Abstract
Number and population size of local chicken breeds in Italy is considered to be critical. Molecular data can be used to provide reliable insight into the diversity of chicken breeds. The first aim of this study was to investigate the maternal genetic origin of five Italian local chicken breeds (Ancona, Livorno, Modenese, Romagnola and Valdarnese bianca) based on mitochondrial DNA (mtDNA) information. Secondly, the extent of the genetic diversity, population structure and the genetic relationships among these chicken populations, by using 27 microsatellite markers, were assessed. To achieve these targets, a 506 bp fragment of the D-loop region was sequenced in 50 chickens of the five breeds. Eighteen variable sites were observed which defined 12 haplotypes. They were assigned to three clades and two maternal lineages. Results indicated that 90% of the haplotypes are related to clade E, which has been described to originate from the Indian subcontinent. For the microsatellite analysis, 137 individual blood samples from the five Italian breeds were included. A total of 147 alleles were detected at 27 microsatellite loci. The five Italian breeds showed a slightly higher degree of inbreeding (FIS=0.08) than the commercial populations that served as reference. Structure analysis showed a separation of the Italian breeds from the reference populations. A further sub-clustering allowed discriminating among the five different Italian breeds. This research provides insight into population structure, relatedness and variability of the five studied breeds.
Introduction
Attention and awareness to genetic conservation has significantly increased in recent years (Allendorf and Luikart, Citation2007). Preservation of genetic variability plays a crucial role in animal science because its decline may reduce populations’ ability to adapt to environmental changes (Lande, Citation1988). Moreover, autochthonous breeds might be an important resource for research purposes and future breeding programmes.
In Italy, the number of native chicken breeds has suffered a dramatic decline leading to the current critical situation. Zanon and Sabbioni (Citation2001) reported the presence in Italy, in the last fifty years, of 90 rural poultry breeds (9 ducks, 11 guinea fowls, 53 chickens, 5 gooses and 12 turkeys): 61.0% of these breeds are extinct, 13.3% are endangered, and only 6.7% are involved in conservation programmes. On the other hand, hybrids based on only few specialized chicken lines provided by globally acting breeding companies are used for industrial production. In Italy, conservation programmes of local chicken breeds are already in place namely: in Veneto region for Ermellinata di Rovigo, Robusta Maculata, Robusta Lionata, Pépoi and Padovana (Baruchello and Cassandro, Citation2003), in Emilia Romagna region for Modenese and Romagnola (Zanon et al., Citation2006) and in Tuscany for Valdarnese bianca (Gualtieri et al., Citation2006).
In this study, five Italian chicken breeds were studied; Ancona from the Marche region, Livornese bianca and Valdarnese bianca, both from Tuscany, Modenese and Romagnola from the Emilia-Romagna region. Ancona breed is renowned as a good layer (about 280 eggs/year) of white shelled eggs and has yellow skin (Mugnai et al., Citation2009), while Livornese bianca (Leghorn Italian type) is supposedly related to the worldwide spread commercial White Leghorn layers (FAO, Citation2010). Valdarnese bianca shows white feathers and dark yellow shank and can be considered as the only traditional Italian meat-type chicken breed (Marelli et al., Citation2006), even the productive performance is far from being economically sustainable when compared to commercial broiler lines. Modenese and Romagnola breeds are two light breeds of Mediterranean-type known to produce eggs and meat for the rural family. The five studied breeds are not used for commercial purposes (Mugnai et al., Citation2009; Sabbioni et al., Citation2006).
The aim of this study is to provide information on the genetic structure and origin of these breeds. In the absence of comprehensive breed characterization data and documentation of the origin of breeding populations, molecular marker information provide the most reliable estimates of genetic diversity within and between a given set of populations. Nonetheless, molecular data should be combined with other information (i.e., adaptive, productive and reproductive performance, extinction probability) in the process of decision-making. Molecular markers can be applied to investigate genetic relationships among populations within a species. In this context, mitochondrial DNA (mtDNA) sequence polymorphism and autosomal microsatellites are two marker types, which have been widely used. Several authors analysed the mtDNA D-Loop region to assess phylogenetic relationships and maternal origin of different chicken populations (Storey et al., Citation2012; Mwacharo et al., Citation2011, Muchadeyi et al., Citation2008; Fu et al., Citation2001). Microsatellite markers have already been successfully applied in different studies to measure the genetic variability among local chicken breeds (Eltanany et al., Citation2011; Mtileni et al., Citation2011; Muchadeyi et al., Citation2007; Hillel et al., Citation2003).
This study provides some first insights into the genetic diversity of the above-mentioned Italian chicken breeds including their unknown genetic origin, the differentiation among them and their present level of diversity. The lack of historical information as well as pedigree data justifies the use of molecular data. For this purpose, sequences of the mitochondrial D-Loop region and microsatellite loci have been analysed with different statistical methods to obtain the most relevant genetic information.
Materials and methods
Animal sampling and DNA extraction
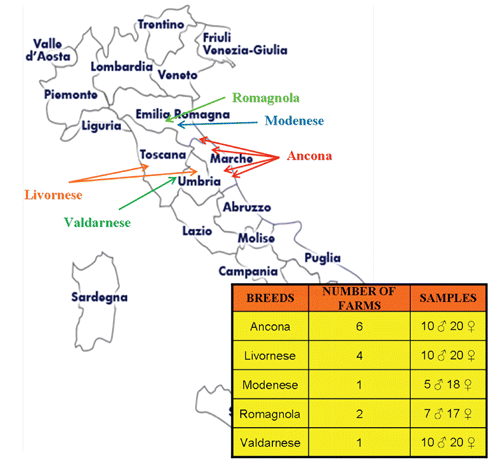
A total of 137 blood samples (2 mL from wing vein of each animal collected in vacutainer tubes, containing EDTA as anticoagulant) were randomly collected from five Italian local chicken breeds: 30 Ancona (AN), 30 Livornese bianca (LI), 23 Modenese (MO), 24 Romagnola (RO), 30 Valdarnese bianca (VA) of both sexes. These breeding animals were selected from different farms to avoid sampling of closely related individuals and to collect a representative sample of each breed. shows the geographical areas, the number of farms and individuals included in the sampling. For VA and MO breeds, a preliminary screening of the farms was carried out to avoid the inclusion of animals, which did not fit to the morphological standard of the breed. As a result, only one farm was suitable for each of these two breeds. Whole blood was stored at -20°C until DNA extraction. DNA was isolated using the GenElute Blood Genomic DNA kit (Sigma Aldrich, St. Louis, MO, USA) and stored at 4°C until genotyping.
Reference populations
Six populations (30 individuals of each population) were used as reference populations. Data were taken from previous studies using the same microsatellite loci that were made available for this project (Muchadeyi et al., Citation2007, Mtileni et al., Citation2011). These populations consisted of broiler dam (BRD) and sire (BRS) lines, two brown-egg layers (BLA and BLC) and two white-egg layers (LSS and WLA). The LSS is an experimental White Leghorn line maintained at the Institute of Farm Animal Genetics (FLI) in Germany as a conservation flock (Hartmann, Citation1997). The other populations are commercial lines.
Mitochondrial DNA analysis
A subset of 50 DNA samples of the five Italian breeds under study was randomly chosen (10 samples for each breed). In relation to the complete mitochondrial sequence of chickens (accession number NC007236; Nishibori et al., Citation2005), mtDNA amplification was performed from nucleotide position (np) 16,750 to np 522 including part of the D-loop region. PCR amplification was performed in a 25 μL volume with 3 mM MgCl2, 50 mM of each dNTP, 1 mM of each primer and 1 unit of Taq® DNA Polymerase (Sigma Aldrich, St. Louis, MO, USA), using a Biometra TGradient 96 Thermocycler at the following conditions: initial denaturation step of 5 min at 95°C, 35 cycles of 30 s at 95°C, 30 s at the 60°C, 75 s at 72°C, and a final extension of 5 min at 72°C. The PCR products were purified using an ExoSAP-IT Purification Kit (USB Corp., Cleveland, OH, USA) and were sequenced with fluorescently labeled primers. The PCR products were sequenced at the Sequencing and Functional Genomics Service (Universidad de Zaragoza, Spain) by means of a Applied Biosystems 3730xl DNA analyzer. A fragment of 506 base pairs in size (from np 1 to np 506 of complete chicken mitochondrial sequence) were used for analysis. Sequences were aligned using the software Sequencher™ 4.10 (Gene Codes Inc., Ann Arbor, MI, USA).
Indexes such as haplotype diversity (Hd), nucleotide diversity (π) and average number of nucleotide differences (k) were estimated by DnaSP 5.10.01 software (Librado and Rozas, Citation2009). ARLEQUIN 3.1 software was applied to carry out a hierarchical analysis of molecular variance (AMOVA) in order to analyze the partitioning of genetic diversity within and among the five Italian chicken breeds (Excoffier et al., Citation2006). The calculations were performed based on 1000 permutations.
Evolutionary relationships of sequences were evaluated through a median-joining network constructed using the software Network 4.6 (www.fluxus-engineering.com). The network also included nine haplotypes representing the main clades (clades A to I) in the Chinese and Eurasian region (Liu et al., Citation2006) as references. Haplotypes from GenBank were aligned with haplotypes observed in this study.
Microsatellite analysis
From a total of 30 microsatellite markers recommended for biodiversity studies of chicken by ISAG/FAO (FAO, Citation2004), 27 markers () were used in this study. The markers were genotyped in standard multiplex PCR amplification using a Biometra TGradient 96. Annealing temperatures were set to values reported at the AVIANDIV (Citation2012) website. Allele calling was adjusted using nine standard DNA samples taken from AVIANDIV project (Weigend et al., Citation1998). Analyses of fragments were performed using an automated DNA sequencer (ABI PRISM 3130xl, Applied Biosystems, Foster City, CA, USA) and the software package GeneMapper version 4.0 (Applied Biosystems).
Analysis of microsatellite genotypes
The observed and expected heterozygosity within breeds was estimated using MICROSATELLITE TOOLKIT 3.1.1 (Park, Citation2001). POPGENE software version 3.2 (Yeh et al., Citation1999) was used to calculate the number of alleles observed at each locus and the mean number of alleles per breed. GENEPOP 4.0 software (Raymond and Rousset, Citation1995) was used to carry out a test for deviation from Hardy-Weinberg equilibrium. A Markov Chain Monte Carlo method (20 batches, 5000 iterations per batch, and a dememorisation number of 10,000) was applied to estimate unbiased exact P-values according to the algorithm described by Guo and Thompson (Citation1992). Weir and Cockerham (Citation1984) estimates of Wright’s fixation indices (FIS, FIT and FST) within and across populations were calculated using FSTAT software version 2.9.3.2 (Goudet, Citation2002). Standard errors were generated by jack-knifing over loci and populations. Fixation index per population (FIS) was estimated, with 1000 bootstraps, using software GENETIX 4.05 (Belkhir et al., 1996-Citation2004). Reynolds weighted genetic distance (Reynolds et al., Citation1983) among the populations was calculated using PHYLIP software 3.6 (Felsenstein, Citation2005). The algorithm implemented in STRUCTURE software, version 2.2 (Pritchard et al., Citation2000) was used to assess genetic clustering of each individual to the various breeds and to reveal possible admixture. The analysis involved an admixture model and correlated allele frequencies.
One hundred independent runs were carried out with 20,000 interactions as burn-in phase followed by 50,000 interactions for sampling from 2≤K≤16 (K=number of assumed clusters). CLUMPP program (Jakobsson and Rosenberg, Citation2007) was used to estimate, per K, the number of identical repeated runs by Greedy algorithm. Further analysis was performed by analyzing the five Italian chicken breeds separately from the population references. The most likely K value describing best the substructure of the populations under study was identified using the dK statistic as described by Evanno et al. (Citation2005). The clustering pattern was visualised using the software DISTRUCT 1.1 (Rosenberg, Citation2004).
Results and discussion
Mitochondrial DNA phylogeny
The present paper represents the first approach to assess the phylogeny of Italian chicken breeds inferred by mtDNA analysis. The sequences of the first 506 bp fragments of the chicken mitochondrial D-loop region were used for analysis. The number of polymorphic sites, the number of haplotypes and haplotype diversity are shown in . In this study, a total of 18 different nucleotide substitutions () were observed forming 12 haplotypes. All the populations, except AN, were polymorphic with a number of haplotypes per population ranging from three (LI, MO and RO) to five (VA). The highest haplotype diversity (Hd), was found in VA chicken (0.8440±0.0800), whereas the lowest value (excluding the monomorphic AN) was observed in RO (0.3780±0.1810). Haplotype diversity estimates of all breeds investigated in this study were similar to what was observed in Hungarian breeds by Revay et al. (Citation2010). The lack of polymorphism in mitochondrial D-loop region of AN breed may be related to higher degree of inbreeding of this breed as also shown later by the microsatellite analysis.
The nucleotide diversity (π) is another parameter than haplotype diversity to estimate the genetic diversity in population and addresses both the frequency of haplotypes and nucleotide differences among haplotypes. The average nucleotide diversity was 0.0045±0.0013 across all the Italian chicken breeds (excluding the monomorphic AN), and ranged from 0.0097±0.0018 in LI to 0.0007± 0.0004 in RO. These values are quite similar to that estimated by Liu et al. (Citation2006) among the clades for chicken sampled in Europe, Middle East, South East and East Asia. Concluding from AMOVA results based on mtDNA sequence polymorphism, the genetic variation among individuals within breeds was 67.83% while genetic variation among breeds (FST) was 32.17% (P<0.001), supporting the hypothesis of a definite separation among the five Italian chicken breeds (). In fact, FST values above 0.25 indicate clear genetic differentiation (Wright, Citation1978).
Median-joining network analysis of the mtDNA D-loop haplotypes using mtDNA sequence polymorphism in the Italian chicken breeds together with reference haplotypes (Liu et al., Citation2006) revealed that Italian breeds clustered in one major and two minor haplogroups, derived from three different lineages (A, B and E) originating from different regions ().
Ninety percent of the animals of the five Italian breeds clustered in the E-lineage derived haplotype LIUE1, while other animals clustered with reference sequences LIUA1 (4%) and LIUB1 (6%), respectively. Interestingly, seven of the eight haplotypes that clustered within haplogroup E were separated from major haplotype E1 by only one mutation. It should be noted that two different sequences from MO and LI were included in haplogroup A (Liu et al., Citation2006). Finally three individual sequences from LI breed clustered in haplogroup B sharing the haplotype with LIUB1.
Haplogroup E has been reported to be widespread in Europe, Middle East and India, while haplogroups A and B are widely distributed in South China and Japan (Liu et al., Citation2006). Other authors (Revay et al., Citation2010; Grimal et al., Citation2011) observed latter haplogroups also in Hungarian and Spanish chicken breeds. In particular, Revay et al. (Citation2010) found two sequences in haplogroup B that were identical to those existing in commercial lines of white egg layer. Therefore, it cannot be excluded that the presence of this haplogroup is a result of introgression from commercial layer lines. No scientific references were found on possible genetic influences of South Eastern Asia chickens to Italian breeds. However, the arrival of these birds to Europe as a result of Romans and Phoenicians activities are documented at least by archaeological finding and can not be disregarded (Mazzorin, Citation2000; Serjeantson, Citation2009).
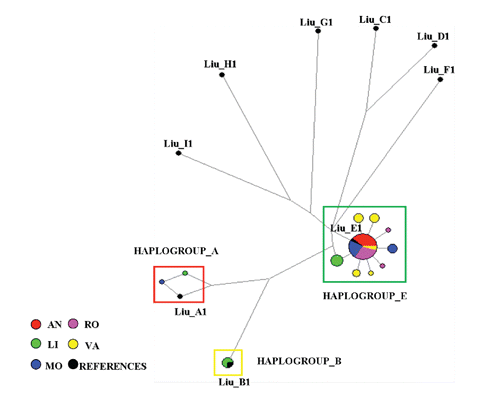
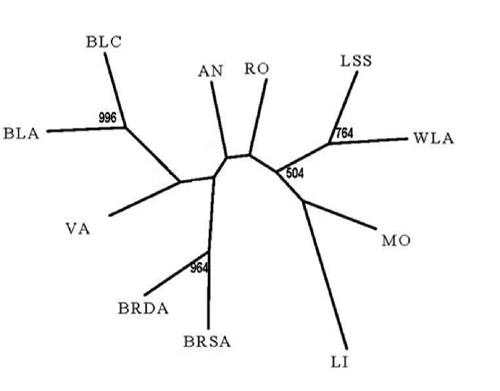
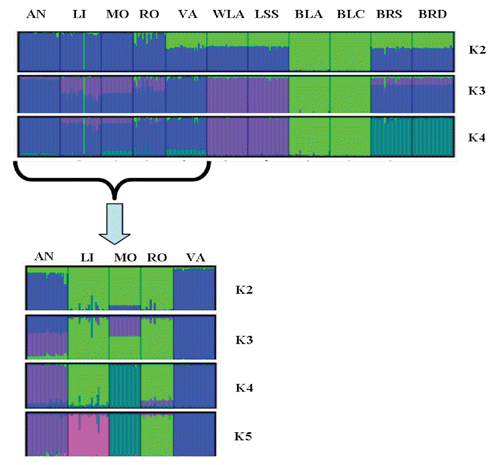
Table 1. Microsatellite loci, chromosomal position, size range and number of alleles observed at each locus.
Table 2. mtDNA diversity indices of the five Italian chicken breeds.
Table 3. Nucleotide polymorphisms observed in the D-loop region of 50 chicken sequences.
Table 4. Results from the hierarchical AMOVA in the five Italian chicken breeds, obtained from mtDNA data.
Table 5. Chicken breeds studied, sample size of each breed, mean number of observed alleles, mean observed and expected heterozygosity, and inbreeding coefficient per breed.
Table 6. Overall population, between-population and within-population inbreeding coefficients and their standard errors of the Italian and commercial populations.
Microsatellites
After the spread of a domestic species in a particular area as a result of one or several immigration events several phenomena, changes in alleles frequencies of autosomal loci usually occur due to several evolutionary forces. Among them, population isolation, natural selection and selection imposed by men for a particular phenotype and especially genetic drift have important effects on allele frequencies of a population and may cause dramatic reductions in the genetic variability and high level of inbreeding (Henson, Citation1992). It is therefore necessary to evaluate the current genetic structure of the autochthonous populations prior to start any conservation or selection programme.
In our study we found 147 alleles in the five Italian breeds across all 27 loci investigated (). The number of alleles at each locus ranged from 2 (MCW0248 and MCW0103) to 11 (MCW0034 and LEI0094) whereas the mean number of alleles per breed () ranged from 2.63 (MO) to 3.67 (VA). These values are similar to those obtained by Zanetti et al. (Citation2010) on a study involving six North Italian chicken breeds (Ermellinata di Rovigo, Robusta Maculata, Robusta Lionata, Pépoi, Padovana and Polverara) using a panel of 20 microsatellite markers, all included in the panel utilized for this study.
It should be noted that all these local breeds are reared in small rural flocks (Dalvit et al., Citation2005). The results show that the genetic diversity is comparable to the diversity found in other European chicken breeds (Granevitze et al., Citation2007). VA displayed the highest value of the observed and expected heterozygosity (0.53 for both of them) while AN and MO the lowest (0.39). The observed and expected values of heterozygosity in each breed showed similar values to that found by Dalvit et al. (Citation2009), Bodzsar et al. (Citation2009) and Granevitze et al. (Citation2007), in different Italian and European poultry breeds, respectively (). A deficiency of heterozygosity (FIS) was observed in both AN (0.19244, P<0.05) and LI (0.10920, oe<0.05) breeds probably due to the mating of related individuals and infrequent exchange of breeding animals among different rural farms (). Observed frequencies of heterozygotes were similar to those expected in MO, RO and VA, and FIS estimates were not significantly different from zero, suggesting that these populations are close to what can be expected under random mating. Therefore these three breeds are well managed in a conservation programme.
The mean FIT, FST and FIS estimates among the five Italian chicken breeds and the six commercial lines respectively, are reported in the . The average inbreeding value at the total sample level (FIT) was 0.349±0.017 (P<0.01) and higher in commercial lines than in Italian breeds. The genetic differentiation (FST) of Italian chicken breeds was lower (0.225±0.019) than the corresponding value of the commercial lines (0.354±0.025), indicating a lower, but still substantial sub-structuring of the Italian breeds. One reason might be the existence of subpopulations within the Italian breeds (Wahlund effect) as samples of each breed were taken from different places (except MO and VA that shown the lowest FIS values). Phylogenetic relationships based on Reynolds genetic distance among the populations were visualised through a Neighbourjoining tree (). The tree showed that the two White Leghorn strains (WLA and LSS) clustered with MO and LI breeds. LI is closely related to the founder breed of White Leghorn used to developed commercial egg layers. The results confirm the common historic origin of White Leghorn strains and LI. As expected, MO and LI appeared in close neighbourhood in the tree because of the historic crossbreeding practices between these two breeds as reported by Mazzon (Citation1932). As stated above, the genetic proximity between MO and LI was also detected in the mitochondrial analysis.
Two more clusters were observed: VA clustered with brown egg layers; genetic introgression of heavier dual-purpose chickens could explain the clustering of VA with brown egg layers (Gualtieri et al., Citation2006, Sacchi, Citation1960). BRDA and BRSA were on one end of the tree, and AN and RO were in a cluster between brown egg layers and white egg layers. Results of STRUCTURE analysis are given in . The analysis was carried out to detect the potential presence of substructures within the breeds. The highest dK values were obtained for K=4. At the lower K values (K=2 and 3) four (BLA, BLC and WLA, LSS) of the six reference populations are separated from the Italian breeds. At K=4, the six commercial lines were divided into three different clusters while the Italian breeds clustered together, even if VA, MO and AN show slightly relation to broilers, and LI and MO to White egg layers as shown in the Neighbour-joining tree. These results may indicate that the five Italian breeds make up a gene pool different from commercial chicken lines, as show the higher FST estimates between the Italian and commercial breeds ().
The five Italian breeds were further subclustered, according to the approach used by Rosenberg et al. (Citation2002), Jakobsson et al. (Citation2008) and Granevitze et al., Citation2009. shows the results of this second step of subclustering. Clustering was carried out from K=2 to K=5. In this approach, the highest dK value was obtained for K=5. At this K-value, the five Italian breeds were discriminated into separate clusters, even if LI and MO are more related to each other than other breeds, as shown in the Neighbour-joining tree. This finding is in agreement with the results of mitochondrial data and FST value. It confirms the genetic differences of the five studied breeds.
Conclusions
Mitochondrial DNA data suggest that the Italian chicken breeds mainly originate from the Indian subcontinent, at least from the maternal lineage standpoint, since most individuals are included in the lineage described by Liu et al. (Citation2006). However, South China and Japan could be a possible origin for the small proportion of birds belonging to the A and B lineages. Another explanation might be crossbreeding with different European breeds.
The results obtained by microsatellite analysis show that the genetic variability of the studied Italian chicken breeds is comparable to other European populations.
Acknowledgments
This research was supported by a scholarship of the Foundation “Ing. Aldo Gini”.
The authors wish to thank Prof. Manuela Gualtieri and Prof. Isabella Romboli for providing the samples of Valdarnese bianca and Livornese bianca breeds and Prof. Francesco Panella for supporting the knowledge of the Italian chicken breeds history.
The authors would also like to acknowledge the use of the General Research Support Service (SAI), Universidad de Zaragoza, Spain.
The authors wish to thank the two anonymous referees for their valuable comments to the manuscript and their constructive suggestions.
References
- AllendorfF.W. LuikartG. 2007. Conservation and the genetics of populations. Blackwell Publ., Malden, MA, USA.
- AVIANDIV, 2012. Available from: http://aviandiv.tzv.fal.de/primer_table.html
- BaruchelloM. CassandroM. 2003. Avicoli veneti. Progetto CO.VA. – Interventi per la conservazione e la valorizzazione di razze avicole locali venete. Available from: http//www.venetoagricoltura.org
- BelkhirK. BorsaP. ChikhiL. RaufasteN. BonhommeF. 2004. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, Université de Montpellier II Ed., Montpellier, France. Available from: http://www.genetix.univmontp2.fr/genetix/genetix.htm
- BodzsarN. EdingH. RevayT. HidasA. WeigendS. 2009. Genetic diversity of Hungarian indigenous chicken breeds based on microsatellite markers. Anim. Genet. 4:516-523.
- DalvitC. De MarchiM. BarcacciaG. ZanonA. SabbioniA. 2005. Genetic similarities among Modenese, Romagnola and three Veneto chicken breeds. pp 106-108 in Proc. 16th Nat. Congr. ASPA, Torino, Italy. Ital. J. Anim. Sci. 4(Suppl.2):106-108.
- DalvitC. ZanettiE. CassandroM. 2009. Estimation of genetic diversity over time in an in-situ marker assisted conservation scheme of local chicken breeds. pp 63-65 in Proc. 18th Nat. Congr. ASPA, Palermo, Italy. Ital. J. Anim. Sci. 8(Suppl.2):63-65.
- EltananyM. PhilippU. WeigendS. DistlO. 2011. Genetic diversity of ten Egyptian chicken strains using 29 microsatellite markers. Anim. Genet. 42:666-669.
- EvannoG. RegnautS. GoudetJ. 2005. Detecting the number of cluster of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14:2611-2620.
- ExcoffierL. LavalG. SchneiderS. 2006. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47-50.
- FAO, 2004. Guideline for development of national management of farm animal genetic diversity. Recommended microsatellite markers. Available from: htpp://dad.fao.org/en/refer/library/guideline/marker.pdf
- FAO, 2010. Domestic animal diversity information system. Available from: http://dad.fao.org/
- FelsensteinJ. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Available from: http://evolution.genetics.washington.edu/phylip.html
- FuY. NiuD. RuanH. 2001. Studies of genetic diversity of Zhejiang native chicken breeds. Acta Genetica Sinica 28:606-613.
- GranevitzeZ. HillelJ. ChenG.H. CucN.T.K. FeldmanM. EdingH. WeigendS. 2007. Genetic diversity within chicken populations from different continents and management histories. Anim. Genet. 38:576-583.
- GranevitzeZ. HillelJ. FeldmanM. SixA. EdingH. WeigendS. 2009. Genetic structure of a wide-spectrum chicken gene pool. Anim. Genet. 40:686-693.
- GrimalA. Viudes de CastroM.P. GómezE.A. GoyacheF. RoyoL.J. 2011. Posible origen materno común de dos poblaciones de gallinas: resultados preliminares del análisis del ADN mitocondrial. pp 482-484 in Proc. 14th Nat. Congr. AIDA on Animal Production, Zaragoza, Spain.
- GoudetJ. 2002. FSTAT 2.9.3.2, a program to estimate and test gene diversities and fixation indices. Available from: http://www.unil.ch/popgen/softwares/fstat.htm
- GualtieriM. PignatelliP. CristalliA. 2006. Pollo di razza Valdarnese bianca. Risorse genetiche animali autoctone della Toscana. ARSIA Ed., Firenze, Italy.
- GuoS.W. ThompsonE.A. 1992. A Monte Carlo method for combined segregation and linkage analysis. Am. J. Hum. Genet. 51:1111-1126.
- HartmannW. 1997. Evaluation of major genes affecting resistance to disease in poultry. World. Poultry Sci. J. 53: 231-251.
- HensonE.L. 1992. In situ conservation of livestock and poultry. Available from: http://www.fao.org/docrep/004/T0559E/T0559E00.htm#TOC>
- HillelJ. GroenenM.A.M. Tixier-BoichardM. KorolA.B. DavidL. KirzhnerV.M. BurkeT. Barre-DirieA. CrooijmansR.P.M.A EloK. FeldmanM.W. FreidlinP.J. Mäki-TanilaA. OortwijnM. ThomsoneP. VignalA. WimmersK. WeigendS. 2003. Biodiversity of 52 chicken populations assessed by microsatellite typing of DNA pools. Genet. Sel. Evol. 35:533-557.
- JakobssonM. RosenbergN.A. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801-1806.
- JakobssonM. ScholzS. W. ScheetP. GibbsJ.R. VanLiereJ.M. FungH-C. SzpiechZ.A. DegnanJ.H. WangK. GuerreiroR. BrasJ.M. SchymickJ.C. HernandezD.G. TraynorB. J. Simon-SanchezJ. MatarinM. BrittonA. van de LeemputJ. RaffertyI. BucanM. CannH.M. HardyJ.A. RosenbergN.A. SingletonA.B. 2008. Genotype, haplotype and copy-number variation in worldwide human population. Nature 451:998-1003.
- LandeR. 1988. Genetics and demography in biological conservation. Science 241:1455-1460.
- LibradoP. RozasJ. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451-1452.
- LiuY.P. WuG.S. YaoY.G. MiaoY.W. LuikartG. BaigM. Beja-PereiraA. DingZ.L. PalanichamyM.G. ZhangY.P. 2006. multiple maternal origins of chickens: Out of the Asian jungles. Mol. Phylogenet. Evol. 38:12-19.
- MarelliS.P. StrillacciM.G. FerranteV. PignatelliP. GualtieriM. Guidobono CavalchiniL. 2006. Genetic variability in Valdarnese Bianca chicken breed using microsatellite markers. World. Poultry Sci. J. 6:207-208.
- MazzonI. 1932. Pollicoltura Padovana. Soc. Coop.Tipografica Ed., Padova, Italy.
- MazzorinJ.D.G. 2000. Introduzione e diffusione del pollame in Italia ed evoluzione delle sue forme di allevamento fino al Medioevo. pp 351-361 in Proc. 3rd Nat. Congr. of Archaeozoology, Siracusa, Italy.
- MtileniB.J. MuchadeyiF.C. MaiwasheA. GroeneveldE. GroeneveldL.F. DzamaK. WeigendS. 2011. Genetic diversity and conservation of South African indigenous chicken populations. J. Anim. Breed Genet. 128:209-218.
- MuchadeyiF.C. EdingH. SimianerH. WollnyC.B.A. GroeneveldE. WeigendS. 2008. Mitochondrial DNA D-loop sequences suggest a Southeast Asian and Indian origin of Zimbabwean village chickens. Anim. Genet. 39:615-622.
- MuchadeyiF.C. EdingH. WollnyC.B.A. GroeneveldE. MakukaS.M. ShamseldinR. SimianerH. WeigendS. 2007. Absence of population substructuring in Zimbabwe chicken ecotypes inferred using microsatellite analysis. Anim. Genet. 38:332-339.
- MugnaiC. Dal BoscoA. CastelliniC. 2009. Effect of rearing system and season on the performance and egg characteristics of Ancona laying hens. Ital. J. Anim. Sci. 8:175-188.
- MwacharoJ.M. BjørnstadG. MobegiV. NomuraK. HanadaH. AmanoT. JianlinH. HanotteO. 2011. Mitochondrial DNA reveals multiple introductions of domestic chicken in East Africa. Mol. Phylogenet. Evol. 58:374-382.
- NishiboriM. ShimogiriT. HayashiT. YasueH. 2005. Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius. Anim. Genet. 36:367-375.
- ParkS.D.E. 2001. Trypanotolerance in West African cattle and the population genetic effects of selection. Degree Diss., University of Dublin, Ireland.
- PritchardJ.K. StephensM. DonnellyP. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945-959.
- RaymondM. RoussetF. 1995. An exact test for population differentiation. Evolution 49:1280-1283.
- RevayT. BodzarN. MobegiV.E. HanotteO. HidasA. 2010. Origin of Hungarian indigenous chicken breeds inferred from mitochondrial DNA D-loop sequences. Anim. Genet. 41:548-550.
- ReynoldsJ. WeirB.S. CockerhamC.C. 1983. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105:767-779.
- RosenbergN.A. 2004. Distruct: a program for the graphical display of population structure. Mol. Ecol. Notes 4:137-138.
- RosenbergN.A. PritchardJ.K. WeberJ.L. CannH.M. KiddK.K. ZhivotovskyL.A. FeldmanM.W. 2002. Genetic structure of human populations. Science 298:2381-2385.
- SabbioniA. ZanonA. BerettiV. SuperchiP. ZambiniE.M. 2006. Carcass yield and meat quality parameters of two Italian autochthonous chicken breeds reared outdoor: Modenese and Romagnola. Paper n. 204 in Proc. 12th Eur. Poultry Conf., Verona, Italy.
- SacchiR. 1960. Il pollo Valdarno ha un avvenire. Avicoltura 11:41-45.
- SerjeantsonD. 2009. Birds. Cambridge University Press, New York, NY, USA.
- StoreyA.A. AthensJ.S. BryantD. CarsonM. EmeryK. deFranceS. HighamC. HuynenL. IntohM. JonesS. KirchP.V. LadefogedT. McCoyP. Morales-MuñizA. QuirozD. ReitzE. RobinsJ. WalterR. Matisoo-SmithE. 2012. Investigating the Global Dispersal of Chickens in Prehistory using ancient mitochondrial DNA signatures. Plos One 7:e39171.
- WeigendS. GroenenM.A.M. Tixier-BoichardM. VignalA. HillelJ. WimmersK. BurkeT. Mäki-TanilaA. 1998. Development of strategy and application of molecular tools to asses biodiversity in chicken genetic resources. Available from: http://aviandiv.tzv.fal.de/
- WeirB.S. CockerhamC.C. 1984. Estimating Fstatistics for the analysis of population structure. Evolution 38:1358-1370.
- WrightS. 1978. Evolution and the genetics of populations, variability within and among natural populations. University of Chicago Press, Chicago, IL, USA.
- YehF.C. YangR.C. BoyleT. 1999. Popgene version 1.31. Microsoft window-based freeware for population genetic analysis. Department of Renewable Resources, University of Alberta Ed., Edmonton, Canada.
- ZanettiE. De MarchiM. DalvitC. CassandroM. 2010. Genetic characterisation of Italian chickens undergoing in situ conservation. Poultry Sci. 89:420-427.
- ZanonA. SabbioniA. 2001. Identificazione e salvaguardia genetica delle razze avicole italiane. Ann. Facoltà Medicina Veterinaria di Parma 21:117-134.
- ZanonA. BerettiV. SuperchiP. ZambiniE.M. SabbioniA. 2006. Psico-chemical characteristics of eggs from two Italian autochthonous chicken breeds: Modenese and Romagnola. Paper n. 203 in Proc. 12th Eur. Poultry Conf., Verona, Italy.