Abstract
The growth hormone (GH) is the main regulator of postnatal growth and metabolism in mammals. The action of GH on target cells depends on the growth hormone receptor (GHR). This is mediated through induced transcription of other genes. GHR gene is one of the candidate genes employed in selection strategy using DNA markers (marker assisted selection). This study was designed to identify the novel single nucleotide polymorphisms (SNPs) in exon 8 and intron 8 of GHR gene that may affect production traits in Bali cattle. A SNP was identified by the direct sequencing technique. Genotypes of the SNPs were identified using PCR-RFLP. The SNP was located in intron 8 of the GHR gene and was caused by an A/G transition. It was identified using the HpyCH4III restriction enzyme. Polymorphism of GHR/HpyCH4III has a significant influence on weaning weight and average daily gain, but not on birth weight of Bali cattle.
Introduction
Growth hormone (GH) is an anabolic hormone which promotes growth and reproduction of cells in humans and other animals. The growth hormone exerts its effects on growth and metabolism by interacting with a growth hormone receptor (GHR) on the surface of the target cells (Listrat et al., Citation2005). Mutation of GHR gene might affect its binding capacity and signaling pathway, thereby altering the GH activity in the target tissues (Di Stasio et al., Citation2005).
In cattle, GHR is encoded by a single gene located in chromosome 20 (Moisio et al., Citation1998). The gene coding for bovine GHR consists of nine exons (from 2 to 10) in the translated part and of a long 5’-noncoding region that includes nine untranslated exons - 1A, 1B, 1C, 1D, 1E, 1F, 1G, 1H, 1I (Jiang and Lucy, Citation2001).
Variation in the GHR gene sequence is associated with a number of performance traits in cattle (Viitala et al., Citation2006; Garrett et al., Citation2008). The single nucleotide polymorphism (SNP) in exon 8 has been shown to be associated with milk yield and composition (Blott et al., Citation2003; Viitala et al., Citation2006), as well as feed intake, feed conversion efficiency and body energy traits (Banos et al., Citation2008).The association between GHR gene polymorphisms with production and carcass trait has been reported in several cattle breeds. For example, SNP sin 5’-noncoding regions have been reported to be associated with the rate of growth and carcass trait in some breed of beef cattle such as Limousin, Charolais and Hereford (Maj et al., Citation2004). Similarly, Ge et al. (Citation2003) reported that a GHR gene polymorphism caused by A/G transitions in the promoter region was associated with the concentration of insulin like growth factor-1 (IGF-1) serum in Angus cattle. This study aims to discover novel single nucleotide polymorphisms (SNPs) in exon 8 and intron 8 of the GHR gene and to determine their effects on birth weight, weaning weight and average daily weight gain in Bali cattle.
Materials and methods
The University of Mataram, Faculty of Medicine, Ethical Committee for Medical Research, Mataram, Indonesia approved all animal procedures for this experiment (Register No. 4/UN18.8/ETIK/2013).
Phenotyping: measurement of the production traits
Two hundred and fifty Bali cattle (aged 1.0-1.5 years) for which phenotypic records were available with respect to body weight and body conformation, were reared at University Field Stations. All animals were ear-tagged. The average daily gain was measured weekly for a period of 3 months. The data on the birth weight and weaning weight of the calves at 6 months old were previously recorded and were used to complement the quantitative data.
Single nucleotide polymorphism discovery
Blood samples for DNA analysis were collected from the jugular vein of each animal. Blood was collected on K2EDTA and stored at 25oC for few weeks or at 75oC up to several months. The isolation of DNA from whole blood of the 250 Bali cattle was performed by the method of Sambrook et al. (Citation1989). Amplification was carried out following the method of Oikonomou et al. (Citation2008). The PCR reaction contained 100 ng DNA, 0.5 μM of each primer, 1×PCR buffer (10 mM Tris-HCl pH 9.0), 1.5 mM MgCl2 and 50 mM KCl, 5% deionized formamide, 200 μM dNTPs, and 0.025 U Taq DNA polymerase (Pharmacia) in a volume of 25 μL. The amplification was performed for 35 cycles using DNA thermal Cycler (Perkin Elmer Cetus Corp.). The first cycle was at 95oC for 5 min followed by 33 subsequent cycles of 94oC x 45 s, then 56oC x 45 s, then 72oC x 60 s and the last cycle at 72oC for 5 min. Sequence and position of the primer can be seen in . The identification of SNPs in the gene fragments was conducted by means of the direct sequencing method (Kwok and Duan, Citation2003). DNA samples (PCR products) were grouped in the form of a DNA Pool for each sub population of cattle. Sequencing of DNA fragments was carried out by the AB3730xl sequencer machine. Sequence alignment was performed using the BioEdit (http://www.mbio.ncsu.edu/bioedit) and Mega4 programme (http://www.megasoftware.net/mega4). The alignments of reverse and forward sequences were applied to produce consensus sequences. The sequences of each individual DNA fragments were aligned with original sequences to identify the presence of SNPs.
Genotyping of the candidate gene
The determination of the genotype of each individual animal was conducted based on the SNPs that were found at the SNPs discovery stage. Genotyping was performed by the PCR-RFLP techniques. PCR reaction used in this step is equal to PCR reaction used at the SNPs discovery stage. A specific restriction enzyme was identified by the webcutter (http://rna.lundberg.gu.se/cutter2) and nebcutter programme (http://tools.neb.com/NEBcutter). The gene fragments containing the SNPs were amplified using PCR, then the gene fragments were digested using a specific restriction enzyme. The size and number of alleles were determined using agarose gel electrophoresis.
Statistical analysis
Genotyping and allele frequency within and among genetic groups were determined by the method of Goodman adapted by Curi et al. (Citation2005). The association analysis was performed using General Linear Model (GLM) and the least square means of the genotypes were compared by t-test (as implemented in the SAS programme). The linear model used was as follows:
where, Yij=production trait, A=overall mean, Gi=fixed effect of the i genotype, and eij=random error.
Results and discussion
Single nucleotide polymorphism discovery inintron 8 - exon 8 of the GHR gene
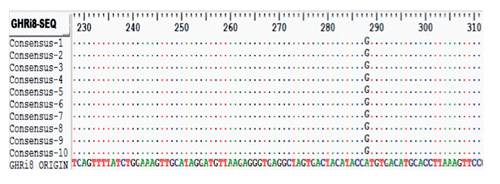
The PCR productis about 341bp, located in exon 8 and part of intron 8 of the GHR gene. DNA samples (PCR products) were grouped in the form of a Pool of DNA for each subpopulation of cattle and then sequenced to identify the mutation. The results of sequence alignment using the Bio Edit and Mega4 programme showed that there is a new mutation in intron 8 of GHR gene ().
Mutations at this locus are transition mutations that change adenine bases in to guanine (A/G transition) at the nucleotide position 241bp (Nt241). These mutations can be identified using restriction enzyme HpyCH4III (ACNiGT). The digestion using HpyCH4III (ACNiGT) produces two alleles, namely A (uncut) and G (cut) alleles. The A allele was not sensitive to HpyCH4III, while G allele was cut by HpyCH4III producing two DNA fragments: 287 bp and 54 bp. The analysis of the GHR locus implies that mutation occured to make it sensitive to HpyCH4III enzyme which recognized the sequence (ACNiGT) as the site of cutting.
The GHR gene SNP has been studied in various cattle breeds, including dairy and beef breeds. Blot et al. (Citation2003) found 10 new SNPs in the GHR gene from the Friesian Holstein and Jersey breeds: one transition in 3’UTR, one indel and one transition in intron 2, one transversion in exon 8 and one in intron 8, one transition in intron 9, and four mutations in exon 10 (one transversion and three transitions). Waters et al. (Citation2010) reported six new SNPs in 5’non coding region and 1 new SNP in intron 2 of GHR gene from the Friesian Holstein dairy cattle. The data reported in this paper add to these reports and extend the work to Bali cattle.
Allelic and genotypic frequencies of the GHR/HpyCH4III gene in Bali cattle
Allelic and genotypic identification of intron 8 GHR gene using the RFLP-HpyCH4III technique produces two alleles namely A and G allele with three genotypes AA, AG and GG (). The distribution of allelic frequency of the A allele was slightly higher than G allele, respectively 0.532 and 0.468, while the frequency distribution of the genotypes AA, AG and GG were respectively: 0.156; 0.752 and 0.092 (). The Chi-square (2) test showed that the genotype distributions of intron 8 GHR gene were not at Hardy-Weinberg equilibrium (H-WE) in Bali cattle. The genotype frequencies at polymorphic loci of intron 8 GHR gene showed a highly significant difference (P<0.01). This contrasts with the exon 10 GHR gene in Polish Holstein Friesian cattle, as reported by Olenski et al. (Citation2010) where the allelic frequencies distributions between the A and G allele differed, the G allele (0.832) being higher than that of the A allele (0.168).
Data in show the results of genetic index measurements in Bali cattle population. These data indicate a genotypic imbalance in the population where genotype heterozygote frequencies are higher than the Hardy-Weinberg expectation. This could be due to intensive selection, resulting in a tendency towards the accumulation of certain genotypes (Tambasco et al., Citation2003) and the possibility of inbreeding (Machado et al.,Citation2003).
The effective population size (Ne) () illustrates that the GHR gene alleles in the population have very different frequencies in which one allele is dominant allele frequencies. Based on the polymorphic information content (PIC) value of 0.37, it can be stated that the genetic diversity of GHR gene within Bali cattle population is at the medium level. This statement is based on PIC levels of polymorphism as determined by Botstein et al. (Citation1980) in which levels of ≤0.25 are classified as low, 0.25≤PIC≤0.5 are classified as medium and PIC≥0.5 are classified as high polymorphism. Zulkharnaim et al. (Citation2010) also reported that Bali cattle have a low genetic diversity at GHR/ Alu1 loci in exon 10 of GHR gene.
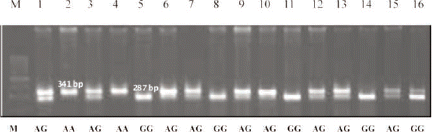
Table 1. Sequence and position of the primer.
Table 2. Allele and genotype frequencies of GHR-HpyCH4III gene in Bali cattle.
Table 3. Genetic index of Bali cattle population.
Table 4. Birth weight, weaning weight and average daily gain in Bali cattle based on the genotype of the GHR- HpyCH4III gene.
Association of GHR gene polymorphism with production traits
The association between the GHR gene genotype and birth weight, weaning weight and average daily gain in Bali cattle can be seen in . The effect of genotype of GHR gene was found to be significant (P<0.01) for weaning weight and average daily gain (ADG) and not significant for birth weight (P>0.05) in Bali cattle. One explanation could be that GHR is a mediator of GH biological activity in target cells through stimulating myogenic signal transduction, while GH is a regulator of growth and metabolism after birth.
The role of GH on target cells mainly depends on the expression of GHR. GH secretion, binding to the receptor and the expression of GHR at the beginning of growth is an important factor in determining the optimal metabolic function in muscle (Katsumata et al., Citation2000). The action of GH precedes the formation of a bond with the transmembrane GHR which is present on the surface of most cells and encourages the activation of GHR. Furthermore, the expression of GHR stimulates the expression of an IGF-1 which is a mediator of GHR activity on the target cell (Locatelli and Bianchi, Citation2014).
The association between weaning weight, ADG and birth weight in Bali cattle and the GHR gene polymorphisms due to a mutation in intron 8 region needs tobe explained. Mutations in intron regions may not lead to changes in the protein sequence of the GHR gene to produce a protein with an unchanged amino acid sequence but it could nontheless result in a change in structural and functional properties (Komar, Citation2007). Mutations in intron 8 regions of GHR gene in Bali cattle may also lead to changes in the amino acid sequence of protein if it occurs at the initial site of the mRNA splicing after transcription. In conclusion this study shows that the A/G SNP in intron 8 of the GHR gene in Bali cattle significantly affects production traits, in particular post natal growth. It remains to be seen if the findings we report also apply to other breeds of cattle.
Conclusions
The SNP analysis of intron 8 of GHR gene reveals A/G transition in the base position of 241 bp. The polymorphisms of nucleotide sequence caused by these mutations were identified using the restriction enzyme HpyCH4III. The A/G transition resulted in three genotypes SNP of GG, AG and AA with a frequency of 0.092, 0.752 and 0.156 respectively and two alleles of A and G with almost equal frequency distribution of 0.532 and 0.468. Genotypic polymorphism of GHR/HpyCH4III has a significant influence on weaning weight and average daily gain, but no significant influence on birth weight of Bali cattle. GG genotype had a weaning weight and average daily gain of: 82.14±10.31 kg and 0.474±0.08 kg/day respectively, higher than the AG and AA genotypes.
References
- Banos G. Woolliams J.A. Woodward B.W. Forbes A.B. Coffey M.P., 2008. Impact of single nucleotide polymorphisms in leptin, leptin receptor, growth hormone receptor, and diacyl glycerol acyl transferase (DGAT1) gene loci on milk production, feed, and body energy traits of UK dairy cows. J. Dairy Sci. 91:3190-3200.
- BioEdit, version 7.2.5., 2013. Available from: http://www.mbio.ncsu.edu/bioedit
- Blott S. Kim J.J. Moisio S. Schmidt-Kuntzel A. Cornet A. Berzi P. Cambisano N. Ford C. Grisart B. Johnson D. Karim L. Simon P. Snell R. Spelman R. Wong J. Vilkki J. Georges M. Farnir F. Coppieters W., 2003. Molecular dissection of a quantitative trait locus. A phenylalanine-to-tyrosine substitution in the transmembrane domain of the bovine growth hormone receptor is associated with a major effect on milk yield and composition. Genetics 163:253-266.
- Botstein D. White R.L. Skolnick M. Davis R.W., 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genetics 32:314-331.
- Curi R.A. De OliveiraH. N. Silveira A.C. Lopes C.R., 2005. Association between IGF-I, IGF-IR and GHRH gene polymorphisms and growth and carcass traits in beef cattle. Livest. Sci. 94:159-167.
- DiStasio L. Destefanis G. Brugiapaglia A. Albera A. Rolando A., 2005. Polymorphism of the GHR gene in cattle and relationship with meat production and quality. Anim. Genetics 36:138-140.
- Garrett A.J. Rincon G. Medrano J.F. Elzo M.A. Silver G.A. Thomas M.G., 2008. Promoter region of the bovine growth hormone receptor gene: single nucleotide polymorphism discovery in cattle and association with performance in Brangus bulls. J. Anim. Sci. 86:3315-3323.
- Ge W.M.E. Davis H.C. Hines K.M. Irvin SimmenR. C.M., 2003. Association of single nucleotide polymorphisms in the growth hormone and growth hormone receptor genes with blood serum insulinlike growth factor I concentration and growth traits in Angus cattle. J. Anim. Sci. 81:641-648.
- Jiang H. Lucy M.C., 2001. Variants of the 5’-untranslated region of the bovine growth hormone receptor mRNA: isolation, expression and effects on translational efficiency. Genetics 265: 45-53.
- Katsumata M.D. Cattaneo W.P. Burton K.A. Dauncey M.J., 2000. Growth hormone receptor gene expression in porcine skeletal and cardiac muscles is selectively regulated by postnatal under nutrition. J. Nutr. 130:2482-2488.
- Komar A.A., 2007. Silent SNPs: impact on gene function and phenotype. Pharmacogenetics 8:1075-1080.
- Kwok P.Y. Duan S., 2003. Single nucleotide polymorphisms methods and protocols. In: KwokP.Y. ( ed.) Methods in molecular biology. Humana Press Inc., Totowa, NJ, USA.
- Listrat A. Hocquette J.F. Picard B. Ménissier F. Djiane J. Jammes H., 2005. Growth hormone receptor gene expression in the skeletal muscle of normal and double-muscled bovines during foetal development. Rep. Nutr. Dev. 45:393-403.
- Locatelli V. Bianchi V.E., 2014. Effect of GH/IGF-1 on bone metabolism and osteoporsosis. Int. J. Endocrinol. 2014: 235060.
- Machado M.B.B. Alencar M.M. Pereira A.P. Oliveira H.N., 2003. QTL affecting body weight in a candidate region of cattle chromosome 5. Genet. Mol. Biol. 26:259-265.
- Maj A. Strzalkowska N. Sloniewski K. Krzyzewski J. Oprzadek L. Zwierzchowski L., 2004. Single nucleotide polymorphism (SNP) in the 5’-noncoding region of the bovine growth hormone receptor gene and its association with dairy production traits in Polish Black and White cattle. Czech J. Anim. Sci. 49:419-429.
- Mega, 2007. Mega version 4. Available from: http://www.megasoftware.net/mega4
- Moisio S. Elo K. Kantanen J. Vilkki J., 1998. Polymorphism within the 3 ’ - flanking region of the bovine growth hormone receptor gene. Anim. Genetics 29:55-57.
- NEB cutter, 2003. NEBcutter ver. 1.0. Available from: http://tools.neb.com/NEBcutter
- Oikonomou G. Angelopoulou K. Arsenos G. Zygoyiannis D. Banos G., 2008. The effects of polymorphisms in the DGAT1, leptin and growth hormone receptor gene loci on body energy, blood metabolic and reproductive traits of Holstein cows. Anim. Genetics 40:10-17.
- Oleński K. Suchocki T. Kamiński S., 2010. Inconsistency of associations between growth hormone receptor gene polymorphism and milk performance traits in Polish Holstein-Friesian cows and bulls. Anim. Sci. P. 28:229-234.
- Sambrook J. Fritsch E.F. Maniatis T., 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA.
- Tambasco D.D. Paz C.C.P. Tambasco-Studart M. Pereira A.P. Alencar M.M. Freitas A.R. Coutinho L.L. Packer I.U. Regitano L.C.A., 2003. Candidate genes for growth traits in beef cattle crosses Bostaurus X Bosindicus. J. Anim. Breed. Genetics 120:51-56.
- Viitala S. Szyda J. Blott S. Schulman N. Lidauer M. Maki-Tanila A. Georges M. Vilkki J., 2006. The role of the bovine growth hormone receptor and prolactin receptor genes in milk, fat and protein production in Finnish Ayrshire dairy cattle. Genetics 173:2151-2164.
- Waters S.M. McCabe M.S. Howard D.J. Giblin L. Magee D.A. MacHugh D.E. Berry D.P., 2010. Associations between newly discovered polymorphisms in the Bostaurus growth hormone receptor gene and performance traits in Holstein– Friesian dairy cattle. Anim. Genetics 42:39-49.
- Webcutter, 1997. Webcutter version 2.0. Available from: http://rna.lundberg.gu.se/cutter2
- Zulkharnaim J. Noor R.R., 2010. Identification of genetic diversity of growth hormone receptor (GHR/Alu1) gene in Bali Cattle. Med. Pet. J. Anim. Sci. Tech. 33:81-87.