Abstract
An important outcome of intensive worldwide Bovine spongiform encephalopathy (BSE) obtained with the surveillance by The National Creutzfeldt-Jakob Disease Surveillance Unit (http://www.cjd.ed.ac.uk/figures.htm), has been the detection of atypical BSE in cattle. The discovery of a prion protein gene (PRNP) E211K variant in an atypical BSE case is particularly remarkable because it is analogous to the most common pathogenic mutation in humans (E200K), which causes hereditary Creutzfeldt-Jakob disease (CJD). Knowledge of the distribution and frequency of PRNP E211K variants in cattle populations is critical for understanding and managing atypical BSE. This study was carried out to investigate the prevalence of the E211K variant in the South-East Asia bovine populations and in the Japanese cattle breeds. It was discovered that E211K variant was monomorphic for a G allele and the GG genotype in the 745 animals analyzed in this study. Therefore, neither the Bos indicus nor the Bos taurus animals analyzed are presently known to harbor the 211K variant predicting that the number of carriers for this variant will also be vanishingly low.
Key Words:
Introduction
There are three forms of bovine spongiform encephalopathy (BSE) thought to afflict cattle. The first one is classical BSE (cBSE, orally acquired BSE) which is associated with ingestion of BSE-contaminated feedstuffs. The largest cBSE outbreak was first recognized in 1986 in Great Britain and reached the peak in the early 1990’s, when the number of confirmed cases rose to over 30,000 in one year (Anderson et al., Citation1996) and has been identified in 24 additional countries around the world (http://www.oie.int). Consumption of beef from cBSE-affected animals was implicated as the most likely cause of human prion disease called variant Creutzfeldt-Jakob disease (vCJD) (Collinge and Rossor, Citation1996; Bruce et al., Citation1997). After regulations were implemented, the number of BSE cases dropped dramatically (http://www.oie.int) resulting to the reduction of the number of vCJD cases as well (http://www.cjd.ed.ac.uk).
The second and the third forms of BSE are associated with either higher (H-form) or lower (L-form) molecular mass of unglycosylated and monoglycosylated isoforms of prion protein (PrP) apparently higher than the cBSE isolates. These two forms differ from cBSE by displaying a different disease phenotype in the central nervous system and they have not been linked to the consumption of contaminated feed (Nicholson et al., Citation2008; Ritch and Hall, Citation2008). The H- and L- forms of BSE are collectively known as atypical BSE. Polymorphisms of the prion protein gene (PRNP) in the non-coding region in cattle have been identified to modulate expression level and influence susceptibility to cBSE but not atypical BSE (Sander et al., Citation2004; Juling et al., Citation2006). According to Capobianco et al. (Citation2007), atypical BSE arising as both genetic and spontaneous disease, in the context of data indicating L-type BSE can convert to cBSE in mice. Recent evidence suggests that specific bovine PRNP variants may represent genetic risk factors for atypical BSE especially in older cattle (Heaton et al., Citation2008). A PRNP mutation variant causing a glutamic acid (E) to lysine (K) substitution at codon 211 (E211K) is thought to be genetically inherited. This variant was discovered in the H-type 12-year BSE case and her 2-year daughter, which were both heterozygous E (GAA) and K (AAA) at PRNP nucleotides 631-633 (Nicholson et al., Citation2008; ). The normal bovine sequence is homozygous GAA/GAA encoding glutamate (E) in both alleles. This polymorphism is an identical pathogenic mutation at the homologous codon position (E200K) in the human PRNP which has been described as the most common cause of genetic CJD. In the human E200K mutation, the normal GAG (E) is replaced by the AAG (K) (Lee et al., Citation1999). Would this be proved true, is an evidence that BSE may be heritable and confirms the existence of all of the three etiological forms of TSEs (spontaneous, hereditary, infection) in cattle. The atypical BSE has been detected in two animals of B. taurus × B. indicus origin in the United States (Nicholson et al., Citation2008; Heaton et al., Citation2008). However, no parental information has been provided. It is therefore unknown whether the corresponding nucleotide change was inherited or the result of spontaneous mutation. If it was inherited, then the E211K allele may have originated in either a B. taurus ancestor or a B. indicus ancestor (USDA, Citation2006). This study was designed to investigate the prevalence of the E211K variant in the South-East Asia bovine populations, which were mainly Bos indicus and the Japanese cattle breeds which were either B. taurus or crosses of the two species.
Materials and methods
We used the genomic DNA from 745 animals representing six bovine populations from a few South-East Asian countries and six Japanese cattle breeds. Some of these animals were analyzed for PRNP polymorphisms at six different loci by Shimogiri et al. (Citation2010). The genomic DNA was obtained as we described previously (Msalya et al., Citation2009). Primers and restriction enzyme were designed from available sequences (accession number EU557971) by dCaps Finder 2.0 Output and Primer 3 output online (Rozen and Skaletsky, Citation2000). The polymerized chain reaction (PCR) included 40 ng of genomic DNA, 10 pmoles of forward primer BE211K3F CCACCACCAAGGGGGAGAACTTC TTC, 10 pmoles of reverse primer BE211KR CTTGCCCCTCGTTGGTAATA, 200 mmol/l of each of the four dNTPs, 0.5 units of Ex-Taq (TaKaRa, Otsu, Shiga, Japan), 2.0 L of Ex-Taq buffer and filled by 12.3 L of ddH2O. The thermocycling conditions were: an initial denaturation of 1:30 min at 94°C, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s and extension at 72°C for 30s and final extension at 72°C for 5 min. Restricted fragment length polymorphism (PCR-RFLP) was performed by digestion with one unit of recognition sequence TTCGAA of the restriction endonuclease (RE) Bacillus species Bsp 1191 (BstBI, Fermentas) in a reaction mixture including 1.0 L buffer Tango, 9.0 L ddH2O and 5.0 L PCR product followed by incubation at 37°C for 16 h. DNA products were identified by PCR and the alleles were resolved on a 2% agarose gel stained with ethidium bromide. Genotypes were readily distinguished on a 10% polyacrylamide gel (PAGE) by visual inspection of molecular mass verified against a 100-bp DNA marker after staining in ethidium bromide. Alleles were calculated by direct counting.
The experiments in this study comply with the current laws of Japan and did not cause harm to human and animals.
Results and discussion
We investigated the presence of an E211K mutation in six South-East Asian bovine populations and in six Japanese cattle breeds. The electrophoresis photograph showed a straight line bundles close to a 100-bp marker in all genomic DNA samples indicating that GG was the only genotype detected at this locus (). The resulting frequencies were 1.00 for the G allele and 0.00 for the K allele in all animals (). Therefore there were no carriers for the K codon in any animal. None of the B. indicus or B. taurus samples evaluated in this study exhibited an E211K amino acid replacement. This means that the animals analyzed in this study have the normal homozygous GAA/GAA bovine sequence, which codes for E at this locus. The positive control (PC) was a KK genotype sample and was detected at a region higher (120-bp) than GG. No bundle was detected at the negative control (NC).
Recent evidence suggests that specific bovine PRNP variants may represent genetic risk factors for atypical BSE in cattle (Clawson et al., Citation2008). Since the US H-type BSE case was reported to be a crossbred beef cow possibly of the B. indicus and B. taurus animals, emphasis was placed on analyzing cattle with B. indicus and B. taurus germplasm. All of the South-East Asia cattle populations constituted the Bos indicus germline and the Bos frontalis population widely known as mythun in some Asian countries including India, Myanmar and Nepal. Although the effective population size was not known, Gibbs et al. (Citation2009) suggests that cattle in the South-East Asia have undergone a rapid decrease in effective population size from a very large ancestral population, possibly due to domestication and artificial selection. However, all of the Japanese cattle breeds used in this study were modern populations representing the B. taurus and an effective population size of 17.2 was reported in the famous JB cattle (Nomura et al., Citation2001). A few analyses conducted earlier on E211K have indicated that the frequency of G/A (K allele) or A/A variants in cattle may be very low (Heaton et al., Citation2008; Hui et al., Citation2010). This was consistent with this study in which none of the 745 animals involved was a carrier of the K allele. On the other hand, our results may also indicate that carriers of the abnormal bovine K allele are absent or extremely low and that the US’s inherited E211K variant might have caused by accidental mutation. Moreover, the discovery of this variant in the open reading frame of cattle PRNP is challenging and predicts that there might be some more polymorphisms in this region of the gene. Although few atypical BSE cases have been identified worldwide, they are significant because of their possible link to sporadic CJD in humans (Brown et al., Citation2006). It is therefore wise to investigate cattle especially in the PRNP-ORF frequently which might be helpful in controlling the BSE disease.
Conclusions
Knowledge of the distribution and frequency of PRNP E211K variants in cattle populations is critical for understanding and managing atypical BSE. For that matter, we carried out a study to investigate the prevalence of the E211K variant in 745 animals representing the South-East Asia bovine populations and the Japanese cattle breeds and discovered that E211K variant was monomorphic for a G allele and the GG genotype. We therefore agree with the previous suggestion that the number of carriers for this variant will be vanishingly low. It is however recommended to carry out evaluations in unanalyzed populations elsewhere to identify the animals harboring the E211K so as to manage them for future development in the BSE crisis.
The authors wish to thank Dr. Kazuaki Tanaka of Azabu University, Azabu city, Japan and Prof. Takashi Amano of Tokyo University of Agriculture for providing the DNA samples for some of the South-East Asian bovine populations.
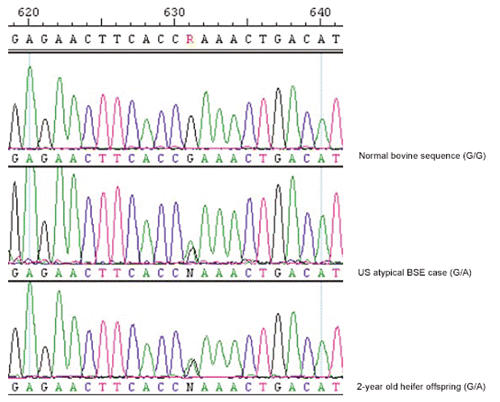
Table 1. Genotypes and allele frequency at the E211K variant.
References
- AndersonR.M. DonnellyC.A. FergusonN.M. WoolhouseM.E.J. WattC.J. UdyH.J. Ma WhinneyS. DunstanS.P. SouthwoodT.R.E. WilesmithJ.W. RyanJ.B. HoinvilleL.J. HillertonJ.E. AustinA.R. WellsG.A. 1996. Transmission dynamics and epidemiology of BSE in British cattle. Nature 382: 779-788.
- BrownP. Mc ShaneL.M. ZanussoG. DetwileL. 2006. On the question of sporadic or atypical bovine spongiform encephalopathy and Creutzfeldt-Jakob disease. Emerg. Infect. Dis. 12 :1816-1821.
- BruceM.E. WillR.G. IronsideJ.W. Mc ConnellI. DrummondD. SuttieA. Mc CardleL. ChreeA. HopeJ. BirkettC. CousensS. FraserH. BostockC.J.1997. Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature 389: 498-501.
- CapobiancoR. CasaloneC. SuardiS. MangieriM. MiccoloC. LimidoL. CataniaM. RossiG. Di FedeG. GiacconeG. BruzzoneM.G. MinatiL. CoronaC. AcutisP. GelmettiD. LombardiG. GroschupM.H. BuschmannA. ZanussoG. MonacoS. CaramelliM. TagliaviniF.2007. Conversion of the BASE prion strain into the BSE strain: the origin of BSE? PLOS Pathog. 3 :e31.
- ClawsonM.L. RichtJ.A. BaronT. BiacabeA.G. CzubS. HeatonM.P. SmithT.P.L. LaegreidW.W. 2008. Association of a bovine prion gene haplotype with atypical BSE. PLoS ONE 3 :e1830.
- CollingeJ. RossorM. 1996. A new variant of prion disease. Lancet 347: 916-917.
- GibbsR.A. TaylorJ.F. Van TassellC.P. BarendseW. EversoleK.A.2009. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324: 528-532.
- HeatonM.P. KeeleJ.W. HarhayG.P. RichtJ.A. KoohmaraieM. WheelerT.L. ShackelfordS.D. CasasE. KingD.A. SonstegardT.S. TassellC.P.V. NeibergsH.L. ChaseC.C.Jr KalbfleischT.S. SmithT.P.L. ClawsonM.L. LaegreidW.W. 2008. Prevalence of the prion protein gene E211K variant in U.S. cattle. BMC Vet. Res. 4 :25.
- HuiZ. Xiao-YanW. WeiZ. Ya-PingZ.2010. Prion protein gene (PRNP) polymorphisms in native Chinese cattle. Genome 53: 138-145.
- JulingR. SchwarzenbacherH. WilliamsJ.L. FriesR.2006. A major genetic component of BSE susceptibility. BMC Biol. 4 :33.
- LeeH.S. SambuughinN. CervenakovaL. ChapmanJ. PocchiariM. LitvakS. QiH.Y. BudkaH. delSer T. FurukawaH. BrownP. GajdusekD.C. LongJ.C. KorczynA.D. GoldfarbL.G.1999. Ancestral origins and worldwide distribution of the PRNP 200K mutation causing familial Creutzfeldt-Jakob disease. Am. J. Hum. Genet. 64 :1063-1070.
- MsalyaG. ShimogiriT. OkamotoS. KawabeK. MinesawaM. NamikawaT. MaedaY.2009. Gene and haplotypes polymorphisms of the Prion gene (PRNP) in Japanese Brown, Japanese native and Holstein cattle. Anim. Sci. J. 80: 520-527.
- NicholsonE.M. BrunelleB.W. RichtJ.A. KehrliM.E.Jr. GreenleeJ.J.2008. Identification of a heritable polymorphism in bovine PRNP associated with genetic transmissible spongiform encephalopathy: evidence of heritable BSE. PLoS ONE 3 :e2912.
- NomuraT. HondaT. MukaiF.2001. Inbreeding and effective population size of Japanese Black cattle. J. Anim. Sci. 79: 366-370.
- RichtJ.A. HallS.M.2008. BSE case associated with prion protein gene mutation. PLoS Pathogens 49 :e1000156.
- RozenS. SkaletskyH.2000. Primer3 on the WWW for general users and for biologist programmers. In:KrawetzS. MisenerS. (eds.) Bioinformatics methods and protocols: methods in molecular biology.Humaua Press, Totowa, NJ, USA, pp 365-386.
- SanderP. HamannH. PfeifferI. WemheuerW. BrenigB. GroschupM. H. ZieglerU. DistlO. LeebT.2004. Analysis of sequence variability of bovine prion protein gene (PRNP) in Germany cattle breeds. Neurogenet. 5: 19-25.
- ShimogiriT. MsalyaG. MyintS. L. OkamotoS. KawabeK. TanakaK. MannenH. MinezawaM. NamikawaT. AmanoT. YamamotoY. MaedaY.2010. Allele distributions and frequencies of the six prion protein gene (PRNP) polymorphisms in Asian native cattle, Japanese breeds, and mythun (Bos frontalis). Biochem. Genet. 48: 829-839.
- United States Department of Agriculture, 2006. Second USDA confirmatory test results positive for BSE. Available from: http://www.usda.gov/wps/portal/usdahome?contentidonly=true&contentid=2006/03/0090.xml
