Abstract
The aim of this study was to characterise the prevalence of class 1 integrons and gene cassettes, tetracycline-resistance genes, phenicol-resistance genes, 16S rRNA methylase genes, extended-spectrum β-lactamase genes and plasmid-mediated fluoroquinolone resistance determinants in 184 Escherichia coli isolates from chickens in Anhui Province, China. Susceptibility to 15 antimicrobials was determined using broth micro-dilution. Polymerase chain reaction and DNA sequencing were used to characterise the molecular basis of the antibiotic resistance. High rates of antimicrobial resistance were observed; 131 out of the 184 (72.3%) isolates were resistant to at least six antimicrobial agents. The prevalences of class 1 integrons, tetracycline-resistance genes, phenicol-resistance genes, 16S rRNA methylase genes, extended-spectrum β-lactamase genes and plasmid-mediated fluoroquinolone resistance determinants were 49.5, 17.4, 15.8, 0.5, 57.6 and 46.2%, respectively. In 82 isolates, 48 different kinds of coexistence of the different genes were identified. Statistical (χ2) analysis showed that the resistance to amoxicillin, doxycycline, florfenicol, ofloxacin and gentamicin had significant differences (P<0.01 or 0.01<P<0.05) among the strains that carried and did not carry the resistance genes, which showed a certain correlation between antimicrobial resistance and the presence of resistance genes.
Introduction
Recently, the frequency and spectrum of antimicrobial-resistance infections have increased in the poultry industry. The emergence of Escherichia coli isolates with multiple antibiotic-resistance phenotypes has been previously reported and is regarded as a serious problem (Maynard et al., Citation2003). Transfer of resistance determinants by mobile genetic elements, including plasmids, transposons, and gene cassettes in integrons (Carattoli, Citation2001; Hall and Collins, Citation1995) are important factors that contribute to the increase in antibiotic resistance in multiple-resistant E. coli. In a previous study (Saénz et al., Citation2004), 17 multiple antimicrobial-resistance nonpathogenic E. coli strains of human, animal, and food origins showed a wide variety of antibiotic resistance genes, many of them carried by class 1 and class 2 integrons. The main purpose of the present study was to investigate the prevalence and characteristics of class 1 integrons, tetracycline-resistant genes, phenicol-resistant genes, 16S rRNA methylase genes, extended-spectrum β-lactamase (ESBL) genes and plasmid-mediated fluoroquinolone resistance (PMQR) genes and their coexistence in 184 E. coli isolates collected from chicken farms.
Materials and methods
Bacterial isolates
In this study, E. coli isolates (n=184) were collected from chicken cloacae at four different farms located in Anhui Province, China, from March to May 2012. The data and location of each farm are as follows: No. 1 chicken farm (n=46, located in Hefei city), No. 2 chicken farm (n=44, located in Changfeng county), No. 3 chicken farm (n=44, located in Feixi county), and No. 4 chicken farm (n=50, located in Feidong county). Sterile cotton swabs were used to collect fecal samples from chicken cloacae. The swabs were immediately transferred to sterile collection containers containing Luria-Bertani (LB) broth and were cultured at 37°C overnight. The cultures were inoculated in E.coli Chromogenic Medium and were grown at 37°C for 18-24 h. Then picked up a single colony which was routinely grown in LB or LB agar at 37°C for 18-24 h.
Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) of 15 antimicrobials [amoxicillin (AMX), ceftriaxome (CRO), ceftiofur (CTF), amikacin (AMI), gentamicin (GEN), apramycin (APR), doxycycline (DC), oxytetracycline (OTC), florfenicol (FFC), enrofloxacin (ERO), ofloxacin (OFX), lomefloxacin (LOM), cefquinome (CQN), sarafloxacin (SAR), sulfamonomethoxine (SUL)] were determined using the broth micro-dilution method, according to the guidelines issued by the Clinical and Laboratory Standards Institute. E. coli ATCC 25922 was used as the reference strain.
Genotypic resistance characterisation
Class 1 integrons and gene cassettes, tetracycline-resistance genes (tetA and tetM), phenicol-resistance genes (floR and fexA), 16S rRNA methylase genes (armA and rmtB), ESBL genes (blaCTX-M, blaTEM and blaSHV) and PMQR genes (qnrA, qnrB, qnrC, qnrD, qnrS, aac(6’)-Ib-cr and qepA) were detected by PCR using the primers listed in . All the PCR amplicons were confirmed by dideoxy DNA sequencing. The DNA sequences obtained were compared with those in GenBank using the BLAST program (http://blast.ncbi.nlm.nih.gov/).
Table 1. Polymerase chain reaction primers and annealing temperatures.
Results
Antimicrobial susceptibility of Escherichia coli isolates
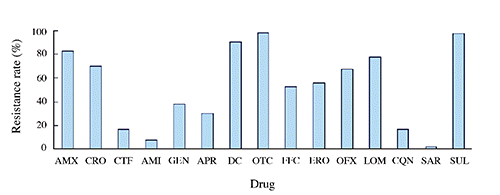
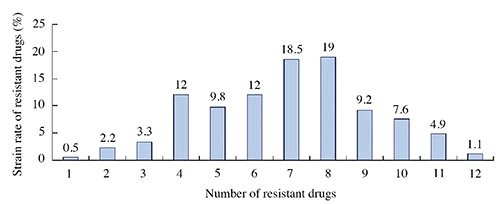
High rates of resistance to OTC (97.8%), SUL (97.3%), DC (90.2%), AMX (82.6%), LOM (77.7%), CRO (70.1%), OFX (67.4%), ERO (55.9%), and FFC (52.7%) were observed among the 184 E. coli isolates. Low rates of resistance to AMI (7.6%) and SAR (2.2%) were observed. The resistance rates of E. coli isolates from four chicken farms to 15 antimicrobials can be seen in . One hundred and thirty-one (72.3%) of the isolates were resistant to at least six antimicrobial agents, while 25 (13.6%) were resistant to at least 10 of these drugs ().
Detection of the six categories of resistance determinants
The PCR results showed that 91 (49.5%) strains harbored class 1 integrons. Seventy of the 91 intl-positive isolates carried gene cassettes, which were dfrA1-tnpAIS26-aadA1 (45/70), dfrA2-aadA12 (16/70) and dfrA1-aadA1 (9/70); 21 isolates did not carry gene cassettes.
Tetracycline-resistance genes were detected in 32 of 184 (17.4%) isolates (tetA, n=27; tetM, n=5). Twenty-nine (15.8%) isolates possessed phenicol-resistance genes (floR, n=29); no fexA genes were detected. Only one isolate carried a 16S rRNA methylase gene in the form of rmtB; no isolates were positive for armA.
ESBL genes were detected in 106 of the 184 (57.6%) isolates (blaCTX-M, n=40; blaTEM-1, n=49; and blaTEM-206, n=17); no isolates were positive for the blaSHV gene. Eighty-five out of the 184 (46.2%) isolates possessed PMQR determinants. Twenty-two (11.9%) isolates carried the qnr determinant and 63 (34.2%) carried aac(6’)-Ib-cr. Among the qnr determinants, only the qnrS-type gene was detected; no isolates were positive for qnrA, qnrB, qnrC, qnrD or qepA genes.
Coexistence of different resistance genes
Coexistence of different resistance genes was identified in 82 E. coli isolates. There were 48 different kinds of coexistence of resistance genes (). Twenty-six isolates carried three different genes, five isolates carried four genes and three carried five genes. Only one isolate harbored six genes, which were dfrA2-aadA12, tetA, tetM, blaTEM-1, qnrS and aac(6’)-Ib-cr. The combination of dfrA1tnpAIS26-aadA1 and aac(6’)-Ib-cr was observed in eight E. coli isolates.
Table 2. Coexistence of different resistance genes.
Relationship between resistance genes and antibiotic resistance of the 184 Escherichia coli isolates
A χ2 test was used to determine the relationship between resistance genes and antimicrobial resistance of the 184 E. coli isolates. Compared the resistance to AMX, DC, FFC, OFX and GEN, the results of strains carrying resistance genes were significantly different (P<0.01 or 0.01<P<0.05) from the strains not carrying resistance genes, while resistance to the remaining 10 antimicrobials showed no difference (P>0.05) ().
Table 3. Comparison of resistance between the strains carrying and not carrying resistance genes.
Discussion
No information is available about the occurrence and distribution of the different resistance determinants, such as integrons and gene cassettes, tetracycline-resistance genes, phenicol-resistance genes, 16S rRNA methylase genes, ESBL and PMQR genes in E. coli from chickens in Anhui Province, China, therefore we screened 184 E. coli isolates for the presence of these resistance determinants. 71.2% of the isolates were resistance to at least six antimicrobial agents, while 13.6% were resistance to at least 10 of these drugs, indicating a high prevalence of multiple antibiotic-resistance E. coli in chickens in Anhui Province. In Henan Province, the proportion of multiple antibiotic resistance isolates in chickens was 76.5% (Yuan et al., Citation2010). In Shanghai, 56.9% E. coli isolates were resistance to 10 to 15 antimicrobial agents (Ma et al., Citation2009). Additionally, 95.1% E. coli isolates from different regions showed multiple antibiotic resistance (Lin et al., Citation2009). The differences in multiple antibiotic resistance could be caused by different antimicrobial usage in these areas.
In this study, 184 E. coli isolates were assayed for the presence of class 1 integrons. 49.5% of isolates carried class 1 integrons, harbouring gene cassettes such as dfrA1-tnpA IS26-aadA1, dfrA2-aadA12 and dfrA1-aadA1, which encode resistance to sulfonamides and aminoglycosides. There are differences in the prevalence of the class 1 integrons in China and abroad. 52% of E. coli isolated from poultry food possessed class 1 integrons (Soufi et al., Citation2011) in Tunisia. In Spain, a study suggested that the frequency of class 1 integrons was 51% (Marchant et al., Citation2013). In China, one study showed that 60.4% of E. coli isolates from chickens had class 1 integrons (Zhang et al., Citation2009). In addition, 89.9% E. coli collected from food-producing animals were positive for class 1 (Lin et al., Citation2011). In this study, the frequency of class 1 integrons is similar to the data obtained abroad, but lower than the domestic data.
In addition, we detected tetracycline-resistance genes, phenicol-resistance genes and 16S rRNA methylase genes. The results showed that in Anhui Province, the frequencies of tetA, tetM, and floR were 14.7, 2.7 and 15.8%, respectively. Only one isolate harbored a 16S rRNA methylase gene (rmtB). No isolates carried fexA and armA. In other studies, the frequencies of tetA and tetM were 87.9 and 15.5%, respectively (Zhang et al., Citation2010), the prevalence of floR was 45.1% (Du et al., Citation2007) and the frequency of rmtB was 30.3% (Zhou et al., Citation2010). Thus the frequencies of these three genes in the current study were lower than those reported previously.
The blaTEM-1 gene was the most common β-lactamase gene among E. coli isolates in Anhui Province. This was in agreement with previous findings (Xia et al., Citation2010). With respect to PMQR genes, qnrS and aac(6’)-Ib-cr were predominant, while other PMQR genes were not detected in this study.
Notably, 82 isolates carried more than two types of genes, resulting in 48 different kinds of coexistence of class 1 integrons and gene cassettes, tetracycline-resistance genes, phenicol-resistance genes, ESBL and PMQR genes. Thus the coexistence of resistance genes was very common and varied in Anhui Province. The most frequent coexistence was dfrA1-tnpAIS26-aadA1 and aac(6’)-Ib-cr, which was found in eight E. coli isolates, while the others were found in ≤ four isolates. Six determinants, dfrA2-aadA12, tetA, tetM, blaTEM-1, qnrS and aac(6’)-Ib-cr, were detected in an individual E. coli isolate. To the best of our knowledge, this is the first report of these six genes coexisting in an E. coli strain in China. In addition, statistical analysis indicated a correlation between antimicrobial resistance and the presence of resistance genes. Except for CQN, the frequency of resistance to the other 14 antimicrobial agents in those isolates that carried resistance genes was higher than those without resistance genes. This was especially the case for resistance to AMX, DC, FFC, OFX and GEN (P<0.01 or 0.01<P<0.05), which suggested that the prevalence of ESBL genes, tetracycline-resistance genes, phenicol-resistance genes, PMQR genes and aadA was related, to a certain extent, to the observed resistance to AMX, DC, FFC, OFX and GEN.
Conclusions
In conclusion, this is the first study describing the prevalence and characteristics of six categories of resistance determinants in E. coli isolates from chickens in Anhui Province, China. The most abundant genes were ESBL genes (57.6%), class 1 integrons (49.5%) and PMQR genes (46.2%). In 82 isolates, 48 different types of coexistence of the different genes were identified. Six genes, dfrA2-aadA12, tetA, tetM, blaTEM-1, qnrS and aac(6’)-Ib-cr, were detected in an individual E. coli isolate for the first time. Resistance to AMX, DC, FFC, OFX and GEN was significantly different (P<0.01 or 0.01<P<0.05) among the strains that carried and did not carry resistance genes, which indicated that resistance genes contributed to the corresponding antimicrobial resistance.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31201957), and the Annual Disciplinary and Academic Degree Construction Projects in 2013 (XKXWD2013006).
References
- CarattoliA., 2001. Importance of integrons in the diffusion of resistance. Vet. Res. 32:243–259.
- DaiL. LuL.M. WuC.M. LiB.B. HuangS.Y. WangS.C. QiY.H. ShenJ.Z., 2008. Characterization of antimicrobial resistance among Escherichia coli isolates from chickens in China between 2001 and 2006. FEMS Microbiol. Lett. 286:178–183.
- del CastilloB.R. VinueL. RomanE.J. GuerraB. CarattoliA. TorresC. Martínez-MartínezL., 2013. Molecular characterization of multiresistant E. coli producing or not producing extended-spectrum β-lactamases. BMC Microbiol. 13:84.
- DuX.D. LiX.S. ZhangS.M. MoJ. WuN.P. CuiB.A., 2007. Detection of the floR genes in florfenicol resistant Escherichia coli isolated from diseased chickens. J. Henan Agr. Univ. 41:188–191.
- GiovanettiE. BrencianiA. LupidiR. RobertsM.C. VaraldoP.E., 2003. Presence of the tet(O) gene in erythromycin and tetracycline-resistant strains of Streptococcus pyogenes and linkage with either the mef(A) or the erm(A) gene. Antimicrob. Agents Ch. 47:2844–2849.
- HallR.M. CollisC.M., 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593–600.
- KehrenbergC. SchwarzS., 2005. Florfenicolchloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob. Agents Ch. 49:813–815.
- LinJ.C. ChenY.L. CaoS.J. ShuG. WenX.T., 2011. Molecular characterization of integron-gene cassettes in multidrugresistant E. coli isolates from food-producing animals. Acta Vet. Zoot. Sinica 42:77–81.
- LinJ.C. ZhuoJ.Z. JiangH.X. LiuJ.H. ZengZ.L., 2009. Surveillance of antimicrobial resistance among Escherichia coli isolates from swine and poultry in different regions. J. South China Agr. Univ. 30:86–88.
- MaQ.C. GuX. ZhangK.Y. JinL.Y. XueF.Q., 2009. Identification, isolation and antimicrobial susceptibility of Escherichia coli from poultry in Shanghai. China Poultry 31:11–13.
- MarchantM. VinueL. TorresC., 2013. Change of integrons over time in E. coli isolates recovered from healthy pigs and chickens. Vet Microbiol. 163:124–132.
- MaynardC.J. FairbrotherM. BekalS. SanschagrinF. LevesqueR.C. BrousseauR. MassonL. LarivièreS. HarelJ., 2003. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob. Agents Ch. 47:3214–3221.
- SáenzY. BriñasL. DomínguezE. RuizJ. ZarazagaM. VilaJ. TorresC., 2004. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal and food origins. Antimicrob. Agents Ch. 48:3996–4001.
- SoufiL. SáenzY. LauraV. AbbassiM.S. RuizE. ZarazagaM. HassenA.B. HammamiS. TorresC., 2011. E. coli of poultry food origin as reservoir of sulphonamide resistance genes and integrons. Int. J. Food Microbiol. 144:497–502.
- WangY. HeT. HanJ. WangJ. FoleyS.L. YangG.Y. WanS.X. ShenJ.Z. WuC.M., 2012. Prevalence of ESBLs and PMQR genes in fecal Escherichia coli isolated from the non-human primates in six zoos in China. Vet Microbiol. 159:53–59.
- XiaL.N. LiL. WuC.M. LiuY.Q. TaoX.Q. DaiL. QiY.H. LuL.M. ShenJ.Z., 2010. A survey of plasmid-mediated fluoroquinolone resistance genes from Escherichia coli isolates and their dissemination in Shandong, China. Foodborne Pathog. Dis. 7:207–215.
- YanJ.J. WuJ.J. KoW.C. TsaiS.H. ChuangC.L. WuH.M. LuY.J. LiJ.D., 2004. Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J. Antimicrob Ch. 54:1007–1012.
- YuanL. LiuJ.H. HuG.Z. PanS.Y. LiuZ.M. MoJ. WeiY.J., 2009. Molecular characterization of extended-spectrum β-lactamase-producing Escherichia coli isolates from chickens in Henan Province, China. J. Med. Microbiol. 58:1449–1453.
- YuanL. WuH. LiuZ.M. FuX.L. HuG.Z., 2010. Surveillance of antimicrobial resistance among strains of Escherichia coli isolated from chickens in Henan. Acta Agric. Jiangxi 22:128–130.
- ZhangC.P. NingY.B. SongL., 2010. Resistance to tetracycline and distribution of tetracycline resistance determinants in commensal Escherichia coli isolated from clinically healthy chickens and pigs. Sci. Agr. Sin. 43:2578–2583.
- ZhangX.Y. DingL.J. YueJ., 2009. Occurrence and characterization of class 1 and 2 integrons in resistent E. coli isolates from animals and farm workers in Northeastem China. Microb. Drug Resist. 15:323–328.
- ZhouY. YuH. GuoQ. XuX. YeX. WuS. GuoY. WangM., 2010. Distribution of 16S rRNA methylases among different species of Gram-negative bacilli with high-level resistance to aminoglycosides. Eur. J. Clin. Microbiol. 29:1349–1353.
