAUT24), suggesting that these loci may be under the effect of balancing selection, in fact as showed in Figure S1 (Appendix), these two loci are placed under the lower confidence limit of 95% (CitationBeaumont and Nichols, 1996; CitationBeaumont and Balding, 2004). These two microsatellites were never reported to be associated with phenotypic traits, but, interestingly, BM1818 maps on bovine 23 very closely (from 140 to 210 kb) to four coding loci (KIF13A (kinesin family member 13A), UMJM31, NUP153 (nucleoporin), FAM8A1). However, the function of the coded proteins is still unknown. The data were then analyzed both including and excluding the two outliers. While no significant variations were observed in the clustering results, some differences were identified in the NJ trees, as discussed below. In order to estimate the number of genetic clusters within the 50 Italian Podolian individuals, a parametric genetic mixture analysis in the Structure 2.3 software (CitationPritchard et al., 2000) was performed. In , a graphical presentation of the estimated membership coefficients to the clusters of the structure analysis obtained with K=4 is displayed. A K value of 4 was chosen because this corresponds to the number of Italian regions where the animals originated from. Clusters are presented as different colors and individuals are depicted as bars partitioned into colored segments whose lengths correspond to the membership coefficients in each subgroup. The analysis did not show a partitioning of genetic variability according to geographical origin since all of the animals showed a membership that was equally distributed between the four clusters, with the notable exception of some animals that belonged to the same single herd. This finding could be explained by the transhumance. Further analysis using the same software was performed in order to estimate the number of genetic clusters among all of the examined European breeds. Between 2 and 15 clusters (K values) were tested using the admixture model, assuming that each individual did not necessarily have a genetic background originating from one of the K populations. Consistent results across runs were obtained and a clear clustering of breeds was observed for each K tested. In order to identify the optimal K value we applied the methodology described in CitationEvanno et al. (2005), and concluded that 9 was the optimal K (Figure S2, Appendix). The most interesting K values were 3, 4, 9 and 15. This latter corresponding to the number of breeds. The corresponding graphics are displayed in . Jersey (1) and N’Dama (15) were well differentiated; also, Chianina (5) and Romagnola (8) individuals were distinguished. At K=4, the Podolian cluster, including the Italian Podolian, Istrian, Chianina and Romagnola breeds, was easily recognized. At K=9 (the best clustering number according to Evanno’s test), the cattle group of known Podolian origin was split and a weak similarity was noted between the Istrian and Romagnola breeds only. The Italian Pezzata Rossa and Simmental were very similar, while Jersey, N’Dama, Holstein Fresian and (to a lesser extent) Limousine were clearly noticeable. A genetic relationship was also noted between two alpine breeds (Swiss Brown and Rendena), and Piemontese, Istrian, Cabannina and Podolian displayed the higher genetic admixture. As envisaged from Evanno’s test, increasing the K above 9 did not add more information, but it is worthwhile noting that at K=15, ITPRI and CHSIM were in the same cluster and the relationship between Brown and Rendena was still detectable, while the Piemontese, Grigio Alpina and Istrian breeds displayed a certain degree of genetic admixture. The tree obtained from Reynold’s genetic distances (CitationReynolds et al., 1983) between the 15 breeds is displayed in Figure S3 (Appendix). Three breed groups are clearly distinguishable in the tree: i) a cluster formed by the Piemontese, Holstein Friesian, Simmental and Pezzata Rossa Italiana breeds; ii) a cluster including the Limousine, Grigio Alpina, Rendena, Cabannina and Swiss Brown breeds; iii) a cluster including the Istrian Podolian, Italian Podolian, Romagnola and Chianina breeds. As expected, belonging all to the same Podolian Type the Italian Podolian, was found to belong to the same branch with Romagnola, Chianina and Istrian Podolian. This result confirms the common origin of these breeds and the tight linkage between the Podolian cattle of different countries, as was also observed by AFLP fingerprinting (CitationNegrini et al., 2007), although the Istrian and Italian Podolian breeds displayed a lower genetic distance than that existing between the Chianina and Romagnola breeds (Figure S3, Appendix). The Piemontese, linked to the Podolian breed, was not positioned in the same branch and this result is consistent with our structure results and with previous studies that investigated other genomic loci (CitationAstolfi et al., 1983; CitationCiampolini et al.,1995; CitationMoioli et al., 2004). The Piemontese breed is also not so close to Limousin as was reported using SNP markers (CitationNegrini et al., 2008). However, considering the mitochondrial DNA haplotypes, the Piemontese has the same polymorphisms as the other Italian Podolian breeds (CitationBeja-Pereira et al., 2006; CitationPellecchia et al., 2007; CitationCiani and Matassino, 2001; Citation2007; CitationMatassino and Ciani, 2009). For this reason, the Piemontese, Italian Podolian and the other Italian Podolian-derived breeds were assigned to the Bos primigenius taurus subspecies (CitationHiendleder et al., 2008; CitationMatassino and Ciani, 2009). Other breed groups can be clearly detected in the tree. Interestingly, when the tree was drawn excluding the two outlier loci (), the cluster formed by the Piemontese, Holstein Friesian, Simmental and Pezzata Rossa Italiana breeds was split and Piemontese was placed alone on a different branch. This latter tree was better fitted to the breed history and illustrated how the inclusion of outlier loci in breed comparisons can influence the results.
Table 1 Diversity measurements for each of the cattle populations analysed in the current study.
Obtained with a 100,000 burn-in, a 100,000 MCMC, under the admixture model, for Podolian individuals belonging to 14 Italian herds. Each individual is represented by a single vertical line broken into K (4) coloured segments, with lengths proportional to each of the four inferred clusters. Each colour represents the proportion of membership (M) of each individual (represented by a vertical line) to the K clusters. The numbers correspond to four Italian regions: 1 Puglia; 2 Basilicata; 3 Molise; 4 Campania.
Conclusions
The results obtained indicated the existence of consistent genetic variability within the Podolian breed that could be the result of a weak selective pressure for performance, and highlight the importance of this breed in the constitution of cattle biodiversity. The microsatellite polymorphisms analyzed did not show a genetic structure within the Italian Podolian populations reared in different regions. It can be concluded that the Italian Podolian breed is genetically homogenous despite its moderate phenotypic variability and the low use of artificial insemination; nonetheless, it must be pointed out that sires are exchanged between herds. However, this data must be completely confirmed by analyzing the consistent sampling of Italian Podolian individuals from Calabria. Compared to the other European breeds, the Podolian showed the highest genetic variability in terms of expected heterozygosity and PIC values. The structure analysis as well as the genetic distance trees highlighted the close relationship between the Italian Podolian breed and the other Podolian breeds from Italy and Croatia (with the exception of the Piemontese). Moreover structure analysis also showed the relation among Alpine breeds (Swiss Brown and Rendena) while even at maximum K value Italian Pezzata Rossa and Simmental where not distinguishable.
Figure S1 Upper (green) and lower (blue) confidence limits of 95% quantiles; median (red) of 50,000 replications of expected FST and heterozygosity
Download TIFF Image (620.6 KB)Figure S2 Evanno’s test results showing K=9 as the best value for cluster description. SD, standard deviation.
Download TIFF Image (1.7 MB)Figure S3 Phylogenetic relationships between the 15 breeds studied.
Download TIFF Image (186.1 KB)References
- ANABIC 2001 Available from: www.anabic.it [accessed 14th April 2011]
- Astolfi P. Pagnacco G. Guglielmino-Matessi C.R. 1983 Phylogenetic analysis of native Italian Cattle breeds Z. Tierz. Zuchtungsbio 100 87 100
- Baker C.M.A. Manwell C. 1980 Chemical classification of cattle. 1 Breed groups Anim. Blood Groups Bi 11 127 150
- Beaumont M.A. Balding D.J. 2004 Identifying adaptive genetic divergence among populations from genome scans Mol. Ecol 13 969 980
- Beaumont M.A. Nichols R.A. 1996 Evaluating loci for use in the genetic analysis of population structure P. R. Soc. B 263 1619 1626
- Beja-Pereira A. Caramelli D. Lalueza-Fox C. Vernesi C. Ferrand N. Casoli A. Goyache F. Royo L.J. Conti S. Lari M. Martini A. Ouragh L. Magid A. Atash A. Zsolnai A. Boscato P. Tiantaphylidis C. Ploumi K. Sineo L. Mallegni F. Taberlet P. Erhard G. Sampietro L. Bertranpetit J. Barbujani G. Luikart G. Bertorelle G. 2006 The origin of European cattle: Evidence from modern and ancient DNA P. Natl. Acad. Sci. USA 103 8113 8118
- Brenneman R. Chase C. Olson T. Riley D. Coleman S. 2007 Genetic diversity among Angus, American Brahman, Senepol and Romosinuano cattle breeds Anim. Genet 38 50 53
- Bruzzone A. Barone C.M.A. Iamartino D. Incoronato C. Pilla F. Matassino D. 2001 Genetic characterisation of the Podolica cattle: preliminary results Proc. 14th Nat. Congr ASPA, Firenze, Italy 2 58 60
- Canon J. Alexandrino P. Bessa I. Carleos C. Carretero Y. Dunner S. Ferran N. Garcia D. Jordana J. Laloe D. Pereira A. Sanchez A. Moazami-Goudarzi K. 2001 Genetic diversity measures of local European beef cattle breeds for conservation purposes Genet. Sel. Evol 33 311 332
- Canon J. Tupac-Yupanqui I. Garcia-Atance M.A. Cortes O. Garcia D. Fernandez J. Dunner S. 2008 Genetic variation within the Lidia bovine breed Anim. Genet 39 439 445
- Ciampolini R. Moazami-Goudarzi K. Vaiman D. Dillman C. Mazzanti E. Foulley J.L. Leveziel H. Cianci D. 1995 Individual multilocus genotypes using microsatellite polymorphisms to permit the analysis of the genetic variability within and between Italian beef cattle breed J. Anim. Sci 73 3259 3268
- Ciani F. Matassino D. 2001 Il bovino grigio allevato in Italia; origine. Nota 1: il bovino Macrocero Taurus speciale 13 89 99
- Ciani F. Matassino D. 2007 Il bovino grigio allevato in Italia: origine ed evoluzione. Nota 2: il bovino Brachicero Taurus speciale 18 69 76
- Dadi H. Tibbo M. Takahashi Y. Nomura K. Hanada H. Amano T. 2008 Microsatellite analysis reveals high genetic diversity but low genetic structure in Ethiopian indigenous cattle population Anim. Genet 39 425 431
- Dalvit C. De Marchi M. Dal Zotto R. Zanetti E. Meuwissen T. Cassandro M. 2008 Genetic characterization of the Burlina cattle breed using microsatellite markers J. Anim. Breed. Genet 125 137 144
- D’Angelo F. Ciani E. Sevi A. Albenzio M. Ciampolini R. Cianci D. 2006 The genetic variability of the Podolica cattle breed from the Gargano area Ital. J. Anim. Sci 5 79 85
- Dario C. Dario M. Ciotola F. Peretti V. Cornicella D. Selvaggi M. 2009 Analysis of STAT5/A va I gene polymorhism in four Italian cattle breeds Biochem. Genet 47 671 679
- Del Bo L. Polli M. Longeri M. Ceriotti G. Looft C. Barre-Dirie A. Dolf G. Zanotti M. 2001 Genetic diversity among some cattle breeds in the Alpine area J. Anim. Breed. Genet 118 317 325
- European Cattle Genetic Diversity Consortium 2006 Marker-assisted conservation of European cattle breeds: an evaluation Anim. Genet 37 475 481
- Evanno G. Regnaut S. Goudet J. 2005 Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study Mol. Ecol 14 2611 2620
- FAO 2011 Fao Domestic animal information system Available from: http://dad.fao.org/ [accessed on 16th June 2011]
- Georgescu S.E. Manea M.A. Zaulet M. Costache M. 2009 Genetic diversity among Romanian cattle breeds with a special focus on the Romanian Grey Steppe Breed Rom. Biotechnol. Lett 14 4194 4200
- Guo S.W. Thompson E.A. 1992 Performing the exact test of Hardy-Weinberg proportion for multiple alleles Biometrics 48 361 372
- Hiendleder S. Lewalski H. Janke A. 2008 Complete mitochondrial genomes of Bos taurus and Bos indicus provide new insights into intra-species variation, taxonomy and domestication Cytogenet. Genome Res 120 150 156
- ISAG/FAO 2004 Standing Committee Recommendations Available from: http://dad.fao.org/cgi-bin/getblob.cgi?sid=-1,50006220 [accessed 7th September 2011]
- Liu K. Muse S.V. 2005 PowerMarker: an integrated analysis environment for genetic marker analysis Bioinformatics 21 2128 2129
- Luikart G. England P.R. Tallmon D. Jordan S. Taberlet P. 2003 The power and promise of population genomics: from genotyping to genome typing Nature Rev. Genet 4 981 994
- Manatrinon S. Fischerleitner F. Baumung R. 2008 Genetic characterization among some Austrian and Hungarian cattle breeds Arch. Anim. Breed 51 426 437
- Martin-Burriel I. Rodellar C. Lenstra J.A. Sanz Cons C. Osta R. Reta M. De Argueullo S. Sanz A. Zaraoza P. 2007 Genetic diversity and relationships of endangered Spanish cattle breeds J. Hered 98 687 691
- Matassino D. Ciani F. 2009 Origine e storia della “Podolica” in Italia Proc. Int. Congr. on the tracks of Grey Podolic cattle Matera, Italy 3 111 124
- McHugh D.E. Loftus R.T. Bradley D.G. Sharp M. Cunningham E.P. 1994 Microsatellite DNA variation within and among European cattle breeds P. R. Soc. B 256 25 31
- Moazami-Goudarzi K. Laloe D. Furet J.P. Grosclaude F. 1997 Analysis of genetic relationship between 10 cattle breeds with 17 microsatellites Anim. Genet 28 338 345
- Moioli B. Napolitano F. Catillo G. 2004 Genetic diversity between Piedmontese, Maremmana, and Podolica cattle breeds J. Hered 95 250 256
- Ndumu D.B. Baumung R. Hanotte O. Wurzinger M. Okeyo M.A. Jailin H. Kibogo H. Solkener J. 2008 Genetic and morphological characterisation of the Ankole Longhorn cattle in the African Great Lakes region Genet. Sel. Evol 40 467 490
- Negrini R. Nicoloso L. Crepaldi P. Milanesi E. Colli L. Chegdani F. Pariset L. Dunner S. Leveziel H. Williams J.L. Ajmone Marsan P. 2008 Assessing SNP markers for assigning individuals to cattle populations Anim. Genet 40 18 26
- Negrini R. Nijman I.J. Milanesi E. Moazami-Goudarzi K. Williams J.L. Erhardt G. Dunner S. Rodellar C. Valentini A. Bradley D.G. Olsaker I. Kantanen J. Ajmone-Marsan P. Lenstra J.A. European Cattle Genetic Diversity Consortium 2007 Differentiation of European cattle by AFLP fingerprinting Anim. Genet 38 60 66
- Page R.D. 1996 TreeView: an application to display phylogenetic trees on personal computer Comput. Appl. Biosci 12 357 358
- Pariset L. Mariotti M. Nardone A. Soysal M.I. Ozkan E. Williams J.L. Dunner S. Leveziel H. Maróti-Agóts A. Bodò I. Valentini A. 2010 Relationships between Podolic cattle breeds assessed by single nucleotide polymorphisms (SNPs) genotyping J. Anim. Breed. Genet 127 481 488
- Pellecchia M. Negrini R. Colli L. Patrini M. Milanesi E. Achilli A. Bertorelle G. Cavalli Sforza L.L. Piazza A. Toroni A. Ajmone-Marsan P. 2007 The mystery of Etruscan origins: novel clues from Bos taurus mitochondrial DNA P. R. Soc. B 274 1377 1385
- Pritchard J.K. Stephens M. Donnelly P. 2000 Inference of population structure using multilocus genotype data Genetics 155 945 959
- Raymond M. Rousset F. 1995 GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism J. Hered 86 248 249
- Reynolds J. Weir B.S. Cockerham C.C. 1983 Estimation of the Coancestry coefficient, basic for a short-term genetic distance Genetics 105 767 779
- Rosenberg N.A. 2004 DISTRUCT – a program for the graphical display of population structure Mol. Ecol. Notes 4 137 138
- Sambrook J. Fritsch E.F. Maniatis T. 1989 Molecular cloning: a laboratory manual Cold Spring Harbor Laboratory Press New York, NY, USA
- Schneider S. Roessli D. Excoffier L. 2000 Arlequin Ver 2.000: a software for population genetics data analysis Genetics and Biometry Laboratory, University of Geneva Switzerland
- Sun W.B. Chen H. Lei C.Z. Lei X.Q. Zhang Y.H. 2008 Genetic variation in eight Chinese cattle breeds based on the analysis of microsatellite Genet. Sel. Evol 40 681 692
- Wiener P. Burton D. Williams J.L. 2004 Breed relationships and definition in British cattle: a genetic analysis Heredity 93 597 602