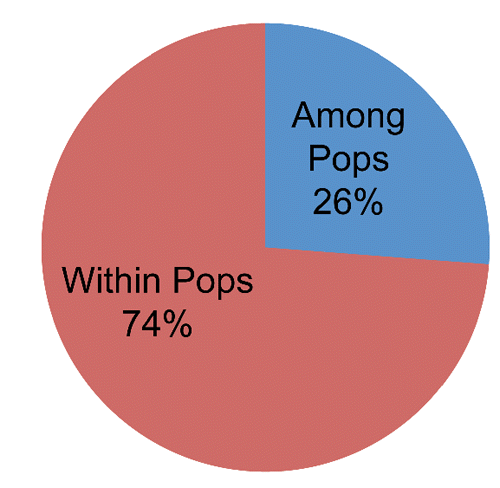Abstract
Since 13rd century, Italian domestic autochthonous donkey population has been characterised by Mediterranean grey mousy cruciate ancestral phenotype, currently typical of Amiata donkey (AD) genetic type. This phenotype persisted up to the 16th century when a marked introduction of Hispanic and French big sized and dark bay or darkish coloured sires occurred. In the context of a safeguard programme of Latial Equide resources, the aim of this research was to evaluate the genetic diversity and similarity between the AD breed and an autochthonous donkey population native from Lazio, the Viterbese donkey (VD), using molecular markers. A total of 135 animals (50 AD and 85 VD) were genetically characterised by using 16 short tandem repeat markers. A high genetic differentiation between populations (FST=0.158; P<.01) and a low between-breeds genetic similarity (0.233±0.085) were observed. Correspondence analysis, the result of STRUCTURE software analysis and analysis of molecular variance would seem to indicate genetically different entities as well. It would be desirable to increase the number of comparison with other breeds to better understand the origin of VD. Moreover, results obtained in this study suggest that the loss of genetic variation observed in VD could mainly derive from unnoticed sub-population structuring (Wahlund effect), rather than to other factors such as inbreeding, null alleles or selection influence.
Introduction
Nowadays in Europe, about 55 breeds or varieties of donkey are still present, 15 of which are present in Italy (FAO, Citation2013). Originally, from the phylogenetic point of view, Italian donkeys were divided into four ancestral breeds: Pugliese, Siciliana, Pantelleria and Sarda (Mascheroni, Citation1929). Besides, within these breeds it was possible to recognise some subdivisions or local breeds; for example, Siciliana breed was divided into two populations, one of the West and the other of the East of the island. The Pugliese breed, the most diffused in Italy in the past, included these populations: Calabrese, Basilicata, Leccese, Martina Franca, Marchigiana and Romagnolo. After a period of abandonment of many donkey populations, during the last few years, due to exploitation of donkey’s products (milk and meat) many local populations are growing in census. Moreover, some small populations are undergoing safeguard programmes (Rizzi et al., Citation2011). The use of donkeys as riding and pack animals has been always important in Central Italy, because of the landscape characterised by rugged hills. In particular, in this area there are two donkey populations, the Amiata donkey (AD) and the Viterbese donkey (VD). Amiata donkey breed was present in the area of the Amiata mountain (a large inactive volcano) since the end of the 19th century and it was characterised by uniform grey mantle with typical striping of the limbs, slim shape, and withers height up to 140 cm. Actually the height of its withers was about 129 cm in females and 132 cm in males (Cecchi et al., Citation2007). After a bottleneck in the years 1945 to 1970 due to the advent of rural motorisation, the population has started to increase again because of specific actions carried out both by private and public organisations. More than 152 farms are active today in Central Italy and they maintain a population of about 600 registered animals (Cecchi et al., Citation2006), with a relatively low genetic variability (Ciampolini et al., Citation2007).
Up to the 18th century, VD was mainly characterised by grey mousy-crusader or dapple gray or sable or dark sable coat, middle size (withers height of 120 cm), rusticity, robustness, and longevity (Mascheroni, Citation1929). In the following years, crosses with Marchigiani or Martinesi stud (Mascheroni, Citation1929; Tortorelli, Citation1973) characterised by dark sable or blackish coat and great size (withers height more than 130 cm) and following selective interventions on the obtained crosses gave rise to some ecotypes denominated Laziale and Ciociaro, which, although characterised by a wide somatic variability, demonstrated to be good sires giving robust pack-mules, particularly suitable for wood hauling (Tortorelli, Citation1973).
Until 2011, VD was considered extinct because it was not registered in Herd Book. From 2001 a safeguard programme of Latial Livestock Resources has been carried out involving VD too. In the context of this programme, thanks to the molecular characterisation, this population was registered in the Zootechnical Book of Equine and Ass populations at limited diffusion with 0009742 (7.05.2012) decree of the Italian Ministry of Agriculture, Food and Forestry Policies with the denomination of Asino Viterbese or Asino di Allumiere. In 2012, 146 heads were estimated distributed throughout about 15 farms.
Genetic typifying is an important preliminary step in any safeguard biodiversity programme (Matassino et al., Citation2010). Literature reports studies on the genetic variability within and between breeds using microsatellite markers in some Spanish breeds (Andaluza, Catalana, Mallorquina, Encartaciones, and Zamorano-Leonesa; Aranguren-Mendez et al., Citation2001, Citation2002), one French donkey breed (Baudet du Poitou; Bellone et al., Citation1998), the Catalonian donkey breed (Jordana et al., Citation1999, Citation2001) and three Croatian donkey populations (Ivankovich et al., Citation2002). In Italy, studies were carried on some donkey Sardinian populations (Cosseddu et al., Citation2001; Colli et al., Citation2013); Martina Franca and Romagnolo breeds (Blasi et al., Citation2005; Colli et al., Citation2013); Ragusano, Pantesco and Grigio Siciliano genetic types (Blasi et al., Citation2005; Guastella et al., Citation2007; Bordonaro et al., Citation2012; Colli et al., Citation2013); Amiata breed (Ciampolini et al., Citation2007; Colli et al., Citation2013) and Asinara (Colli et al., Citation2013).
Given the breeding area of AD and VD and the similarity in the coat colour, it could be argued that AD can be an ancestor of VD. Hence, the aim of this study, inserted in a wide safeguard programme of Equide Latial biodiversity (with particular reference to the Regional Law n. 15 of 1 March 2000), was to contribute to the evaluation of genetic diversity and population genetic structure of VD compared with AD, using 16 microsatellite markers.
Materials and methods
Animals
A total of 135 animals – 50 ADs (15♂♂, 35♀♀) and 85 (34♂♂, 51♀♀) VDs – were sampled from different flocks trying to avoid closely related individuals. In particular, 15 farms located in the provinces of Viterbo, Roma and Frosinone for VD, and 35 farms located in different provinces of Tuscany and Lazio for AD.
DNA, short tandem repeat and statistical analyses
Genomic DNA was extracted from 5 mL of peripheral blood samples and DNA was isolated using GenElute Blood Mammalian Genomic DNA Minipreps Kit® (Sigma-Aldrich, St. Louis, MO, USA). Sixteen microsatellites were investigated (ATH04, HTG07, HTG10, HMS02, VHL20, COR71, ASB23, HMS07, VHL209, COT58, HMS03, HTG04, HTG06, HMS06, HMS045, SGC28) and they were amplified in multiplexes in 4 independent PCR reactions. Primer sequences for the microsatellites are available at the Domestic Animal Diversity Information System web site (FAO, Citation2013). Genotyping was carried out by using an Applied Biosystems 310 DNA sequencer (Applied Biosystems, Foster City, CA, USA). Data analysis was performed using the GENEMAPPER software (ABI, v. 4.0, 2005).
Number of alleles (Na), allelic frequencies, expected and observed heterozigosity were estimated using Genepop v.3.3 (Raymond and Rousset, Citation1995). Exact tests for deviation from the Hardy-Weinberg (HW) equilibrium were performed using the ARLEQUIN package v. 3.11 (Excoffier et al., Citation2005).
The following parameters were computed at the population level by using the programme MolKin (Gutièrrez et al., Citation2005): molecular coancestry coefficients (Caballero and Toro, Citation2002), kinship distance, and F-statistics. The molecular coancestry between two individuals – i and j (fij) – is the probability that two randomly sampled alleles from the same locus in two individuals are identical by state (Caballero and Toro, Citation2002). The molecular coancestry of an individual i with itself is self-coancestry (si), which is related to the coefficient of inbreeding of an individual i (Fi) by the formula Fi=2si–1. In turn, the kinship distance (Dk) between two individuals i and j is Dk=[(si+sj)/2]-fij (Caballero and Toro, Citation2002). MolKin computes within- and between-breed molecular coancestry and Dk by simply averaging the corresponding values for all the within or between-population pairs of individuals.
Genetic similarities were investigated by comparing the individual multilocus genotype of each individual with each other (Ciampolini et al., Citation1995). Genetic similarity is defined as P=A/2L, where P is the proportion of common alleles (A) in relation to the 2L possibilities (L=number of considered loci). The similarities between each pair of individuals were then averaged over the whole population. In addition, the factorial correspondence analysis performed with the option 3D over populations was estimated using Genetix software (Belkhir et al., Citation2000). Moreover, a likelihood test of breed assignment was performed using the ARLEQUIN package v. 3.11 (Excoffier et al., Citation2005).
Further assignment test and analysis of molecular variance were carried using GenAlEx software v. 6.5 (Peakall and Smouse, Citation2012) which offers both frequency-based and distance-based analyses.
Breed differentiation was also investigated using the Bayesian clustering algorithm implemented in the STRUCTURE software v. 2.2 (Falush et al., Citation2007). The test was run for K from 2 to 6 and for each K value five repetitions were performed. The test was carried: i) using an admixture model; ii) using 100.000 burn in period; iii) using 100.000 Markov Chain Monte Carlo iterations; iv) giving no prior information; v) choosing the best fitting K value by the likelihood score [LnP(D)] and Evanno method (Evanno et al., Citation2005).
Results and discussion
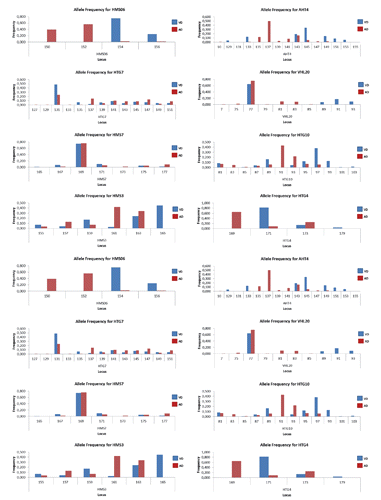
All 16 microsatellite markers resulted to be polymorphic in the whole sample and in each breed. A total of 100 alleles were found in VD and 95 in AD, with Na ranging from 2 to 10 in VD [mean value 6.25±2.57; coefficient of variation (CV)=41%] and from 3 to 13 in AD (mean value 5.94±2.89; CV=49%). The mean effective number of alleles (Ne) was 2.99 (±1.20; CV=40%) in VD and 3.00 (±1.49; CV=50%) in AD. The most polymorphic loci were: HTG7 in both genetic types (13 alleles in AD and 10 in VD), HTG10 (9 alleles in AD and 10 alleles in VD) and AHT4 (10 alleles in AD and 9 alleles in VD); the less polymorphic loci were: HMS06 in both breeds (2 alleles in VD and 3 in AD); HTG4 (3 alleles in both populations), HMN045 (3 alleles in AD and 4 alleles in VD) and VHL209 in AD (3 alleles) ().
Only 10 markers are common to those used by other authors (ATH4, HTG07, HTG10, HMS02, VHL20, HMS07, HMS03, HTG04, HTG06, HMS06) (Aranguren-Mendez et al., Citation2001, Citation2002; Jordana et al., Citation2001; Bordonaro et al., Citation2012; Colli et al., Citation2013); ASB23 marker has also been used by Bordonaro et al. (Citation2012), while Ivankovic et al. (Citation2002) used a completely different set of markers. Ciampolini et al. (Citation2007) used two markers more than that reported in this research (ASB2 and HMS1). So, although a comparison with other breeds can be biased due to the different marker sets used by different authors, we highlight that in terms of mean Na the genetic variability observed in VD was higher than that reported in three Sicilian donkey breeds (Bordonaro et al., Citation2012). On the contrary, AD showed the lowest Na. Furthermore, both populations had an Na lower than that reported by Aranguren-Mendez et al. (Citation2001) on five Spanish donkey breeds (7.0-7.5). In both populations AHT4 and HTG7, markers have the highest Na as reported by Aranguren-Mendez et al. (Citation2001) and by Bordonaro et al. (Citation2012), while HTG4 marker has only three alleles as reported by Bordonaro et al. (Citation2012).
The two populations VD and AD were characterised by a different allelic pattern within locus (). A total of 52 breed-specific alleles were observed (28 in VD and 24 in AD), most of them at a frequency lower than 10%. On the contrary, 7 breed-specific alleles (5 in AD and 2 in VD) had a frequency higher than 20%: allele 137 of ATH4 (50%), allele 231 of HMS02 (42%), allele 169 of HTG04 (65%), allele 150 and 152 of marker HMS06 (39 and 56%, respectively) in AD, and allele 165 of HMS03 (45%) and allele 159 of ASB23 (23%) in VD ().
The significant overall loci FST index was 0.158 (P<.01). This value reveals that 15.8% of the genetic variation observed in the sample is explained by population differences, whereas the remaining is due to the differences within population. This value is generally higher than those observed in studies carried out on European donkey breeds (Arangures-Mendez et al., Citation2002; Ivankovich et al., Citation2002; Bordonaro et al., Citation2012; Colli et al., Citation2013) and in other species such as for example Italian horse breed (Di Stasio et al., Citation2008), Italian cattle breeds (Cecchi et al., Citation2012), and Italian sheep breeds (Ciani et al., Citation2013).
Observed heterozigosity () ranges from a minimum value=0.400 for the locus HMS7 to a maximum value=0.785 for locus ASB23, with an average value of 0.578±0.155 in AD and from 0.380 for locus HMS06 to 0.798 for locus ASB23 in VD with an average value of 0.607±0.162. Expected heterozigosity ranges from a minimum value=0.310 for locus HTG4 to a maximum=0.797 for locus ASB23 with an average value of 0.607±0.163 in VD and from 0.335 for locus VHL209 to 0.874 for locus HTG7 in AD with an average value of 0.602±0.165 (not showed data). The results revealed moderate degree of heterozigosity in the two populations. The expected mean heterozigosity was lower in both populations than that reported in a study on Catalonian donkeys (0.712; Jordana et al., Citation2001), on three donkey populations of the Croatian coastal region (0.66-0.70; Ivankovic et al., Citation2002), and on the Baudet du Poitou donkey population (0.623) (Bellone et al., Citation1998), but higher than that reported on three Sicilian donkey breeds (0.581) (Bordonaro et al., Citation2012).
Of the total 16 analysed microsatellite, 4 markers were in significant HW disequilibrium in VD (AHT4, VHL209, HMS7 and ASB23) and 5 markers in AD (AHT4, HMS2, HTG10, HTG07 and ASB23) (data not shown); the disequilibrium is given by an excess of homozygotes. The polymorphism information content (PIC) per locus showed nine markers with values over 50% in both populations and an average value of 55.52% in AD and 56.81% in VD, respectively (). The PIC was originally introduced by Botstein et al. (Citation1980). It refers to the value of a marker for detecting polymorphism within a population, depending on the number of detectable alleles and the distribution of their frequency and it has been proved to be a general measure of how informative a marker is; the higher is the PIC value, more informative a marker is. In the present study, ATH4 and HTG07 and HTG10 microsatellites appeared informative, as reported also by Bordonaro et al. (Citation2012) on three Sicilian donkey breeds and by Aranguren-Mendez et al. (Citation2001) on five Spanish donkey breeds.
The highest values of FIS was observed in VD (0.070) and the lowest was observed in AD (0.056). Molecular coancestry (fij), Dk and Fi are similar in the two breeds ranging from 0.392 (VD) to 0.397 (AD) for fij, from 0.317 in AD to 0.324 in VD for Dk, and from 0.428 in AD to 0.434 in VD for Fi. These values are so close to each other that the difference could be due to stochastic effects rather than to an actual difference between the two populations.
The genetic similarity within the populations were 0.467 (±0.086) and 0.473 (±0.107) respectively in VD and AD. These values were higher than those reported in other species such as cattle [0.281; D’Angelo et al. (Citation2006); 0.374-0.420; Cecchi et al. (Citation2012)], sheep [0.318-0.370; D’Angelo et al. (Citation2009)], and dog [0.455; Ciampolini et al. (Citation2011); 0.412; Ciampolini et al. (Citation2013)], showing a low genetic variability in the two populations.
Also within-breed mean coancestry and Kinship distances show that both populations are homogeneous; in fact, the values observed in our study for fij and for Fi were clearly greater than that reported in literature on other species such as horse (Marletta et al., Citation2006), sheep (Dalvit et al., Citation2008, Citation2009; D’Angelo et al., Citation2009; Ciani et al., Citation2013) and cattle (Cecchi et al., Citation2012), while Dk was smaller than reported in literature. The observed values might suggest that the low level of genetic variability have arisen as a possible consequence of mating among relatives.
The between-breed Dk and the mean fij values were 0.323 and 0.336, respectively, while the between-breeds genetic similarity was 0.233 (±0.085), showing a low genetic similarity between these two populations.
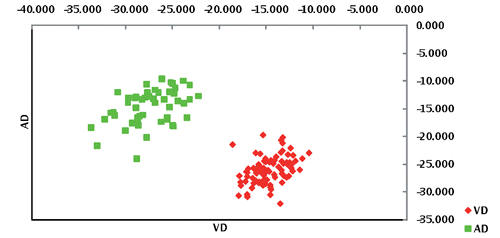
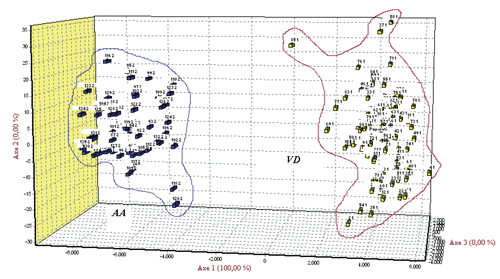
By adopting a likelihood test of population assignment, it emerged that all the animals were correctly allocated to the true breed of origin as showed in , thus highlighting a clear differentiation between the two donkey populations. This result was supported by FCA () and by the bayesian clustering analysis ().
Correspondence analysis highlighted a clear absence of transvariation area between the two genetic types and an apparent greater dispersion of VD population against AD; indeed, the surface leakage concerning the two populations was higher in the VD with a value equal to 1.41 (41%). Analysing the results derived from STRUCTURE () using K=2, it would seem that the two populations are separated in different groups as observed from plot 1 of ; for K=3 and K=4 it would seem that VD population was constituted by two sub-populations. Moreover, for K=5 and K=6 both AD and VD would seem divided into different sub-populations. From LnP(D) showed in and Evanno method reported in the Appendix the best K value is found to be 6; in fact, the K6 plot shows VD population seemingly characterised by more genetic heterogeneity as it was marked by four clusters, while AD population was marked by two clusters.
The results of STRUCTURE software analyses, together with FIS value make us hypothesise the presence of a Wahlund effect that affects the calculation of the inbreeding coefficient.
Even the analysis of molecular variance allows to define those two populations as different; in fact, the 26% (P= 0.001) of molecular variance is explained by the differences between them ().
Conclusions
In the limits of the observation field, the results show that actually a high genotypic diversity between the two populations exists. Indeed, the two populations, although having the same level of heterozygosity, are characterised by quanti-qualitative allelic diversity, consisting into a different allelic pattern within locus. All distance measures (FST and genetic similarities) would seem to indicate genetically different entities. It would be desirable to increase the number of comparison with other breeds to better understand the origin of VD. In particular, research will continue comparing VD with Ragusano, Martina Franca and Asino Romagnolo donkey breeds. Concerning the low genetic variability observed in VD, inbreeding can be one cause but we must remember that there are other factors that can also cause a lack of heterozygotes in the population. Firstly, null alleles (i.e. non-amplifying alleles) may be present and lead to a false observation of excess homozygotes. Despite the fact that pedigrees were not available for analysis, it was not possible to demonstrate the presence of null alleles (usually caused by a mutation in the primerbinding site leading to an allele that will not amplify). Secondly, the presence of population substructure within the breed may lead to a Wahlund effect. The selection influence could not be proved because production data were not available. Results obtained in this study suggest that the loss of genetic variation in VD could mainly derive from unnoticed sub-population structuring (Wahlund effect). It would be very interesting to increase the number of VD samples genotypically typed using a larger number of microsatellite markers. In fact, the analysis of a large number of loci increases the power of detecting population substructure because each locus will contain an independent history of the population depending on the amount of random drift, mutation, and migration that has occurred. An important contribution to better understand its division into sub-populations could be provided using mitochondrial DNA analysis.
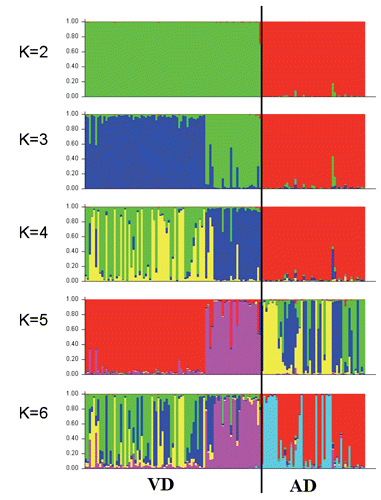
Table 1. Number of alleles, observed heterozigosity, polymorphism information content, and effective allele number within each microsatellite locus in AD, VD and the total.
Table 2. STRUCTURE: likelihood scores.
References
- Aranguren-MandezJ. JordanaJ. GomezM., 2001. Genetic diversity in Spanish donkey breeds using microsatellite DNA markers. Genet. Sel. Evol. 33 :433-442.
- Aranguren-MendezJ. JordanaJ. GomezM., 2002. Genetic conservation of five endangered Spanish donkey breeds. J. Anim. Breed Genet. 119 :256-263.
- BelkhirK. BorsaP. Chikhi Raufaste N. BonhommeF., 2000. Genetix, logiciel sous Windows pour la génétique des populations. Laboratoire Génome et Populations, CNRS UPR 9060, Universitéde Montpellier II ed., Montpellier, France.
- Bellone R.R. Cothran E.G. Ketchum M.S., 1998. Genetic variation in the rare donkey breed, Baudet du Poitou. Anim. Genet. 29(Suppl.1):17.
- BlasiM. PerrottaG. LanzaA. IamartinoD. PillaF., 2005. Genetic diversity in three Italian donkey population assessed by microsatellite markers. Page 127 in Proc. 16th Nat. Congr. ASPA, Torino, Italy. Ital. J. Anim. Sci. 4(Suppl.2):127 (abstr.).
- BordonaroS. Guastell A.M. CriscioneA. ZuccaroA. MarlettaD., 2012. Genetic diversity and variability in endangered Pantesco and two other Sicilian donkey breed assessed by microsatellite markers. TheScientificWorldJo. 2012 :648-427.
- BotsteinD. White R.L. SkolnickM. Davis R.W., 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32 :314-331.
- CaballeroA. Toro M.A., 2002. Analysis of genetic diversity for the management of conserved subdivided populations. Conserv. Genet. 3 :289-299.
- CecchiF. CiampoliniR. CastellanaE. CianiE., 2012. Genetic diversity within and among endangered local cattle breeds from Tuscany (Italy). Large Anim. Rev. 18 :79-85.
- CecchiF. CiampoliniR. CianiE. MatteoliB. MazzantiE. TancrediM. PresciuttiniS., 2006. Demographic genetics of the endangered Amiata donkey breed. Ital. J. Anim. Sci. 5 :387-391.
- CecchiF. CiampoliniR. CianiE. MazzantiE. TancrediM. PresciuttiniS., 2007. Morphological characterization of the Amiata donkey breed through the data reported in the anagraphic register. Page 70 in Proc. 17th Nat. Congr. ASPA, Alghero, Italy. Ital. J. Anim. Sci. 6(Suppl.1):70 (abstr.).
- CiampoliniR. CecchiF. BramanteA. CasettiF. PresciuttiniS., 2011. Genetic variability of the Bracco Italiano dog breed based on microsatellite polimorphysm. Ital. J. Anim. Sci. 10 :267-270.
- CiampoliniR. CecchiF. MazzantiE. CianiE. TancrediM. De Sanctis B., 2007. The genetic variability analysis of the Amiata donkey breed by molecular data. pp 78-80 in Proc. 17th Nat. Congr. ASPA, Alghero, Italy. Ital. J. Anim. Sci. 6(Suppl.1):78-80.
- CiampoliniR. CecchiF. PaciG. PolicardoC. SpaternaA., 2013. Investigation on the genetic variability of the American Pit Bull Terrier dogs belonging to an Italian breeder using microsatellite markers and genealogical data. Cytol. Genet. 47 :217-221.
- CiampoliniR. Moazami-GoudarziK. VaimanD. DillmannC. MazzantiE. Foulley J.L. LevezielH. CianciD., 1995. Individual multilocus genotypes using microsatellite polymorphisms to permit the analysis of the genetic variability within and between Italian beef cattle breeds. J. Anim. Sci. 73 :3259-3268.
- CianiE. CiampoliniR. Andrea M.S. CastellanaE. CecchiF. IncoronatoC. D’AngeloF. AlbenzioM. PillaF. MatassinoD. CianciD., 2013. Analysis of genetic variability within and among Italian sheep breeds reveals population stratification and suggests the presence of a phylogeographic gradient. Small Ruminant Res. 112 :21-27.
- ColliL. PerrottaG. NegriniR. BombaL. BigiD. ZambonelliP. Verini Supplizzi A. LiottaL. , Ajmone-MarsanP., 2013. Detecting population structure and recent demographic history in endangered livestock breeds: the case of the Italian autochthonous donkeys. Anim. Genet. 44 :69-78.
- Cosseddu G.M. FraghìA. MuraL. CartaA. CherchiR. PauS., 2001. Relazioni genetiche tra le popolazioni asinine della Sardegna. Analisi con marcatori molecolari. Ippologia 12 :25-33.
- DalvitC. De Marchi M. ZanettiE. CassandroM., 2009. Genetic variation and population structure of Italian native sheep breeds undergoing in situ conservation. J. Anim. Sci. 87 :3837-3844.
- DalvitC. SaccaE. CassandroM. GervasoM. PastoreE. PiasentierE., 2008. Genetic diversity and variability in Alpine sheep breeds. Small Ruminant Res. 80 :45-51.
- D’AngeloF. AlbenzioM. SeviA. CiampoliniR. CecchiF. CianiE. MuscioA., 2009. Genetic variability of the Gentile di Puglia sheep breed based on microsatellite polymorphism. J. Anim. Sci. 87 :1025-1029.
- D’AngeloF. CianiE. SeviA. AlbenzioM. CiampoliniR. CianciD., 2006. The genetic variability of the Podolica cattle breed from the Gargano area. Preliminary results. Ital. J. Anim. Sci. 5 :79-85.
- DiStasioL. PerrottaG. BlasiM. ClaudioL., 2008. Genetic characterization of the Bardigiano horse using microsatellite markers. Ital. J. Anim. Sci. 7 :243-250.
- EvannoG. RegnautS. GoudetJ., 2005. Detecting the number of cluster of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14 :2611-2620.
- ExcoffierL. LavalG. SchneiderS., 2005. S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1 :47-50.
- FalushD. StephensM. Pritchard J.K., 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7 :574-578.
- FAO,2013. Available from: http://dad.fao.org
- Guastella A.M. ZuccaroA. BordonaroS. CrisioneA. MarlettaD. D’UrsoG., 2007. Genetic diversity and relationship among the three autochthonous Sicilian donkey populations assessed by microsatellite markers. Page 143 in Proc. 17th Nat. Congr. ASPA, Alghero, Italy. Ital. J. Anim. Sci. 6(Suppl.1):143 (abstr.).
- Gutiérrez, J.P. Royo L.J. AlvarezI. GoyacheF., 2005. MolKin v2.0: a computer program for genetic analysis of populations using molecular coancestry information. J. Hered. 96 :718-721.
- IvankovicA. KavarT. CaputP. MiocB. PavicV. DovcP., 2002. Genetic diversity of three donkey populations in the Croatian coastal region. Anim. Genet. 33 :169-177.
- JordanaJ. FolchP. Aranguren J.A., 2001. Microsatellite analysis of genetic diversity in the Catalonian donkey breed. J. Anim. Breed Genet. 118 :57-63.
- JordanaJ. FolchP. SanchezA., 1999. Genetic variation (protein markers and microsatellites) in endangered Catalonian donkeys. Biochem. Syst. Ecol. 27 :791-798.
- MarlettaD. , Tupac-YupanquiI. BordonaroS. GarciaD. Guastella A.M. CrisioneA. Cañon, J. DunnerS., 2006. Analysis of genetic diversity and determination of relationships among western Mediterranean horse breeds using microsatellite markers. J. Anim. Breed Genet. 123 :315-325.
- MascheroniE., 1929. Zootecnia speciale. Utet, Torino, Italy.
- MatassinoD. Costanza M.T. IncoronatoC. OccidenteM. PaneF. PaolettiF. PasquarielloR. CianiF., 2010. Analysis of genomic component of Maremmano ‘Romano’ horse by means of microsatellite markers. pp 37-43 in Proc. 12th Nat. Conf. on New Findings in Equine Practice, Druento, Italy.
- PeakallR. Smouse P.E., 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research: an update. Bioinformatics 28 :2537-2539.
- RaymondM. RoussetF., 1995. GENEPOP: population genetics software for exact tests and ecumenicism. J. Hered. 86 :248-249.
- RizziR. TulloE. Cito A.M. CaroliA. PieragostiniE., 2011. Monitoring of genetic diversity in the endangered Martina Franca donkey population. J. Anim. Sci. 89 :1304-1311.
- TortorelliN., 1973. Zootecnica speciale. Edagricole, Bologna, Italy.