Abstract
Two coat phenotypes exist in Alpaca, Huacaya and Suri. The two coats show different fleece structure, textile characteristics and prices on the market. Although present scientific knowledge suggests a simple genetic model of inheritance, there is a tendency to manage and consider the two phenotypes as two different breeds. A 13 microsatellite panel was used in this study to assess genetic distance between Suri and Huacaya alpacas in a sample of non-related animals from two phenotypically pure flocks at the Illpa-Puno experimental station in Quimsachata, Peru. The animals are part of a germplasm established approximately 20 years ago and have been bred separately according to their coat type since then. Genetic variability parameters were also calculated. The data were statistically analyzed using the software Genalex 6.3, Phylip 3.69 and Fstat 2.9.3.2. The sample was tested for Hardy-Weinberg equilibrium (HWE) and after strict Bonferroni correction only one locus (LCA37) showed deviation from equilibrium (P<0.05). Linkage disequilibrium (LD) was also tested and 9 loci associations showed significant disequilibrium. Observed heterozygosis (Ho= 0.766; SE=0.044), expected heterozygosis (He=0.769; SE=0.033), number of alleles (Na=9.667, SE=0.772) and Fixation index (F=0.004; SE=0.036) are comparable to data from previous studies. Measures of genetic distance were 0.06 for Nei’s and 0.03 for Cavalli-Sforza’s. The analysis of molecular variance reported no existing variance between populations.
Considering the origin of the animals, their post domestication evolution and the reproductive practices in place, the results do not show genetic differentiation between the two populations for the studied loci.
Introduction
Two coat phenotypes exist in Alpaca, Huacaya and Suri. They differ phenotypically in terms of fleece structure. The first is a classically built fleece: compact, bulky and with high fibre crimp, similar to a merino fleece. The second is a fleece with longer and lustrous fibre organized in defined, hanging locks, more similar to a Lincoln or an Angora type of fleece.
These two phenotypes also differ with regard to fibre structure and characteristics (CitationAntonini et al., 2001; CitationRenieri et al., 2004; CitationFrank et al., 2006). In terms of demography, in the Peruvian population there is a predominance of the Huacaya type which represents 90% of the alpaca fleece processed in Peru (CitationHoffman and Fowler, 1995). This occurs despite the fact that the Suri trait seems to segregate in a way closely similar to a single dominant gene or a haplotype (CitationPonzoni et al., 1997).
A different distribution of phenotypes can be observed in North America, Europe and Australia, where growing interest of both the textile industry and breeder associations for the Suri fibre has led to increasing Suri/Huacaya ratios and often to a paid premium for Suri fibre in the same fineness range as Huacaya (CitationMcGregor, 2006). Over the years this trend has generated two different schools of thought about the way the two phenotypes should be bred, classified and genetically managed. This is mainly true at the level of farming and breeder associations in North America and in Australia who often recommend considering and managing the two phenotypes as two separate breeds in order to keep the Suri line pure (CitationHolt, 2005; CitationWalker, 2006; CitationBaychelier, 2002). This is not supported by present scientific knowledge, which suggests that the phenotype fleece type is a qualitative trait determined by 1 or 2 loci (CitationPonzoni et al., 1997; CitationRenieri et al., 2009b; CitationPresciuttini et al., 2010).
Post domestication evolution and secondary breed structure
CitationWheeler et al. (2006) recently confirmed the unanimous agreement that alpacas were domesticated from vicuñas (Vicugna vicugna mensalis) but the species’ post domestication evolution has been strongly affected by historical events. In fact, its evolution is divided into three main periods: i) pre-Conquest (from the domestication to the arrival of the conquerors); ii) the great Conquest crisis (lasting only a few decades, but with devastating consequences); and iii) the subsequent development (new expansion of domesticated camelids to the present day). The potential existence of breeds in the first period is difficult to study and remains unknown; however, it is important to stress that the Conquest caused a drastic decline in population size (90% according to CitationFlores Ochoa, 1982), the geographic marginalization of the animals and the loss of any reproductive barrier. The complete disruption of the breeding system led to the mixing of species because four species co-existed within the same area: llamas, alpacas, vicuna and guanaco. The genetic consequences of these events were drastic and permanently changed the animals in comparison to the preceding specialization of pre-Conquest times. In this context, natural selection regained a dominant role over artificial selection and the specialization that had been established was lost. Since the Conquest, there has been a steady and slow demographic increase. At present, the demographic structure of the Andean alpaca population seems to be based on primary populations, also called primitive breeds, in which natural selection is favored over artificial selection. The mating system is panmictic and the phenotypic variation inside the flocks is high (CitationMason 1973; CitationDenis 1982). For the moment, secondary standardized breeds do not seem to exist, apart from some experiences on very large private farms (Pacomarca, Mallkini). Nevertheless, the Andean alpaca population is subject to substantial genetic erosion through the diffusion of the full white phenotype. Suri is considered a phenotype with reduced fitness; for this reason, it is generally reared separately from Huacaya (CitationRenieri et al., 2009a).
Population genetics by means of microsatellite analysis
A 10 microsatellite markers panel has been used by the ARI (Alpaca Registry Inc.) since 1998, mainly for parentage verification, and has found other applications throughout the years in other fields such as genome mapping, population structure and comparable genome analysis, as reviewed by CitationMunyard et al. (2009). The continuous and ongoing effort in sequencing the alpaca genome will rapidly lead to a much larger set of markers, as demonstrated by a number of studies published in the past decade (CitationObreque et al., 1999; CitationPenedo et al., 1999; CitationMcPartlan et al., 1998). CitationReed and Chaves (2008) report an additional 1516 potential loci by blasting bos taurus CitationSSRs and Munyard et al. (2009) have recently found a set of 9 tetranucleotide markers. Some of these markers have already been used to calculate genetic distances among different species of South American Camelids (CitationWheeler et al., 2006; CitationBustamante et al., 2002). Since CitationGoldstein et al. (1995) evaluated their use for the calculation of genetic distances, they have been used in an increasingly large number of species.
Given the unusual post domestication evolutionary history of the species and the increasing interest in Suri fibre, the aim of this study was, therefore, to use a microsatellite panel to study the genetic distance between Suri and Huacaya alpacas and to assess the amplitude of genetic variability in the Peruvian alpaca population. The studied population belongs to a germplasm established approximately 20 years ago in Quimsachata (Puno province, Peru) and has been bred separately according to their coat type since then.
Materials and methods
Material collection and sample structure
Alpaca blood samples were collected in spring 2008 within a larger sampling program including skin biopsies and fibre. Animals were kept and managed in the Illpa-Puno Experimental Station in Quimsachata (Puno province, Peru) at an altitude of approximately 4200 m a.s.l. The animals belonged to two geographically separate and phenotypically pure flocks (one Suri and one Huacaya) that have been managed and bred separately since the alpaca germplasm was created at the experimental station 20 years ago. It is important to emphasize that common breeding practices in local Peruvian communities involve rearing Suri and Huacaya animals separately.
Blood samples were taken from a subset of 65 non-related animals selected for the microsatellite analysis. The sample was structured in order to include an equal number of Suri and Huacaya individuals and to respect the sex ratio as much as possible. It included all the available males (n=15) and a subset of females (n=50): in total 32 Huacaya (7 males and 25 females) and 33 Suri (8 males and 25 females).
For convenience and due to the lack of basic facilities, blood was collected by spotting a total of 100 µL of blood on Whatman FTA Nucleic Acid Collection cards (# WB120205). Possibly due to the non-sterile and difficult conditions of facilities on the Peruvian plateau, not all samples allowed a sufficient quality/quantity of DNA to be amplified for all microsatellites, and in order to minimize the number of missing data, the dataset was rearranged to obtain a final number of 49 individuals: 10 males (5 Huacaya and 5 Suri) and 39 females (19 Huacaya and 20 Suri), and 13 microsatellites; LCA 19 was not included in the analysis.
Microsatellite amplification details
All samples were processed in Italy and genomic amplification was carried out by LGS genetic laboratories (Cremona, Italy). The markers, dyes utilized, primer sequences, multiplex associations and allele sizes for the 14 microsatellites are shown in . The panel has been optimized to be amplified in two multiplex reactions of 7 primer pairs each.
Table 1 Markers and multiplex reaction data.
Amplifications were carried out in 20 µL reactions with final concentrations of 0.2 µM of each primer, 1X Ampli-Taq Gold Buffer, 1.5 mM MgCl2, 200 µM of each dNTP and a total of 2 units of Ampli-Taq Gold (#4398833, Applied Biosystems, Monza, italy). After a first denaturation step at 94°C for 10 min, reactions were cycled 30 times (45 s at 94°C; 30 s at 58°C; 30 s at 72°C) with a final 10 min elongation step at 72°C.
Software and statistical analysis
All microsatellite data were first checked with the software micro-checker (CitationOosterhout et al., 2004) in order to spot null alleles and wrong size detections. The statistical analysis of the microsatellite data for the genetic variability measures, including the analysis of molecular variance (AMOVA) and the principal coordinate analysis (PCA), was performed using the latest version of the software Genalex 6.3 (CitationPeakall and Smouse, 2006), while the Excel Microsatellite Toolkit (CitationPark 2001) was used for calculating the polymorphism information content (PIC) for each allele.
A series of indices and parameters were calculated, such as allele frequencies and number of alleles, number of effective alleles, private alleles, expected and observed heterozygosis and fixation indices. The Fixation index (Wright’s inbreeding coefficient) was calculated using the software Genalex as (He - Ho)/He. CitationArcos-Burgos and Muenke (2002) gives an exhaustive review of this measure which, when applied to microsatellite data, describes the probability that a given locus becomes fixed. Populations showing high levels of homozygosity will have a Fixation index significantly different from zero.
All these parameters were calculated for the whole dataset and for the two populations: Suri and Huacaya. In order to assess if there were statistically significant differences between these results, the null hypothesis was tested by means of one way analysis of variance (ANOVA) for observed heterozygosis (Ho), expected heterozygosis (He), Fixation index (F), number of effective alleles (Ne) and PIC and by means of a non-parametric Mann- Whitney Test for number of alleles (Na).
The statistical population genetics package Fstat 2.9.3.2 (CitationGoudet, 1995) was used to calculate deviation from Hardy-Weinberg equilibrium (HWE) and genotypic disequilibrium among loci applying a strict Bonferroni correction for multiple comparisons. The test for HWE and the test for linkage disequilibrium were carried out using 1300 randomizations: 1% table wide level was significant.
Cavalli-Sforza’s chord distance and Reynolds-Weir Cockerham distance were calculated using Gendist, an application of the software package Phylip version 3.69 (CitationFelsenstein, 1989). The first measure is assuming a stepwise mutation model in an infinite allele model with equilibrium between mutation and genetic drift, whereas the second and the third measures are dimensional models assuming only genetic drift.
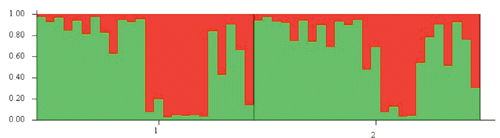
Finally, the genetic structure of the sample was investigated with the Structure 2.3.3 software (CitationPritchard, 2000) and the indentification of the most likely number of clusters (K) was made by the Evanno method (CitationEvanno, 2005) using the online version of Structure Harvester (CitationEarl, 2011). The burning period was set to 50,000 and repetitions of the MCMC chain to 106; the ancestry model chosen was the admixture model. Four replicates for each tested value of K (1–6) were performed.
Results
Hardy-Weinberg equilibrium and linkage disequilibrium
A number of indices have been calculated only on 12 microsatellites, excluding YWLL43, which is linked to the X sexual chromosome. Only one locus (LCA37) was found not to be in Hardy-Weinberg equilibrium after strict Bonferroni correction, showing excess of homozygosis (P<0.05). There was no difference in results when the sample was split into two different populations.
The test for genotypic disequilibrium between pairs of loci showed 9 loci associations out of 65 to be in some degree of linkage disequilibrium. The loci LCA8, LCA66 and LCA65 appear in 8 of the 9 associations showing linkage disequilibrium.
Heterozygosis, polymorphism information content and fixation index
When the dataset was considered as a single population, the average Ho for the 12 markers was high (Ho=0.766; SE=0.044), extremely close to the average He (He=0.769; SE=0.033), and the unbiased expected heterozygosis (UHe=0.778; SE=0.033), with an overall average fixation index of 0.004 (SE 0.036). The high mean number of alleles (Na=9.667; SE=0.77), effective alleles (Ne=4.89; SE=0.39) and the low fixation indices (F) confirm such high values of heterozygosis and genetic variability. The polymorphism information content (PIC) for each locus is in line with previous findings and ranges from 0.411 for locus YWLL46 to 0.826 for locus YWLL44. shows Ho, He, UHe, PIC and F for each locus and as a mean for all loci with the relative standard errors.
Table 2 Population genetic parameters for a single population.
There was no significant difference in results when the sample was analyzed as two separate populations (Huacaya and Suri). The null hypothesis was tested for Ho (P=0.69), He (P=0.61), F (P=0.95), Ne (P=0.69), PIC (p=0.61) and Na (P=0.37). Genetic variability parameters for the two populations are listed in . A number of private alleles were detected between the two phenotypes and a summary is given in according to loci and phenotypes.
Table 3 Population genetic parameters for the two populations.
Table 4 List of private alleles in each population.
Genetic distance, AMOVA, PCA and genetic structure
Genetic distance calculated by Pairwise Population Matrix of Nei’s Genetic Distance and Unbiased Nei’s Genetic Distance (CitationNei, 1978) were 0.062 and <0.0001, respectively. When calculated as the Cavalli-Sforza’s chord distance (CitationCavalli-Sforza and Edwards, 1967) results showed 0.03. Reynolds-Weir Cockerham distance (CitationReynolds et al., 1983), suggested to be more precise in the calculation of genetic distances between closely related species and breeds (CitationLaval et al., 2002), was 0.04.
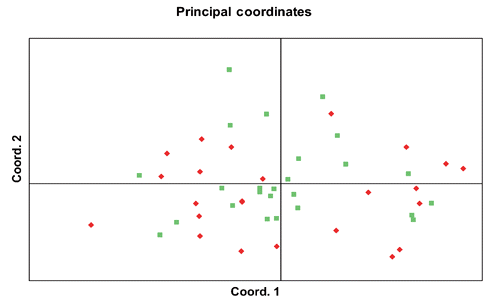
The PCA calculated on the distance matrix among the individuals in graphically shows how samples from the Suri and Huacaya datasets overlap and do not segregate into different groups. The first 3 dimensions of the PCA explain 64.64% of the total variance. All the variance observed in the two populations with the AMOVA test, calculated both by Fst and Rst values, was due to variation within populations (100%) and not to variation between populations (0%). The software Structure rendered a maximum likelihood for K=2 both with the classic method by CitationPritchard (2000) and with that by CitationEvanno (2005), therefore suggesting a possible dual ancestry for the analyzed sample. However, individuals from both phenotypes were assigned in equal proportions to the two clusters without a clear-cut distinction between the two groups, as shown in .
Discussion
In terms of genetic variability within the Peruvian alpaca sample analyzed, all parameters and findings, such as number of alleles (Na=9.667; SE=0.772), number of effective alleles (Ne=4.89; SE=0.388), observed and expected heterozygosis (Ho=0.766, SE=0.044; He=0.769, SE= 0.033), show that the Peruvian population is still conserving high genetic variability and does not show any sign of artificial selection pressure for the studied loci. The low fixation indices for these loci confirm this interpretation of the data (F=0.004; SE=0.036) and suggest that the microsatellite panel used is suitable for genetic diversity studies. In comparison with the data collected and analyzed by the ARI since 1998 and by several other authors, the sample in this study shows the presence of a high number of alleles, matching the whole allelic range described in previous publications (CitationObreque et al., 1999; CitationPenedo et al., 1999; CitationMcPartlan et al., 1998). Values for the polymorphic information content (PIC=0.746, SE=0.033) were also in line with previous findings. Only one locus (YWLL46) showed a PIC value less than 0.7 (PIC=0.411), which reflects the lower than average Na and Ne found for this specific locus (Na=6; Ne=1.753) in previous studies. Nevertheless, this value is higher than that from previous bibliographic data (CitationLang et al., 1996).
There was no significant difference between the two phenotypic groups when these parameters were evaluated separately. In terms of genetic distance and differentiation between the two phenotypes the PCA analysis shows no separate segregation or grouping of Suri and Huacaya individuals.
To our knowledge, no previous studies have investigated genetic distance between Suri and Huacaya alpaca by means of codominant markers, although microsatellite markers have been used in the past to investigate the phylogeny of a wider group of South American Camelids (CitationWheeler et al., 2006, CitationBustamante et al., 2002).
When ANOVA was carried out considering the two phenotypes as two separate populations, it clearly identified the source of all variance in the component within populations, excluding any source of variance to be found between populations. This result is supported by Nei’s index of genetic distance, Cavalli-Sforza’s chord distance and Reynolds-Weir Cockerham distance, which also show no differentiation between the two populations. There are two important factors to be taken into consideration while interpreting these results. Firstly, the germplasm established 20 years ago at the experimental station of Quimsachata is not subjected to genetic selection and was created for the sole purpose of conserving the genetic diversity of the species. Secondly, although Suri and Huacaya alpacas at the experimental station have been bred and managed separately since the creation of the germplasm, the time interval of 20 years is unlikely to generate genetic differentiation between the two phenotypes, especially considering the absence of selection, the reproductive physiology of the species and its generation time (CitationMason, 1973).
Conclusions
Given these considerations, if a secondary breed structure had been present within the species at the time of the creation of the germplasm, it would have been preserved by the breeding practices in place at the experimental station. Nevertheless, the data obtained from the 13 loci suggest no genetic divergence between the two phenotypes and do not support the idea of two distinct populations of Peruvian Suri and Huacaya alpacas. Furthermore, the two phenotypes have similar genetic parameters in terms of allelic frequencies and genetic variability, showing high values both in terms of allelic richness and heterozygosis.
Acknowledgment:
the authors would like to thank the LGS laboratories for carrying out the multiplex amplifications and band scoring.
References
- AntoniniM. PerdomiciF. CatalanoS. FrankE.N. GonzalesM. HickM.V.H. CastrignanòF. 2001 Cuticular cell mean scale frequency in different types of fleece of domestic South American camelids (SAC) EAAP Publ 105 110 116
- Arcos-BurgosM. MuenkeM. 2002 Genetics of population isolates Clin. Genet 61 233 247
- BaychelierP. 2002 What is a pure Suri? Alpacas Australia 39 30 33
- BustamanteA.V. ZambelliA. De LamoD.A. von ThungenJ. Vidal-RiojaL. 2002 Genetic variability of guanaco and llama populations in Argentina Small Ruminant Res 44 97 101
- Cavalli-SforzaL.L. EdwardsA.W. 1967 Phylogenetic analysis. Models and estimation procedures Am. J. Hum. Genet 19 233 257
- DenisB. 1982 Consequences génétiques de l'evolution des races INRA Publ 20 12 18
- EarlD.A. 2011 Structure Harvester v0.6.6 Available from: http://users.soe.ucsc.edu/~dearl/software/structureHarvester/
- EvannoG. RegnautS. GoudetJ. 2005 Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study Mol. Ecol 14 2611 2620
- FelsensteinJ. 1989 Notices; PHYLIP Phylogeny Inference Package (ver. 3.2) Cladistics 5 164 166
- Flores OchoaJ.A. 1982 Causas que originaron la actual distribución espacial de las Alpacas y Llama Universidad de San Antonio Abad, Cusco, Peru, Revista del Museo e Instituto de Arqueología 23 223 250
- FrankE.N. HickM.V.H. GaunaC.D. LamasH.E. RenieriC. AntoniniM. 2006 Phenotypic and genetic description of fibre traits in South American domestic camelids (llamas and alpacas) Small Ruminant Res 61 113 129
- GoldsteinD.B. LinaresA.R. Cavalli-SforzaL.L. FeldmanM.W. 1995 An Evaluation of Genetic Distances for Use With Microsatellite Loci Genetics 139 463 471
- GoudetJ. 1995 FSTAT, ver. 1.2: A Computer Program to Calculate F-Statistics J. Hered 86 485 486
- HoffmanE. FowlerM.E. 1995 The alpaca book Clay Press Herald, CA, USA
- HoltC. 2005 The Spin on Suris - and how they differ from Huacayas Alpacas Australia 47 36 46
- LangK.D.M. WangY. PlanteY. 1996 Fifteen polymorphic dinucleotide microsatellites in llamas and alpacas Anim. Genet 27 293
- LavalG. San CristobalM. ChevaletC. 2002 Measuring genetic distances between breeds: use of some distances in various short term evolution models Genet. Sel. Evol 34 481 507
- MasonI.L. 1973 The role of natural and artificial selection in the origin of breeds of farm animals Z. Tierz. Zuchtungsbio 90 229 244
- McGregorB.A. 2006 Production, attributes and relative value of alpaca fleeces in southern Australia and implications for industry development Small Ruminant Res 61 93 111
- McPartlanH.C. MatthewsM.E. RobinsonN.A. 1998 Alpaca microsatellites at the VIAS A1 and VIAS A2 loci Anim. Genet 29 158 159
- MunyardK.A. LedgerJ.M. LeeC.Y. BabraC. GrothD.M. 2009 Characterization and multiplex genotyping of alpaca tetranucleotide microsatellite markers Small Ruminant Res 85 153 156
- NeiM. 1978 Estimation of average heterozygosity and genetic distance from a small number of individuals Genetics 89 583 590
- ObrequeV. MancillaR. Garcia-HuidobroJ. CothranE.G. HinrichsenP. 1999 Thirteen new dinucleotide microsatellites in Alpaca Anim. Genet 30 397 398
- OosterhoutC.V. HutchinsonW.F. WillsD.P.M. ShipleyP. 2004 Micro-checker: software for identifying and correcting genotyping errors in microsatellite data Mol. Ecol. Notes 4 535 538
- ParkS.D.E. 2001 Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection PhD. Diss University of Dublin Ireland
- PeakallR. SmouseP.E. 2006 Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research Mol. Ecol. Notes 6 288 295
- PenedoM.C. CaetanoA.R. CordovaK. 1999 Eight microsatellite markers for South American camelids Anim. Genet 30 166 167
- PonzoniR.W. HubbardD.J. KenyonR.V. TuckwellC.D. McGregorB.A. HowseA. CarmichaelI. JudsonG.J. 1997 Phenotypes resulting from Huacaya by Huacaya, Suri by Huacaya and Suri by Suri crossings 136 139 Proc. 12th Nat. Conf. of the Association for the Advancement of Animal Breeding and Genetics Armidale, Australia
- PresciuttiniS. ValbonesiA. ApazaN. AntoniniM. HuancaT. RenieriC. 2010 Fleece variation in alpaca (Vicugna pacos): a two-locus model for the Suri/Huacaya phenotype BMC Genet 11 70 77
- PritchardJ.K. StephensM. DonnellyP. 2000 Inference of population structure using multilocus genotype data Genetics 155 945 959
- ReedK.M. ChavesL.D. 2008 Simple sequence repeats for genetic studies of alpaca Anim. Biotechnol 19 243 309
- RenieriC. AntoniniM. FrankE. 2004 Fibre recording systems in camelids CardellinoR. RosatiA. MosconiC. ICAR Technical Series no 11 131 143
- RenieriC. FrankE.N. RosatiA.Y. AntoniniM. 2009a Definición de razas en llamas y alpacas Animal Genetic Resources Information 45 45 54
- RenieriC. ValbonesiA. La MannaV. AntoniniM. AsparrinM. 2009b Inheritance of Suri and Huacaya type of fleece in Alpaca Ital. J. Anim. Sci 8 83 91
- ReynoldsJ. WeirB.S. CockerhamC.C. 1983 Estimation of the coancestry coefficient: basis for a short-term genetic distance Genetics 105 767 779
- WalkerD. 2006 The Suri Alpaca: A Unique Breed Purelysury Herdsire Magazine 1 98 100
- WheelerJ.C. ChikhiL. BrufordM.W. 2006 Genetic Analysis of the Origins of Domestic South American Camelids ZederM.A. BradleyD.G. EmshwillerE. SmithB.D. Documenting domestication: new genetic and archaeological paradigms University of California Press Berkeley, CA, USA 331 343