Abstract
A 14.2 kDa protein isolated from Bali cattle (Bos sondaicus/javanicus) saliva has been reported to have a bactericidal activity. The aim of this study was to analyse the nature of a 14.2 kDa protein using single dimension (1-D) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and matrix-assisted laser desorption ionization time-of-flight tandem mass spectrometer (MALDI TOF/TOF). The protein was isolated by means of polyethylene glycol (PEG)/sodium sulfate aqueous two-phase system, and then determined by 12.5% SDS-PAGE. A band of 14.2 kDa was sliced and analysed by MALDI TOF/TOF mass spectrometer using a 5800 proteomics analyzer. Mascot search and National Center for Biotechnology Information (NCBI) Blast search revealed that the spot of 1-D SDS-PAGE consisted of three proteins: zymogen granule protein 16 homologue B, pancreatic adenocarcinoma upregulated factor-like, and prolactin-inducible protein homologue precursor. The three proteins have been observed in Bos taurus and other species such as mouse. The actual nature of the proteins and their function in Bali cattle (Bos sondaicus/javanicus), as well as the connection with the evolution of bovines need further analysis.
Introduction
Saliva is one of the most important body fluid because its position and function is directly at the forefront, dealing with various elements of pathogenic opportunities, before the pathogens go further into the animal’s body through the oral cavity. Consequently, besides supplying its components involved in the process of food digestion, saliva also plays an important role in maintaining the body’s immunity by providing multiple immunogenic components (Malamud et al., Citation2011). In addition, saliva can also be used as a diagnostic medium for non-invasive examination of the animal condition (Kaufman and Lamster, Citation2002; Lamy and Mau, Citation2012).
Studies on saliva have been conducted much more on human saliva, while they have just recently begun to develop on ruminants (Beauchemin et al., Citation2008; Lamy et al., Citation2009, Citation2010; Wheeler et al., Citation1998, Citation2002, Citation2007, Citation2011; Lamy and Mau, Citation2012). There are many different types of proteins in the ruminant saliva with quite a large range of molecular mass reported elsewhere (Lamy et al., Citation2009). However, the saliva proteome information in cattle (bovine), especially in Bali cattle (Bos sondaicus/javanicus) is still limited.
A protein with an estimated molecular mass of about 14.2 kDa, determined by a single dimension (1D) SDS-PAGE, has been isolated from Bali cattle (Bos sondaicus/javanicus) saliva. From the molecular weight characteristics, and its ability to lyse Staphylococcus aureus, it was suggested that the protein was included in the lysozyme-like proteins (Depamede et al., Citation2012).
Bali cattle are native of Indonesia and derived from domesticated banteng, Bos javanicus, Bos banteng (Nijman et al., Citation2003; Mohamad et al., Citation2009). They are one of the main pedestals of beef cattle, and are exploited as a source of labor to plow the land in Indonesian rural areas. Compared to those from other breeds – although they are fed on poor-quality fodder –, they have several advantages, such as high fertility, remarkable resistance to heat stress as well as to most diseases (McCool, Citation1992). The superior properties of Bali cattle are thus very suitable for tropical regions such as Indonesia and may be for other tropical countries too. In 2010, there were approximately 3.271 million Bali cattle in Indonesia, representing more than 27% of the total cattle population (General Directorate of Livestock, Citation2010; Purwantara et al., Citation2012).
In order to determine the 14.2 kDa protein or peptide, we carried out a further study based on the proteomic analysis approach using 1-D SDS-PAGE and MALDI TOF/TOF mass spectrometer. These methods had been used for proteomic analysis with satisfactory results (Garcia et al., Citation2005; Schipper et al., Citation2007; Lamy et al., Citation2009; Lisitsa et al., Citation2010; Petushkova et al., Citation2012). The information reported in this study is expected to provide additional information about salivary proteome in cattle, as well as to explore the uniqueness of Bali cattle as the native cattle of Indonesia.
Materials and methods
Collection of saliva
Mixed saliva was obtained from oral cavity of 4 healthy, non-pregnant and non-lactating female Bali cattle (Bos sondaicus/javanicus) by means of sponge gags (Muscher et al., Citation2010; Yisehak et al., Citation2011). The cattle were raised in the Teaching Farm of the Faculty of Animal Sciences, Mataram University, Indonesia. The animals were fed 25-30 kg King grass (Pennisetum purpureum) supplemented with 4-5 kg concentrate (67% corn, 30% soybean meal, 1.5% mineral mix, and 1.5% calcium) per head, twice a day, and with water ad libitum. Upon collection, the saliva was centrifuged 3000 x g for 5 min at 4°C (Depamede et al., Citation2012). Supernatant was collected, aliquoated and kept at -60°C until used for further analysis.
Sample preparation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis
Proteins of the saliva were separated by means of polyethylene glycol (PEG)/sodium sulfate aqueous two-phase system as mentioned by Su and Chiang (Citation2006) and Depamede et al. (Citation2012), with slight modifications. The stock solution of PEG 6000 (50%, w/w) and sodium sulfate (30%, w/w) was prepared in double distillated water (ddW). The polymers, buffer, salt and ddW were mixed prior to the addition of saliva. The pH of the system was 9 and the total weight was 10 g. The phases were mixed gently for 60 min at room temperature followed by centrifugation at 1500 x g for 20 min still at room temperature. The formed fractions were extensively dialysed separately against ddW. The protein concentration was determined by Bradford reagent (Sigma, St. Louis, MO, USA) using hen white eggs lysozyme (Calbiochem, La Jolla, CA, USA) as a standard.
A SDS-PAGE (Nacalai Tesque, Kyoto, Japan; Laemmli, Citation1970) was used in this study. One hundred micrograms of each fraction (40 μL) were mixed with 10 μL of 5 x SDS-PAGE loading buffer (0.5 M Tris, pH 6.8, 10% SDS, 38% glycerol, and 0.1% bromophenol blue). The samples were immediately heated to 95°C for 5 min, allowed to cool to room temperature. Each sample was loaded onto a 10% SDS-PAGE separation gel (size: 50 mm x 80 mm x 1 mm, Mini V80-10; Life Technologies, Carlsbad, CA, USA). Electrophoresis was carried out at 45 mV until the bromophenol blue front reached the bottom of the gel. The proteins were stained with commassie brilliant blue (CBB) Stain One (Nacalai Tesque) according to the manufacturer’s procedure. Following protein staining, the band corresponding to the molecular weight of around 14 kDa (molecular weight marker PageRuler; Thermo Scientific, Waltham, MA, USA) was cut and put in a microtube containing 50 μL deionised water before MALDI TOF/TOF mass spectrometer was performed.
Matrix-assisted laser desorption ionization time-of-flight tandem mass spectrometer
The protein sample was reduced, alkylated and trypsin digested. Peptides were extracted according to standard techniques (Bringans et al., Citation2008) and analysed by MALDI TOF/TOF mass spectrometer using a 5800 Proteomics Analyzer (AB Sciex, Framingham, MA, USA). Spectra were analysed to identify protein of interest using Mascot sequence matching software (Matrix Science, Boston, MA, USA) with Ludwig NR Database.
Search parameters used were MS/MS ion search, with carbamidometyl as fixed modification, oxidation as variable modification, and monoisotopic with unrestricted protein mass as mass value. Peptide and fragment mass tolerance were ±0.4 Da (mix missed cleavages= 1, and number of quires=40).
Results and discussion
In the present study we attempted to identify proteins/peptide contained in the 14 kDa correspondence band () using MALDI TOF/TOF mass spectrometer. The protein was reported to have bactericidal activity (Depamede et al., Citation2012), still unresolved for its proteomic. The 14 kDa band was selected, excised, and analysed by MALDI TOF/TOF mass spectrometer. By using this approach and using Mascot search, three main protein hits were identified with sequence ID of G3MZ19, F1N1Z8, and F1MCV8 (). Mascot search results also revealed that the protein sequence coverage was 48, 25 and 13% of G3MZ19, F1MCV8 and F1N1Z8, respectively.
Searching in UniProtKB/TrEMBL showed that G3MZ19 – observed in Bos taurus (bovine) – belongs to the uncharacterised protein with the nominal mass (Mr) 17.708 kDa, and with the gene name BT.106170 (G3M19_BOVIN; Zimin et al., Citation2009). Although G3MZ19 was categorised as uncharacterised protein, NCBI Blast search of G3MZ19 showed 100% maximum identity with HRPE773-like and the predicted zymogen granule protein 16 homologue B both for bovine Bos taurus, with accession numbers gi|296473588|DAA15703.1 and gi|359079571|XP_002697975.2, respectively.
Additionally, from the UniProt 12_11, it was mentioned that the sequence ID G3MZ19 is a member of the mannose-binding protein or mannose-binding lectin (MBL) family. Mannose-binding lectin is known to give an important contribution to innate immune defense (van Asbeck et al., Citation2008; Lambourne et al., Citation2009). Although less information is available regarding MBL and its role in the saliva of cattle, especially the Bali breed, the presence of MBL-like family in the saliva of Bali cattle possibly contributes to the innate immunity that works together with epithelial barriers around the oral cavity as the first line protection system against pathogens.
The second Mascot hit of sequence ID F1N1Z8 – which also belongs to the uncharacterised protein – was categorised as a member of the Jacalin-like superfamily. The sequence ID F1N1Z8 has a molecular mass around 22.091 kDa of Bos taurus and its gene name is BT.45760 (Zimin et al., Citation2009). Blast results showed that the F1N1Z8 sequence for Bos taurus was 100% maximum identity to pancreatic adenocarcinoma upregulated factor-like, with the accession number gi|296473587|DAA 15702.1. Even though the Mascot hit positioned the sequence ID F1N1Z8 in rank number two, its molecular mass 22.091 kDa was quite distinct from the molecular mass of protein attempted to be identified in this study, i.e. 14.2 kDa (Depamede et al., Citation2012).
The third protein hit was the sequence ID F1MCV8, which belongs to prolactin-inducible protein (PIP) homologue (fragment) with molecular mass around 9.007 kDa, and its gene ID is 613853 PIP. It was reported that F1MCV8 belongs to seminal vesicle autoantigen (SVA); this family consists of seminal vesicle autoantigen and PIP with maximum identity 99% to prolactin-inducible protein homologue precursor (Bos taurus) >gi|465766 with the accession number gi|124249234|NP_001074 382.1.
The PIP is a prolactin and androgen regulated product of human breast tumor cell line T-47D (Mirels et al., Citation1998). It is also known as gross cystic disease fluid protein-15 (GCDFP-15), which associates with secretory cell differentiation and is believed to originate from a limited set of tissues, including breast and salivary glands. PIP/GCDFP-15 has been reported to be expressed in rat salivary glands (Mirels et al., Citation1998). Expression of GCDFP-15/PIP homologue was also reported in mouse salivary gland, although its function is still doubtful (Lamy et al., Citation2010; Lamy and Mau, Citation2012), and the expression PIP homologue was also reported in Bos taurus cattle (Zimin et al., Citation2009). In this present study, PIP homologue was observed in the saliva of Bali cattle (Bos sondaicus or Bos javanicus).
From the above-mentioned search results, it can be clearly seen that the analysis of a sliced 14.2 kDa 1D SDS-PAGE band of Bali cattle saliva using MALDI TOF/TOF mass spectrometer resulted in 3 main proteins of 9.007 kDa, 17.708 kDa and 22.091 kDa. According to Lokhov et al. (Citation2004) and Lisitsa et al. (Citation2010), a mixture of 2-5 proteins was commonly observed in the SDS-PAGE slice band when analysed using peptide mass fingerprinting. Similar results were also observed by Petushkova et al. (Citation2012): separation of protein by SDS-PAGE resulted in more than one protein per gel band.
In this study, the slicing gel used was of one slice from 1-D SDS-PAGE gel of about 5x3x1 mm size. Hence, it can be understood that the three different molecular weight peptides, which were likely to come, were fragmentations of the three identified proteins.
As previously reported by Depamede et al. (Citation2012), a protein from Bali cattle saliva, with Mr 14.2 kDa in SDS-PAGE gel was able to inhibit Staphylococcus aureus. The results of the present study emphasise that the 14.2 kDarelated proteins in Bali cattle saliva possibly are not a single protein, but rather two or three proteins. Of the three proteins, based on their molecular mass, the protein with ID F1MCV8 (9.007 kDa) or PIP homologue and G3MZ19 (17.708 kDa) or zymogen possibly are closely related to the 14.2 kDa band identified in this study as well as by Depamede et al., (Citation2012), while F1N1Z8 (22,091 kDa) was quite distinct. Lamy et al. (Citation2010) reported that the mass of 14.4 kDa protein isolated from 12% SDS-PAGE spot of mouse whole saliva and analysed with mass spectrometry was identic to PIP homologue. The results of Lamy et al. (Citation2010) strengthen our result that the 14.2 kDa Bali cattle saliva protein was closely identic to PIP homologue. Furthermore, observation of the bactericidal activity of the 14.2 kDa proteins (Depamede et al., Citation2012) also strengthens the similarity of these proteins with PIP, since PIP was reported to have affinity to Staphylococcus mutant in human saliva (Ambatipudi et al., Citation2010). Furthermore, Schenkels et al. (Citation1997) also reported that PIP might modulate the colonisation of oral and non-oral bacteria such as those of the genera Streptococcus as well as Gemella, and Staphylococcus hominis isolated from human skin and ear canal. Since the three proteins were observed in Bos taurus as well as in other mammals such as rats, mice, and ovine, it would be interesting to check whether proteins can be used for evolution studies on Bali cattle (Bos sondaicus/javanicus) and ruminants in general.
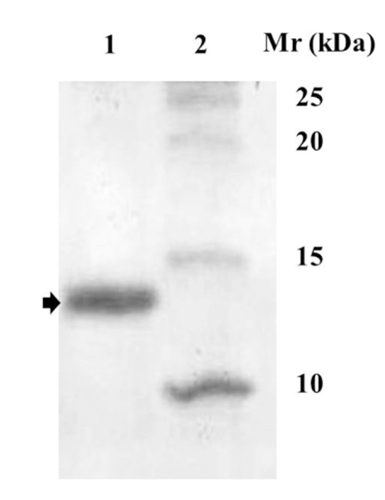
Table 1. Mascot search results of protein spot (single dimension 12.5%, sodium dodecyl sulphate-polyacrylamide gel electrophoresis of a 14.2 kDa isolated from Bali cattle saliva) identification by matrix-assisted laser desorption ionization time-of-flight tandem mass spectrometry.
Conclusions
Proteomic analysis of a 14.2 kDa protein isolated from Bali cattle saliva using 1-D SDS-PAGE gel and MALDI TOF/TOF mass spectrometer shown that the protein consisted of 3 main proteins with molecular mass varying between 9.007 and 22.091 kDa. To reveal more in depth the real nature and the role of the three proteins in Bali cattle saliva, a further study needs to be done. Similarly, a deeper characterisation study on the proteins should also be carried out using 2-D SDS-PAGE and MALDI TOF/TOF mass spectrometer, since, as presented in this study, redundancy results were observed from one slice of 1-D SDS-PAGE gel.
Acknowledgements
This study was supported by a grant from the General Directorate of Higher Education, Ministry of Education and Culture, Republic of Indonesia (Research competency grant no. 126/SP2H/PL/Dit. Litabmas/III/2012). The author is grateful to Dr. Dahlanuddin who provided Bali cattle as part of an Australian Centre for International Agricultural Research (ACIAR)-funded collaborative project (LPS 2008- 038), as well as to Mr. Suparman, Mr. Khalid and Mr. Fahrul Irawan for their technical assistance.
References
- AmbatipudiK.S. HagenF.K. DelahuntyC.M. HanX. ShafiR. HryhorenkoJ. GregoireS. MarquisR.E. MelvinJ.E. KooH. YatesJ.R.3rd 2010. Human common salivary protein 1 (CSP-1) promotes binding of Streptococcus mutants to experimental salivary pellicle and glucans formed on hydroxyapatite surface. J. Proteome Res. 9:6605-6614.
- BeaucheminK.A. EriksenL. NørgaardP. RodeL.M. 2008. Short communication: salivary secretion during meals in lactating dairy cattle. J. Dairy Sci. 91:2077-2081.
- BringansS. EriksenS. KendrickT. GopalakrishnakoneP. LivkA. LockR. LipscombeR. 2008. Proteomic analyses of the venom of Heterometrus longimanus (Asian black scorpion). Proteomics 8:1081-1096.
- DepamedeS.N. AsriN. JulisaniahN.I. SuryadiB.F. KisworoD. 2012. Isolation and partial purification of lysozyme from saliva of Bali cattle (Bos sondaicus) using an aqueous mixture of polyethylene glycol (PEG) with sodium sulfate. Afr. J. Biotechnol. 11:1977-1980.
- GarciaB.A. SmalleyD.M. ChoH.J. ShabanowitzJ. LeyK. HuntD.F. 2005. The platelet microparticle proteome. J. Proteome Res. 4:1516-1521.
- General Directorate of Livestock, 2010. Statistik Peternakan 2010. General Directorate of Livestock, Ministry of Agriculture of the Republic of Indonesia ed., Jakarta, Indonesia.
- KaufmanE. LamsterI.B. 2002. The diagnostic applications of saliva: a review. Crit. Rev. Oral. Biol. M. 13:197-212.
- LaemmliU.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685.
- LambourneJ. AgranoffD. HerbrechtD. TrokeP.F. BuchbinderA. WillisF. Letscher-BruV. AgrawalS. DoffmanS. JohnsonE. WhiteP.L. BarnesR.A. GriffinG. LindsayJ.A. HarrisonT.A. 2009. Association of mannosebinding lectin deficiency with acute invasive aspergillosis in immunocompromised patients. Clin. Infect. Dis. 49:1486-1491.
- LamyE. da CostaG. SantosR. Capela e SilvaF. PotesJ. PereiraA. CoelhoA.V. Sales BaptistaE. 2009. Sheep and goat saliva proteome analysis: a useful tool for ingestive behavior research? Physiol. Behav. 98:393-401.
- LamyE. GraçaG. da CostaG. FrancoC. Capela e SilvaF. BaptistaE.S. CoelhoA.V. 2010. Changes in mouse whole saliva soluble proteome induced by tannin-enriched diet. Proteome Sci. 8:65.
- LamyE. MauM. 2012. Saliva proteomics as an emerging, non-invasive tool to study livestock physiology, nutrition and diseases. J. Proteomics 75:4251-4258.
- LisitsaA.V. PetushkovaN.A. ThieleH. MoshkovskiiS.A. ZgodaV.G. KaruzinaI.I. ChemobrovkinA.L. SkipenkoO.G. ArchakovA.I. 2010. Application of slicing of one-dimensional gels with subsequent slice-by-slice mass spectrometry for the proteomic profiling of human liver cytochromes P450. J. Proteome Res. 9:95-103.
- LokhovP.G. TikhonovaO.V. MoshkovskiiS.A. SerebriakovaM.V. MaksimovB.I. ToropiguineI.Y. ZgodaV.G. GonovorunV.M. ArchakovA.I. 2004. Database search post-processing by neural network: advanced facilities for identification of components in protein mixtures using mass spectrometric peptide mapping. Proteomics 4:633-642.
- MalamudD. AbramsW.R. BarberC.A. WeissmanD. RehtanzM. GolubE. 2011. Antiviral activities in human saliva. Adv. Dent. Res. 23:34-37.
- McCoolC. 1992. Buffalo and Bali cattle-exploiting their reproducive behaviour and physiology. Trop. Anim. Health Pro. 24:165-172.
- MirelsL. HandA.R. BraninH.J. 1998. Expression of gross cystic disease fluid protein-15/prolactininducible protein in rat salivary glands. J. Histochem. Cytochem. 46:1061-1071.
- MohamadK. OlssonM. van TolH.T. MikkoS. VlamingsB.H. AnderssonG. Rodríguez-MartínezH. PurwantaraB. PalingR.W. ColenbranderB. LenstraJ.A. 2009. On the origin of Indonesian cattle. PLoS One 4:e5490.
- MuscherA.S. SchröderB. BrevesG. HuberK. 2010. Dietary nitrogen reduction enhances urea transport across goat rumen epithelium. J. Anim. Sci. 88:3390-3398.
- NijmanI.J. OtsenM. VerkaarE.L.C. de RuijterC. HanekampE. OchiengJ.W. ShamshadS. RegeJ.E.O. HanotteO. BarwegenM.W. SulawatiT. LenstraJ.A. 2003. Hybridization of banteng (Bos javanicus) and zebu (Bos indicus) revealed by mitochondrial DNA, satellite DNA, AFLP and microsatellites. Heredity 90:10-16.
- PetushkovaN.A. LarinaO.V. ChernobrovkinA.L. TrifonovaO.P. KisrievaY.S. LisitsaA.V. KaruzinaI.I. ArchakovA.I. 2012. Optimization of the SDS-PAGE gel slicing approach for identification of human liver microsomal proteins via MALDI-TOF mass spectrometry. J Proteomics Bioinform. 5:40-49.
- PurwantaraB. NoorR. AnderssonG. Rodriguez-MartinezH. 2012. Banteng and Bali cattle in Indonesia: status and forecasts. Reprod. Domest. Anim. 47:2-6. (abstr.).
- SchenkelsL.C. Walgreen-WeteringsE. OomenL.C. BolscherJ.G. VeermanE.C. Nieuw AmerongenA.V. 1997. In vivo binding of the salivary glycoprotein EP-GP (identical to GCDFP-15) to oral and non-oral bacteria. Detection and identification of EP-GP binding species. Biol. Chem. 378:83-88.
- SchipperR.G. SillettiE. VingerhoedsM.H. 2007. Saliva as research material: biochemical, physicochemical and practical aspects. Arch. Oral. Biol. 52:1114-1135.
- SuC.-K. ChiangB.H. 2006. Partitioning and purification of lysozyme from chicken egg white using aqueous two-phase system. Process Biochem. 41:257-263.
- van AsbeckE.C. HoepelmanA.I.M. ScharringaJ. HerpersB.L. VerhoefJ. 2008. Mannose binding lectin plays a crucial role in innate immunity against yeast by enhanced complement activation and enhanced uptake of polymorphonuclear cells. BMC Microbiol. 8:229.
- WheelerT.T. HaighB.J. BroadhurstM.K. HoodK.A. MaqboolN.J. 2011. The BPI-like/ PLUNC family proteins in cattle. Biochem. Soc. T. 39:106-111.
- WheelerT.T. HaighB.J. McCrackenJ.Y. WilkinsR.J. MorrisC.A. GrigorM.R. 2002. The BSP30 salivary proteins from cattle, LUNX/PLUNC and von Ebner’s minor salivary gland protein are members of the PSP/LBP superfamily of proteins. Biochim. Biophys. Acta 1579:92-100.
- WheelerT.T. HaighB.J. WilkinsR.J. McCrackenR.J. MorrisC.A. 1998. A candidate gene marker for bloat susceptibility in cattle? Proc. New Zeal. Soc. An. 58:10-12.
- WheelerT.T. HoodK.A. MaqboolN.J. McEwanJ.C. BingleC.D. ZhaoS. 2007. Expansion of the bactericidal/permeability increasinglike (BPI-like) protein locus in cattle. BMC Genomics 8:75.
- YisehakK. BeckerA. BelayD. BoschG. HendriksW.H. ClaussM. JanssensG.P.J. 2011. Salivary amino acid concentrations in zebus (Bos indicus) and zebu hybrids (Bos indicus × Bos taurus) fed a tannin-rich diet. Belg. J. Zool. 141:93-96.
- ZiminA.V DelcherA.L. FloreaL. KelleyD.R. SchatzM.C. PuiuD. HanrahanF. PerteaG. Van TassellC.V. SonstegardT.S. MarçaisG. RobertsM. SubramanianP. YorkeJ.A. SalzbergS.L. 2009. A wholegenome assembly of the domestic cow, Bos taurus. Genome Biol. 10:R42.