Abstract
In this study, animals from the Kıvırcık breed of sheep and two closely related breeds (Pırlak and Karacabey Merino) were analysed by using 15 microsatellite markers to evaluate the genetic diversity within the Kıvırcık breed and to elucidate its relationship with the other two breeds. A total of 244 alleles were detected on all of the 15 microsatellite loci studied. While the Kıvırcık population of sheep showed a significant deviation from the Hardy-Weinberg equilibrium (HWE) for all 15 loci, both the Pırlak and Karacabey Merino breeds showed a significant deviation from the HWE for 10 loci. The mean observed heterozygosity and FIS exhibited ranges of 0.63 to 0.72 and 0.0672 to 0.2139, respectively. The highest mean number of alleles value and the greatest number of private alleles were observed in the Kıvırcık population. While microsatellite analysis revealed high genetic diversity in breeds investigated, a high level of inbreeding was also observed between different Kıvırcık populations and other two breeds. The genetic differentiation between breeds was found to below. The Kıvırcık from Istanbul and Bursa were the most closely related genetically, while the Kıvırcık from Manisa differed from the other populations. In addition, the Karacabey Merino and Pırlak breeds grouped together.
Introduction
Kıvırcık is one of the most commonly used sheep breeds in Turkey and it constitutes 6 to 7% of the total sheep population in Turkey. Although the breed is reared for multipurpose production, it has higher meat and wool quality than the other native sheep breeds (Kaymakçı et al., Citation2001).
Today it is well known that sheep were domesticated in the region named the Fertile Crescent, which includes a portion of Turkey (Zeder, Citation2008). The Kıvırcık sheep originated from Romanian Tsigai sheep breed and arrived in Central Europe during the second half of the 18th century (Kusza et al., Citation2008). According to Drăgănescu (Citation2007), the Kıvırcık sheep originated from Romania Tsigai and has relatives in the Balkan countries. Although it is reared in the Thrace, Marmara and Aegean regions of Turkey, its breeding extended to the interior of the country. Despite its numerically large population, uncontrolled crossbreeding pressure threatens the existence of purebred Kıvırcık populations. In Turkey, Merino sheep were used extensively in breeding programmes with almost every native Turkish sheep breed, including Kıvırcık. For several years, a conversion effort was maintained to all of the native breeds to Merino. Karacabey Merino is a crossbred sheep that was obtained by crossbreeding Kıvırcık and Merino sheep. Pırlak is thought to be a local sheep breed. However, there are some doubts about whether this breed was originally reared in some internal parts of Turkey or if it is a crossbred sheep obtained by crossbreeding Kıvırcık with other breeds.
Even near the Kıvırcık’s own geographic regions, it is difficult to find individuals that belong to a pure bred of sheep. For this reason, both in situ and ex situ conservation programmes have begun under the direction of the General Directorate of Agricultural Research of Turkey. The in situ conservation and pure breeding herds are reared in the Thrace, Marmara, and Aegean regions of Turkey. There are many studies aimed at understanding the genetic relationship between Kıvırcık and the other native sheep breed populations of Turkey; however, there has not yet been a study designed to reveal the genetic diversity within the breed. Although there is increasing consciousness and effort to protect the breed, until now, the current level of genetic diversity within the breed, the number of subpopulations and the degree of gene flow between these subpopulations has remained unknown. It is not possible to establish effective conservation and breeding programmes without this type of information for certain breeds.
In this study, the aim was to measure genetic diversity within the Kıvırcık breed and between the various Kıvırcık populations raised in different geographic regions of Turkey. This study also aimed to examine the genetic similarity between the Kıvırcık, Karacabey Merino and Pırlak sheep breeds because it is well known that intensive uncontrolled matings have been done over generations between these three breeds.
Materials and methods
Animal resources and DNA isolation
DNA was isolated from blood samples that were collected from 165 animals belonging to three Turkish sheep breeds (Kıvırcık, Karacabey Merino and Pırlak), which were reared on 23 distinct farms in different regions of Turkey (). Three populations of the Kıvırcık breed were collected from different geographic regions: in Bursa (n=30) from six farms, in Istanbul (n=36) from one farm (where the population originated in Thrace and collected different breeders at different times), and in Manisa (n=34) from six farms. Karacabey Merino (n=34) and Pırlak (n=31) samples were obtained from four and five farms in Bursa and Eskişehir, respectively. The Kıvırcık population from Istanbul was of special importance and was specifically sampled because this population is believed to include the purest individuals of this breed. Total DNA was extracted using a genomic DNA purification kit (K0512; Fermentas, Hanover, MD, USA) according to the manufacturer’s instructions.
Polymerase chain reaction amplifications and fragment analysis
To examine the genetic diversity within the Kıvırcık breed and similarities between the Karacabey Merino and Pırlak breeds, 15 microsatellite markers were chosen according to FAO (Citation2011). Two multiplex groups were created for amplification. The amplification reactions were carried out on a Veriti® 96-Well Thermal Cycler (Applied Biosystems, Carlsbad, CA, USA) by the Touchdown polymerase chain reaction (PCR) technique in 25 L reactions containing 50 to 100 ng of genomic DNA, 0.20 mM dNTPs (Applied Biological Materials Inc., Richmond, Canada), 2.0 mM MgCl2, 1X PCR buffer, 1 U of Taq DNA polymerase (Applied Biological Materials Inc.) and 0.10 M of each fluorescently labeled primer. The temperature profiles of the Touchdown PCR were programmed as indicated in Appendix Table; PCR products were visualised on Beckman Coulter GeXP (Beckman Coulter Inc., Brea, CA, USA) analyser™. DNA Size Standard Kit-400 was also included in each run. The allele sizes of each locus were detected using the Beckman Coulter GeXP software (Beckman Coulter Inc.).
Statistical analysis
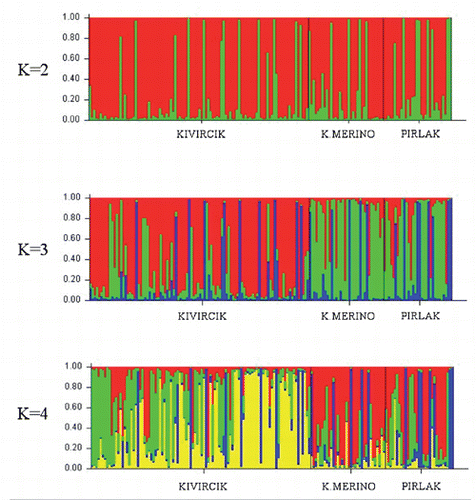
The results obtained by fragment analysis were used to check whether each breed was in Hardy-Weinberg equilibrium (HWE), the polymorphism statistics [observed heterozygosity (Ho), expected heterozygosity (He) and mean number of alleles (MNA)] and the F statistics (FST, FIS, FIT) (Weir and Cockerham, Citation1984) were estimated using GenAlEx (Peakall and Smouse, Citation2006), Fstat ver. 2.9.3 (Goudet, Citation2001), POPGENE (Yeh et al., Citation1997) and MEGA 4 (Tamura et al., Citation2007). A bootstrap-supported (1000 replications) dendrogram was constructed according to Nei’s (Citation1972) minimum distances. The population structures were analysed by cluster techniques based on the Bayesian approach, using the STRUCTURE 2.1 software (Pritchard et al., Citation2000). The burn-in and Markov Chain Monte Carlo lengths were 20,000 and 100,000, respectively. The analyses were realised at different K values (2 to 4). The most appropriate cluster number (cluster-K) was detected using the method (ΔK=m|L’’(K)|/s[L(K)]) reported by Evanno et al. (Citation2005). The optimal K was determined using Structure Harvester Web version 0.6.93 (Earl, Citation2012). Population structure analysis was repeated to reveal genetic diversity within and between the Kıvırcık populations.
Results
A total of 244 alleles were detected over all of the 15 microsatellite loci studied. The number of detected alleles ranged from 12 (MCM0527) to 22 (OarFCB20). The MNA for all loci and per locus found were 16.27 and 11.89, respectively. The polymorphic information content (PIC) value was found to be between 0.49 (BM1818) and 0.86 (OarFCB304 and OarJMP29). The genetic variability measures corresponding to the 15 investigated loci are listed in . The highest MNA value (14.33) was observed in the Kıvırcık breed in general (Kıvırcık-Bursa, Kıvırcık-Istanbul and Kıvırcık-Manisa together). In all investigated breeds, a total of 60 private alleles were found. Among the breeds studied, the highest number of private alleles (n=40) was observed in the Kıvırcık breed (Kıvırcık-Bursa, Kıvırcık-Istanbul and Kıvırcık-Manisa together). Despite the high number of private alleles, only five of these showed a frequency above 0.50 ().
The mean value of average heterozygosity (Ĥ) for all loci investigated was 0.78. While the highest Ho value found was for the OarCP34 (0.95) locus, the lowest values found were at the MCM0527 and HSC (0.51) loci. The highest and lowest He values were 0.56 (BM1818) and 0.88 (OarFCB304), respectively. The FIS values ranged between -0.185 (MAF214) and 0.405 (HSC). The BM1818 (0.063) locus had the highest FST value. The FIT value indicates that the general heterozygosity deficiency was observed between -0.130 (MAF214) and 0.417 (HSC). Genetic diversity values (DST), gene diversity coefficient (GST) and Nei’s mean gene diversity (HT) were 0.010, 0.014 and 0.803, respectively. All diversity parameters are given in .
While the Kıvırcık population showed significant deviations from HWE for all 15 loci, both the Pırlak and Karacabey Merino breeds showed significant deviations from HWE for 10 loci. The highest value of observed heterozygosity (Ho) was almost equal for the Karacabey Merino (0.72) and Kıvırcık breeds (0.71). The observed heterozygosity (Ho) was lower for Pırlak sheep breed (0.63) (). According to the dendrogram constructed following Nei’s (Citation1972) minimum distances, the Karacabey Merino and Pırlak were grouped in the same cluster; while the Kıvırcık population remained separate (). These findings are supported by STRUCTURE () and FCA (data not shown). K is ranging from 2 to 4 in order to decide the optimal K value. The results and graphics obtained from the analysis with STRUCTURE are given in . The ΔK statistics peaked at K=2 as shown in . When the dendrogram was constructed for only the Kıvırcık populations, it seemed that the Kıvırcık Bursa and Istanbul were grouped together and the Kıvırcık-Manisa population was separate from them (). The dendrogram was in concordance with the genetic similarities calculated for the Kıvırcık Bursa compared to the Kıvırcık Istanbul (0.9462), the Kıvırcık Bursa compared to the Kıvırcık Manisa (0.9015) and the Kıvırcık Istanbul compared to the Kıvırcık Manisa (0.8937). Genetic identity and genetic distance matrix is given in . The graphics obtained from STRUCTURE analysis for Kıvırcık population are not shown.
Discussion
In the present study, 93% of the markers used were highly informative with a high (0.78) PIC value. Only one marker was moderately informative with 0.49 (BM1818) of PIC value. The observed mean heterozygosity and the mean alleles per locus supported this high genetic variability. The mean number of alleles per locus was higher for all breeds investigated than was reported for the Laxta (7.5) and Merino (9.9) breeds (Arranz et al., Citation1998), but only the Kıvırcık breed had a higher mean allele per locus value than that reported for the Manchega breed (12.4) (Calvo et al., Citation2006).
In the present study, the FIS values were positive, which indicates that there is a general risk of inbreeding. According to the FIS values, the Karacabey Merino is the least inbred population among the breeds investigated. Kuzsca et al. (Citation2008) found notably higher FIS values in the Kıvırcık sheep populations from Thrace and Marmara (0.374 and 0.282, respectively) than the FIS value found in the present study (). The FIS values found for the Slovak and Hungarian Tsigai sheep breeds were also higher than those of the present study (0.252 and 0226, respectively) (Kusza et al., Citation2009, Citation2010). In contrast, the FIS values found in our study are quite high compared to the FIS values found in the Merino populations from different countries (Diez-Tascón et al., Citation2000), Muzzafarnagri (Arora and Bhatia, Citation2004), Manchega and Churra (Calvo et al. Citation2006, Citation2011), Romanian Tsigai (Zăhan et al., Citation2011). The Pırlak sheep breed population showed reduced heterozygosity (0.63). Kuzsa et al. (Citation2009) also found low heterozygosity in the several groups of Slovak and Hungarian Tsigai sheep populations, and they stated that the least variable populations are usually the most distinct. In this sense, the restricted heterozygosity could support the idea that Pırlak is a separate breed, as believed. In contrast, the Kıvırcık-Bursa populations represented the lowest value (0.68) among the Kıvırcık populations investigated. The mean observed heterozygosity found for the Kıvırcık and the Karacabey Merino in this study were similar to the results obtained for the New Zealand Merino, German Mutton (Diez-Tascón et al., Citation2000) and Romanian Tsigai (Zăhan et al., Citation2011) and higher than those of some reported in previous studies of the Slovak and Hungarian Tsigai sheep breeds (Kusza et al., Citation2009, Citation2010). Although the Kıvırcık breed exhibited high genetic variation, its high FISvalue indicates that greater selective pressure would constitute a threat and would increase inbreeding within the population (FIS=0.1196).
Deviations from HWE can have several causes. The populations investigated are breeding herds, so selective pressure may lead to deviations from the HWE. The high FIS values estimated, which reflect an excess of homozygotes, can also cause deviations from HWE. The departure from the HWE could also be attributed to the presence of null alleles. Unfortunately, the extent of null alleles could not be estimated due to the lack of pedigree records. In addition, subdivision is not possible due to the high level of admixture between populations. The low number of private alleles showed a frequency above 0.50; along with the GST and Nm values, this result may indicate that the gene flow between populations is quite high. STRUCTURE software analysis illustrated that the Kıvırcık sheep breed presents a complicated population structure for the loci investigated (). Similarly, the results obtained by STRUCTURE software analysis are in concordance with the dendrograms representing genetic similarities between the Pırlak and Karacabey Merino breeds, as well as between the Kıvırcık-Bursa and Kıvırcık-Istanbul populations. Results from STRUCTURE analysis also reveals a high level of admixture of these three sheep breeds.
The FST value estimated for all loci indicates that the genetic differentiation between the breeds is weak. The GST value (0.014) shows that the variation between the populations consists of 1.4% of total variation. Overall, 98.6% of the total variation results from the current variation between breeds. This situation is in agreement with the studies carried out in the Turkish native sheep breeds by using molecular markers. Tapio et al. (Citation2010) reported that breeds of domestic sheep near this region were more variable but less genetically differentiated compared with more northern populations. When these findings are evaluated together, they support the claim that the genetic differentiation between these breeds is quite weak.
Kusza et al. (Citation2008, Citation2011) investigated 13 local sheep breeds from Southern and Eastern Europe, including the Thrace Kıvırcık population and the Marmara Kıvırcık populations, and they were grouped separately. Interestingly, the Thrace Kıvırcık population was grouped together with the Gökçeada sheep breed in other studies carried out by the same authors. The present study also observed interesting clustering, in which the Pırlak and Karacabey Merino populations were grouped together. Although these results seemed puzzling initially, they are most likely the consequence of the utilisation of Karacabey Merino rams in the Pırlak breeding systems due to higher production value of Karacabey Merino. These two breeds have been investigated for the first time by using microsatellite markers in this study and more study is needed to reveal genetic structure and genetic relationship of these breeds.
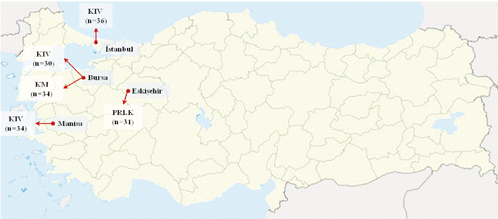
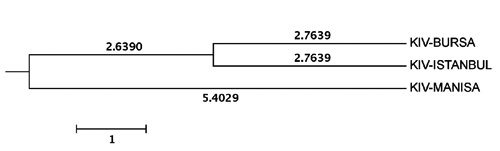
Table 1. Genetic variability measures of the fifteeen investigated loci.
Table 2. Some diversity parameters for each breed across fifteen loci.
Table 3. Estimated posterior probabilities for different numbers of inferred clusters and AK statistics.
Table 4. Nei’s unbiased measures of genetic identity and genetic distance.
Conclusions
The results obtained from the present study are in accordance with the known breeding history of the sheep populations investigated. Better clustering and a deeper understanding of the population structure and demographic history of the Kıvırcık and related populations could be obtained by analysing other Kıvırcık populations from more distant geographic regions. Moreover, for more effective selection and conservation programmes, systematic studies with molecular markers should be used.
Acknowledgements
this work was financially supported by the Scientific Research Council of Uludag University (Project number: KUAP 2012/49). We acknowledge the General Directorate for Agricultural Research and Policies (GDAR) and Dr. Bekir Ankaralı, who supplied animal materials for the study. This manuscript was edited by American Journal Experts (AJE).
References
- AroraR. BhatiaS., 2004. Genetic structure of Muzzafarnagri sheep based on microsatellite analysis. Small Ruminant Res. 54:227-230.
- ArranzJ.J. BayOnY. San PrimitivoF., 1998. Genetic relationships among Spanish sheep using microsatellites. Anim. Genet. 29:435-440.
- CalvoJ.H. Alvarez-RodriguezaJ. Marcos-CarcavillaA. SerranoM. SanzaA., 2011. Genetic diversity in the Churra tensina and Churra lebrijana endangered Spanish sheep breeds and relationship with other Churra group breeds and Spanish mouflon. Small Ruminant Res. 95:34-39.
- CalvoJ.H. BouzadaJ.A. JuradoJ.J. SerranoM., 2006. Genetic substructure of the Spanish Manchega sheep breed. Small Ruminant Res. 64:116-125.
- Diez-TascónC. LittlejohnR.P. AlmeidaP.A.R. CrawfordA.M., 2000. Genetic variation within the Merino sheep breed: analysis of closely related populations using microsatellites. Anim. Genet. 31:243-251.
- DrägänescuC., 2007. A note on Balkan sheep breeds origin and their taxonomy. Arch. Zootec. 10:1-12.
- EarlD.A., 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. 4:359-361.
- EvannoG. RegnautS. GoudetJ., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14:2611-2620.
- FAO, 2011. Molecular genetic characterization of animal genetic resources. Food and Agriculture Organization Publ., Rome, Italy.
- GoudetJ., 2001. FSTAT: a program to estimate and test gene diversities and fixation indices (Version 2.9.3). Available from: http://www2.unil.ch/popgen/softwares/fstat.htm
- KaymakçıM. OǧuzI. ÜnC. BilgenG. Ta§kınT., 2001. Basic characteristics of some Turkish indigenous sheep breeds. Pak. J. Biol. Sci. 4:916-919.
- KuszaS. GyarmathyE. DubravskaJ. NagyI. JávorA. KukovicsS., 2009. Study of genetic differences among Slovak Tsigai populations using microsatellite markers. Czech J. Anim. Sci. 54:468-474.
- KuszaS. IvankovicA. RamljakJ. NagyI. JávorA. KukovicsS., 2011. Genetic structure of Tsigai, Ruda, Pramenka and other local sheep in Southern and Eastern Europe. Small Ruminant Res. 99:130-134.
- KuszaS. NagyI. NémethT. MolnárA. JávorA. KukovicsS., 2010. The genetic variability of Hungarian Tsigai sheep. Arch. Tierzucht 53:309-317.
- KuszaS. NagyI. SasváriZ.S. StágelA. NémethT. MolnárA. KumeK. BõszeZ. JávorA. KukovicsS., 2008. Genetic diversity and population structure of Tsigai and Zackel type of sheep breeds in the Central Eastern and Southern European regions. Small Ruminant Res. 78:13-23.
- NeiM., 1972. Genetic distance between populations. Am. Nat. 106:283-292.
- PeakallR. SmouseP.E., 2006. GENALEX 6:genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6:288-295.
- PritchardJ.K. StephensM. DonnellyP., 2000. Inference of population structure using multilocus genotype data. Genetics 155:945-959.
- TamuraK. DudleyJ. NeiM. KumarS., 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599.
- TapioM. OzerovM. TapioI. ToroM.A. MarzanovN. CinkulovM. GoncharenkoG. KiselyovaT. MurawskiM. KantanenJ., 2010. Microsatellite-based genetic diversity and population structure of domestic sheep in northern Eurasia. BMC Genet. 11:76.
- WeirB.S. CockerhamC.C., 1984. Estimating F-statistics for the analysis of population, structure. Evolution 38:1358-1370.
- YehF.C. YangR.C. BoyleT.B.J. YeZ.H. MaoJ.X., 1997. POPGENE the user-friendly shareware for population genetic analysis. University of Alberta Publ., Alberta, Canada.
- ZǎhanM. MicleaV. RaicaP. MicleaI. IlisiuaE., 2011. Analysis of genetic variation within Tsigai population from Romania using microsatellite marker. Bull. Univ. Agric. Sci. 68:396-400.
- ZederM.A., 2008. Domestication and early agriculture in the Mediterranean basin: origins, diffusion, and impact. P. Natl. Acad. Sci. USA 105:11597-11604.