Abstract
Aim: During infection development in the host, Paracoccidioides spp. faces the deprivation of micronutrients, a mechanism called nutritional immunity. This condition induces the remodeling of proteins present in different metabolic pathways. Therefore, we attempted to identify membrane proteins and their regulation by zinc in Paracoccidioides lutzii. Materials & methods: Membranes enriched fraction of yeast cells of P. lutzii were isolated, purified and identified by 2D LC–MS/MS detection and database search. Results & conclusion: Zinc deprivation suppressed the expression of membrane proteins such as glycoproteins, those involved in cell wall synthesis and those related to oxidative phosphorylation. This is the first study describing membrane proteins and the effect of zinc deficiency in their regulation in one member of the genus Paracoccidioides.
Lay abstract: The methodology of protein identification allows the characterization of biological processes performed by those molecules. Therefore, we performed a membrane proteomic analysis of Paracoccidioides lutzii and further evaluated the responses of the fungus to zinc deprivation. The results obtained in the work allowed the characterization of membrane proteins present in organelles that are related to different cellular functions. Zinc deprivation changes processes related to cellular physiology and metabolism. These results help us to understand the process of pathogen–host interaction, since zinc deprivation is a condition present during infection.
The genus Paracoccidioides comprises the etiologic agents of paracoccidioidomycosis, the most widespread systemic mycosis in Latin America. As other thermodimorphic fungi, members of the genus Paracoccidioides grow as mycelia in the environment and as yeast cells at 36°C and in host tissues [Citation1]. The infection occurs through the host respiratory route [Citation2], in which inhalation of conidia or mycelial propagules allows these structures to reach the host pulmonary alveoli, in which they perform the dimorphic transition and differentiate into yeast cells [Citation3]. This mycosis represents an important public health problem due the quantity of premature deaths, particularly in certain segments of society, such as rural workers [Citation4].
Zinc is a metal of importance for the development of microorganisms. It serves as a structural or catalytic cofactor for many proteins, participating in several processes, such as cell division and differentiation [Citation5]. Zinc homeostasis, a critical process to cells, is maintained at both transcriptional and post-translational levels occurring in response to changes in intracellular zinc levels [Citation6]. The uptake of zinc in Saccharomyces cerevisiae is mediated by two systems: the high-affinity uptake system, which is active in zinc-limited conditions [Citation7]; and the low-affinity uptake system, which is active in the presence of sufficient zinc concentrations [Citation8]. The expression and activity of the high-affinity zinc transporter (Zrt1) and low-affinity zinc transporter (Zrt2) are regulated by the transcription factor, Zap1 [Citation9,Citation10]. During zinc limitation, Zap1 induces the expression of Zrt1 and Zrt2 [Citation11]. The regulation at post-translational level occurs when the cell, under low zinc concentration, is exposed to high extracellular levels of zinc. This exposure causes the internalization of Zrt1p protein via endocytosis and subsequent degradation in the vacuole [Citation10]. Another mechanism of zinc uptake, at the intracellular level, includes the Msc2p and Zrg17p transporters, which are localized at the endoplasmic reticulum and induced under zinc deficiency [Citation12,Citation13]. The vacuole is the main site of zinc sequestration and detoxification in the yeast S. cerevisiae [Citation14]. Two transporters of the cation diffusion facilitator family named Zrc1p and Cot1p are responsible for vacuole zinc accumulation [Citation15]. Zrt3p is another transporter localized at vacuolar membrane, which is responsible for the release of zinc into the cytoplasm upon metal deficiency in S. cerevisiae [Citation15,Citation16].
In Paracoccidioides spp., in silico analyses identified zinc transporters homologs to those described in S. cerevisiae such as Zrt1p, Zrt2p, Zrc1p, Cot1p and Msc2p. Moreover, a homolog to the transcriptional factor Zap1 is also present in the genome of Paracoccidioides spp. [Citation17]. Studies demonstrated the induction of Zrt1 and Zrt2 expression in yeast cells upon zinc deprivation [Citation18,Citation19] and induction of Zrt2 in neutral to alkaline pH during zinc deficiency, as described to ZrfC in Aspergillus fumigatus [Citation18]. Moreover, studies on Paracoccidioides sp., after contact with human plasma and liver of mice revealed the induction of high-affinity zinc transporter under these conditions [Citation20,Citation21].
Plasma membrane plays an important role in cells, acting as a physical barrier, regulating the exchange of information, ions and metabolites between the cell and the environment [Citation22]. Moreover, membranes are responsible for the intracellular compartmentalization of organelles [Citation23]. Membrane proteins perform a variety of functions such as transport, cell adhesion, signal receptors and nutrient uptake. Zinc transporters (Zrt1p and Zrt2p) are located at the plasma membrane [Citation9] on dependence of the concentration of zinc in the medium [Citation10,Citation23]. Around 30% of the open-reading frames in eukaryote genomes encode for integral membrane proteins [Citation24]. However, analysis of membrane proteins is underrepresented in proteomic analyzes, owing to the heterogeneous nature of those molecules [Citation23]. Insolubility in aqueous buffers, hindering the extraction steps of the lipid bilayer [Citation25] and inhibition of the tryptic activity in the segments of the integral membrane proteins [Citation26] make those molecules underrepresented in proteomic analysis. Therefore, the establishment of a methodology able to encompass all these characteristics is a challenge in proteomic sciences [Citation27].
Although membrane proteins perform cellular processes essential to cell survival, the knowledge about this class of proteins is still limited in Paracoccidioides spp. To our knowledge, Paracoccidioides spp. membrane proteome has not been analyzed yet. Therefore, the current study is intended to investigate, using nanoUPLC-MSE and label-free approach, proteins in membranes of Paracoccidioides lutzii and their responses to zinc deprivation. This study allowed the identification of proteins belonging to the whole membrane system, comprising plasma membrane and membranes of cytoplasmic organelles, such as mitochondria, peroxisome, vacuole and endoplasmic reticulum. The biological processes performed by zinc-regulated proteins include composition of cell wall, vesicles traffic, oxidative phosphorylation, zinc uptake in the vacuole and glycosylation in the endoplasmic reticulum. Besides depicting the first large-scale depository of membrane proteins in the genus Paracoccidioides, this article highlights protein candidates potentially involved in the response to zinc limitation, a condition found in the host.
Materials & methods
Fungal strain, growth conditions & zinc deprivation experiments
The experiments were performed with P. lutzii, Pb01 (ATCC MYA-826). The yeast phase was maintained at 36°C in Fava Netto’s medium (1% [w/v] peptone, 0.5% [w/v] yeast extract, 0.3% [w/v] proteose peptone, 0.5% [w/v] beef extract, 0.5% [w/v] NaCl, 4% [w/v] glucose, 1.2% [w/v] agar, pH 7.2) [Citation28] supplemented with 4% (w/v) glucose. Yeast cells were inoculated in Fava Netto’s liquid medium for 72 h at 36°C, 150 rpm, in order to obtain cells at the exponential growth phase. Afterward, the cells were centrifuged at 1200 × g for 10 min at 4°C and washed in phosphate-buffered saline (PBS 1X). The supernatant was discarded, the cells were resuspended in PBS 1× and inoculated at the concentration of 106 cells/ml in McVeigh/Morton’ liquid medium (MMcM) containing: 4% (w/v) glucose, 0.15% (w/v) KH2PO4, 0.05% (w/v) MgSO4.7H2O, 0.015% (w/v) CaCl2.2H2O, 0.2% (w/v) (NH4)2SO4, 0.2% (w/v) L-asparagine, 0.02% (w/v) L-cystine, 1% (v/v) of vitamin supplement (0.006% [w/v] thiamine, 0.006% [w/v] niacin B3, 0.006% [w/v] Ca+2 pantothenate, 0.001% [w/v] inositol B7, 0.0001% [w/v] biotin B8, 0.001% [w/v] riboflavin, 0.01% [w/v] folic acid B9, 0.01% [w/v] choline chloride, 0.01% [w/v] pyridoxine) and 0.1% (v/v) of trace elements supplement (0.0057% [w/v] H3BO3, 0.0081% [w/v] MnSO4.14H2O, 0.0036% [w/v] (NH4)6MO7O24.4H2O, 0.0157% [w/v] CuSO4.H2O, 0.1404% [w/v] Fe(NH4)2(SO4)2.6H2O) [Citation29], pH 7.0, for 24 h.
Following 24 h of cultivation in MMcM medium, 106 cells/ml were transferred to the same medium containing 30 μM ZnSO4.7H2O, or depleted of zinc. The depleted medium was prepared with no addition of ZnSO4 and was supplemented with the zinc chelator N,N,N,N-tetrakis(2-pyridyl-methyl) ethylenediamine (Sigma-Aldrich, MO, USA) at a concentration of 0.05 mM. The cultures were incubated with gentle shaking at 36°C for 24 h, 150 rpm, as previously described [Citation19].
Extraction of membrane proteins
The protocol described by Vidakovics et al. [Citation30] was used with some modifications. Experiments were performed in biological triplicates as three independent experiments. Yeast cells cultured in MMcM medium were centrifuged at 1200× g for 10 min at 4°C, frozen in liquid nitrogen and disrupted by maceration using a gral and pestle until a fine powder was obtained [Citation31]. After this step, the sample was transferred to a conical tube and resuspended in 50 mM Tris-HCl, pH 7.5. Glass beads were added to the conical tube and after agitation for 20 min at 4°C, the sample was subjected to centrifugation at 8000 × g for 10 min at 4°C. The supernatant was diluted in 10 ml of 0.1 M sodium carbonate (Na2CO3) pH 11 for 1 h. The carbonate treated sample was submitted to ultracentrifugation in a Beckman Coulter Optima L-90K centrifuge at 115,000 × g for 1 h, at 4°C. The supernatant was discarded and the pellet containing the membranes fraction was resuspended in 50 mM Tris-HCL pH 7.5 using a glass homogenizer (grinder) on ice, to solubilize the membranes fraction. The sample was submitted to another step of ultracentrifugation at 115,000× g for 1 h at 4°C. An aliquot of the pellet was used for the transmission electron microscopy (TEM) analysis while the remainder was resuspended in 50 mM ammonium bicarbonate pH 8.0 and stored at -80°C.
Transmission electron microscopy
TEM was performed to evaluate the quality of the sample. TEM of the membranes fraction was performed according to standard protocols [Citation32]. The samples of the membrane fraction and intact cells of P. lutzii were fixed in 2% (w/v) paraformaldehyde and 2% (v/v) glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.2, for 2 h, at room temperature. After washing in 0.1 M sodium cacodylate buffer pH 7.2, the samples were postfixed in 2% (w/v) osmium tetroxide, 1.6% (w/v) potassium fericyanide (1:1) and 5 mM CaCl2 in sodium cacodylate buffer, pH 7.2, for 1 h, at room temperature, followed by washing in 0.1 M sodium cacodylate buffer, pH 7.2. The samples were maintained for 12 h in an aqueous solution of 0.5% (v/v) uranyl acetate at 4°C, washed in distilled water and dehydrated in an ascending series of acetone (v/v) (30, 50, 70, 90 and 100%). The material was embedded in a mixture of (3:1) acetone/Spurr resin (Eletron Microscopy Sciences, Co.) for 6 h, (2:1) acetone/Spurr resin overnight, (1:1) acetone/Spurr resin for 6 h, (1:2) acetone/Spurr resin overnight and finally in pure resin for 6 h. The samples were imbibed in Spurr resin for 3 days in an incubator, at 60°C. The ultrathin sections were contrasted with 3% (v/v) aqueous uranyl acetate and 10% (v/v) lead citrate. The samples were analyzed in a TEM, JEM 1011 (Electron Microscopy Sciences, Co, Jeol, Tokyo, Japan.)
Preparation of membrane proteins for nanoUPLC-MSE analysis
The amount of protein in the membrane extract was determined using the Bradford reagent (Sigma-Aldrich) [Citation33]. The samples were analyzed using nanoscale LC–MS/MS. Sample aliquots (100 μg) were prepared for nanoUPLC-MSE as previously described [Citation34]. Briefly, ammonium bicarbonate pH 8.5 at 50 mM and 75 μl of 0.2% (v/v) RapiGEST (Waters Corp, MA, USA) were sequentially added to the samples. After, the solution was vortexed and incubated in dry bath for 15 min at 80°C. After incubation, the samples were centrifuged and the proteins reduced by adding 2.5 μl of a 100 mM DTT solution, followed by incubation for 30 min at 60°C. The alkylation of proteins was performed by addition of 2.5 μl of 300 mM iodoacetamide and incubation in a dark room for 30 min. An aliquot of 40 μl of trypsin (Promega, WI, USA) 50 ng/μl in 50 mM ammonium bicarbonate was added. The sample was vortexed slightly and digested at 37°C for 16 h. Following digestion, the hydrolysis of RapiGEST was performed by addition of 10 μl of 5% (v/v) trifluoroacetic acid and incubation at 37°C for 90 min. The sample was centrifuged at 20,000 × g at 6°C for 30 min, and the supernatant was transferred to microfuge tubes and dried in a speed vacuum. Peptides were solubilized in 30 μl of ultrapure water, submitted to purification and concentration using a pipette tip with a bed of chromatographic media (ZipTips® C18 Pipette Tips, MA, USA) and dried in a speed vacuum. The peptides were resuspended in a solution of 1 pmol/μl MassPREP Digestion Standard (rabbit phosphorilase B; Waters Corp) to prepare the final concentration of 200 fmol/μl of the rabbit phosphorilase B. The buffer solution of 20 mM ammonium formate was used to increase the pH. After solubilization, peptides were transferred to a Waters Total Recovery vial (Waters Corp).
NanoUPLC-MSE analysis
The proteomic analysis was performed using a label-free nanoUPLC-MSE technology. Briefly, tryptic peptides were separated by RP-RP-HPLC using a nanoACQUITYTM system (Waters Corp), as described before [Citation35]. The first column was loaded with 5 μg digests and sequentially, separated in ten fractions in the mobile phase at pH 10. Each fraction was subjected to the second dimension of RP chromatography with a mobile phase at pH 2.5. Label-free data-independent scanning (MSE) experiments were performed with a Synapt HDMS mass spectrometer (Waters, Manchester, UK), which switched between low collision energy (3 eV) and elevated collision energy (12–40 eV) applied to the trap ‘T-wave’ CID cell with argon gas, as described [Citation36].
The protein identifications and quantitative packaging were generated using specific algorithms [Citation37,Citation38] and search was performed against an in-house P. lutzii-specific database. The ProteinLynx Global server v.2.5.2 (PLGS) was used to perform the spectral processing, database searching conditions and quantitative comparisons. The specific database was randomized in order to access the false-positive rate of identification (4%). The intensity measurements were typically adjusted for these components, in other words, the deisotoped and charge state-reduced exact mass-retention times that were replicated throughout the entire experiment for the analysis at the exact mass retention-time cluster level. Components were typically clustered with a 10 ppm mass precision and a 0.25 min time tolerance against the database-generated theoretical peptide ion masses with a minimum of one matched peptide. The alignment of elevated energy ions with low-energy precursor peptide ions was performed with an approximate precision of 0.05 min. The precursor and fragmentation tolerances were determined automatically. The search parameters used were: trypsin as digest reagent, 1 missed cleavage, carbamidomethyl as fixed modification and phosphorylation STY and oxidation M were used as variable modifications. The minimum fragment ion matches per peptide, the minimum fragment ion matches per protein and the minimum peptide matches per protein were, respectively, set as 2, 5 and 1. The mass variation tolerance was set to 50 ppm. A protein detected in all replicates, presenting a variance coefficient less than 10% (PAAG_08059), was used to normalize the expression data to compare the protein levels between control and zinc-limiting conditions. Protein and peptides tables generated by PLGS were merged and the dynamic range of the experiments, peptides detection type and mass accuracy were determined for each condition [Citation39]. Software FBAT [Citation40], MassPivot (kindly provided by AM Murad), Spotfire® (© TIBCO Software Inc.) and Microsoft Office Excel (Microsoft©) were used.
In silico analysis
The data obtained after analysis by nanoUPLC-MSE and identification in ProteinLynx were submitted to in silico search in database in order to determine proteins subcellular location and association with cell membranes. The subcellular location was determined by using WoLF PSORT (www.genscript.com/psort/wolf_psort.html) [Citation41] and Gene Ontology (GO; http:/pedant.gsf.de/) databases [Citation42]. The protein association with membranes evaluated the presence of transmembrane domains and prenylation, myristoylation, palmitoylation and GPI anchor. The TMHMM program, version 2.0 (www.cbs.dtu.dk/services/TMHMM) [Citation43] was used to predict transmembrane regions. The big-PI Fungal Predictor program (http://mendel.imp.ac.at/gpi/fungi_server.html) [Citation44] was used to predict glycosylphosphatidylinositol anchors. The search for myristoylated proteins was performed using the programs, Myristoylator (http://web.expasy.org/myristoylator/) [Citation45] and TermiNator (www.isv.cnrs-gif.fr/terminator3/index.html) [Citation26]. The last was also utilized for the prediction of palmitoylated proteins, while the prediction of prenylation was achieved with the PrePS-Prenylation Prediction Suite (http://mendel.imp.ac.at/sat/PrePS/index.html) software [Citation46]. The search for signal peptide was performed using the SignalP program, version 4.1 (www.cbs.dtu.dk/services/SignalP/) [Citation47]. Loctree program (https://rostlab.org/services/loctree2/) [Citation48] was employed in order to determine in which organelle membrane proteins were localized. The functional classification of proteins present in membranes fraction was accomplished using Funcat2 database (http://pedant.gsf.de/pedant3htmlview/pedant3view?Method=analysis&Db=p3_r48325_Par_brasi_Pb01).
Analysis of chitin amount in the cell wall of P. lutzii
Calcoflour white (CFW, Sigma Biochemical) was utilized to stain P. lutzii (Pb01) yeast cells in order to evaluate the effect of zinc deprivation at the cell wall. P. lutzii yeast cells were grown under zinc deprivation or in presence of this metal for 24 h. For analysis of chitin amount, the cells were fixed in 100% methanol at -80°C for 20 min, at -20°C for additional 20 min and subsequently washed by centrifugation. The cells were collected, stained with CFW (100 μg/ml in PBS 1×) for 30 min and washed with PBS 1×. The specimens were analyzed under a fluorescence microscope (Zeiss Axiocam MRc – Scope A1) [Citation49]. CFW fluorescence intensity was measured using the AxioVision Software (Carl Zeiss AG, Germany). The minimum of 100 cells for each microscope slides, in triplicates, were used to evaluate fluorescence intensity of the cells upon zinc presence or deprivation, for 24 h. The software provided the fluorescence intensity (in pixels) and the standard error of each analysis. Statistical comparisons were performed using the Student’s t test and p-values ≤0.05 were considered statistically significant.
Schiff periodic acid staining of proteins
The staining with Schiff periodic acid was performed in order to evaluate the profile of glycosylated proteins during zinc deficiency. After separation of membrane proteins by electrophoresis, the proteins were fixed in 10% (v/v) acetic acid and 25% (v/v) isopropanol by 18 h at 4°C. After this step, the polyacrylamide gel was washed twice during 15 min with 7.5% (v/v) acetic acid and immersed in a solution of 0.4% (v/v) periodic acid for 1 h at 4°C. Afterward, the gel was immersed in Schiff reagent for 1 h at 4°C in the dark, and subsequently washed three times, 10 min each, in 0.5% (v/v) potassium metabisulfite in the dark. The gel was washed with 7.5% (v/v) acetic acid for 5 min and stored in the same solution at 4°C [Citation50].
Analysis of glycosylated proteins & glucans by fluorescence microscopy
The profile of glycosylated proteins during zinc deprivation was evaluated by staining with Concanavalin A (ConA) TYPE VI conjugated to FITC (Sigma, catalog n.C7642). The yeast cells cultured in the presence or absence of zinc were stained with ConA+FITC at a final concentration of 100 μg/ml [Citation51]. ConA+FITC in a volume of 200 μl was incubated with yeast cells of P. lutzii at 37°C, for 30 min, under stirring. After, the cells were washed with PBS 1×, twice. The cells were incubated with aniline blue solution 100% (v/v) (Sigma, catalog n.B8563) for 5 min under stirring and subsequently washed twice with PBS 1× [Citation52]. Both samples stained with ConA+FITC or aniline blue were visualized under a fluorescence microscope (Zeiss Axiocam MRc-Scope A1). Fluorescence intensity was calculated as described above, for both dyes.
Results
TEM of Pb01 membranes fraction
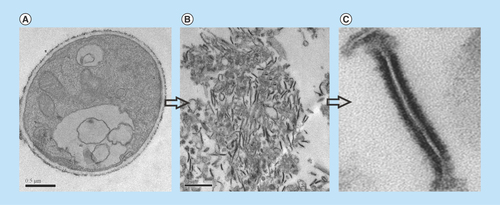
The membrane proteins of Pb01 were obtained according to the steps shown in the workflow chart (Supplementary Figure 1). TEM was performed in order to confirm the enrichment of samples with cell membranes of P. lutzii. As shown in , the membrane fragments detected in the electron micrograph corroborate the enrichment of the extract.
Proteomic data
NanoUPLC-MSE is a method that improves protein and proteome coverage compared with the conventional LC–MS/MS approach [Citation34]. The results presented here are from merged data of three replicates, leading to the identification of 746 proteins from the total membranes preparation of P. lutzii. Of the 746 proteins, 717 were identified with two or more peptides (data not shown). The resulting nanoUPLC-MSE protein and peptide data generated by PLGS process are shown in Supplementary Figures 2 and 3. The experiments resulted in 2938 and 2237 identified peptides; 55 and 59% were obtained from peptide match-type data in the first pass to control and zinc deprivation conditions, respectively, and 16% were obtained in the second pass in both conditions (Supplementary Figure 2). A total of 14% of the peptides were identified by a missed trypsin cleavage in both experimental conditions, whereas in source fragmentation rates of 4 and 1% were obtained to control and zinc deprivation, respectively (Supplementary Figure 2). The results obtained from dynamic range detection are presented in Supplementary Figure 3. This graphic represents the concentration of proteins identified in their detection range.
Subcellular localization of P. lutzii membrane proteins
The identified proteins in the whole membrane system in number of 746 were evaluated regarding to the subcellular localization using the cellular component information of GO and WoLF PSORT softwares (Table 1). The proteins were present in at least two replicates, to be included in the analysis (data not shown). In silico analysis revealed that 27.61% (206 proteins) of the identified proteins are presumably not present in membranes, but located in the cytoplasmic region; 25.2% (188 proteins) of the identified proteins were present in membranes, including plasma membrane, mitochondria, endoplasmic reticulum, Golgi complex, peroxisome and vacuole; 24.66% (184 proteins) were presumably inside mitochondria (Supplementary Figure 4).
Proteins in the whole membrane system of P. lutzii
The criteria for considering proteins as associated with membranes were the presence of transmembrane domains and post-transcriptional modifications (prenylation, myristoylation, palmitoylation and GPI anchor). In addition, proteins described in GO terms as belonging to cell membranes with score higher than 50 were classified as membrane proteins. From the total of 746 identified proteins (Supplementary Table 1), 25.2% (188 proteins) were classified as belonging to the membranes of P. lutzii (Supplementary Table 2).
Regarding to the 188 proteins identified as members of the membranes in yeast cells of P. lutzii, we analyzed the type of cellular membrane that they belong to. According to Loctree software annotations, the identified membrane proteins originated from various organelles, and this finding confirms the ability of the used method to access all cellular membranes. According to our data depicted in Supplementary Table 5, from the whole membrane system, 38% (71 proteins) represent mitochondrial membrane proteins, 24% (46 proteins) and 16% (30 proteins) represent endoplasmic reticulum and plasma membrane proteins, respectively. Besides, Golgi complex membrane proteins represent 5% (10 proteins), vacuole membrane proteins comprise 3% (6 proteins) and nucleus membrane proteins represent 2% (4 proteins). Some proteins exhibited transmembrane domains or post-translational modifications such as prenylation and myristoylation, although they were not classified by the Loctree software. depicts the ten most abundant membrane proteins in this study. Those proteins are present in membranes of different organelles such as mitochondria, peroxissome, endoplasmic reticulum and plasma membrane.
Table 1. The most abundant membrane proteins.
Supplementary Table 3 shows the cell membranes in which proteins are localized. Proteins present at the plasma membrane of yeast cells were represented by those with known functions, such as plasma membrane ATPAse (PAAG_08082), chitin synthase B (PAAG_03391), osmosensor protein (PAAG_04025), a zinc transporter (PAAG_00105) of the Zip family, as well as by proteins with unknown functions. The proteins associated with the endoplasmic reticulum comprise in high number enzymes of the protein glycosylation pathway. Nevertheless, proteins with known mitochondrial localization and functions were detected, such as those involved in import of proteins, enzymes of the electron transport chain and of ATP synthesis. Proteins involved in vesicles transport present in Golgi apparatus were identified, such as ADP ribosylation factor (PAAG_07702), SNARE Ykt6 (PAAG_01588). Furthemore, proteins involved in transport in vacuole and peroxisomes were also identified.
As summarized in Supplementary Table 3, eight proteins present in the endoplasmic reticulum and one Golgi membrane protein were involved in protein glycosylation. Oligosacharyltransferase (PAAG_04719/PAAG_01037) represented with two subunits in the proteome of the endoplasmic reticulum membrane, catalyzes the initial step of N-glycosylation, with the transfer of N-glycan precursor to nascent polypeptide [Citation53]. In addition, calnexin (PAAG_07037), a protein involved in the correct folding of glycoproteins in mammalian cells [Citation54] and fungus [Citation55], was identified as well. The addition of the outer chain to the N-linked core oligosacharide at Golgi is performed by mannosyltransferases; two α-1,2-mannosyltransferases (PAAG_02462/PAAG_07238) were found in membranes of the endoplasmic reticulum and Golgi, as depicted in Supplementary Table 3. In addition, dolichol phosphate mannosyltransferases (PAAG_01874), which perform protein N-glycosylation/O-glycosylation [Citation56], and members of this family were also detected in this study. Proteins related to the glycosylation pathway, such as α-1,2-mannosyltransferase (kre5p, ktr4), mannosyltransferases (PMTs), Stt3p and calnexin [Citation57], were also identified in the membrane proteome of A. fumigatus.
Regulation of P. lutzii membrane proteins by zinc
Proteomic membrane analyses revealed that whole membrane system proteins alter their abundance in response to different stress conditions [Citation30,Citation58]. Therefore, we investigated zinc regulation of membrane proteins in yeast cells grown upon zinc deficiency. Previous studies demonstrated a remodeling of P. lutzii metabolism in response to oxidative stress induced during zinc deprivation [Citation19]. A 1.5-fold change was used as a threshold to determine the up- and downregulated membrane proteins under zinc deprivation. A total of 115 proteins, corresponding to 60.85% of the classified membrane proteins, depicted in Supplementary Table 2, were downregulated upon zinc deprivation (Supplementary Table 4). Eighty one of those proteins were not detectable in the membranes of yeast cells deprived of zinc, only in the control. The functional classification of downregulated membrane proteins revealed that most of them in a percentage of 31% (36 proteins) were involved in transport events (). Of special note, proteins of mitochondrial membrane functionally related to electron transport and oxidative phosphorylation were downregulated, suggesting that energy production by the respiratory chain was inhibited during zinc deprivation. Additionally, the glycosylation of proteins seems to be affected by zinc deprivation, as demonstrated by the downregulation of several enzymes/proteins involved in the process (Supplementary Table 4).
Figure 2. Functional classification of membrane proteins regulated by zinc availability.
The database FunCat2 was used to perform this classification. Light gray bars represent downregulated proteins and dark gray bars represent upregulated proteins. Proteins upregulated: Transport – 56% (10 proteins), Cell cycle and DNA processing – 0% (0 protein), Metabolism – 0% (0 protein), Cellular communication – 0% (0 protein), Protein synthesis – 6% (1 protein), Protein fate – 6% (1 protein), Biogenesis of cellular components – 0% (0 protein), Cell rescue – 6% (1 protein), Energy – 6% (1 protein), Unclassified – 17% (3 proteins), Cell-type differentiation – 0% (0 protein), Cell fate – 6% (1 protein). Proteins downregulated: Transport – 31% (36 proteins), Cell cycle and DNA processing – 2% (2 proteins), Metabolism – 14% (16 proteins), Cellular communication – 5% (6 proteins), Protein synthesis – 1% (1 protein), Protein fate – 10% (11 proteins), Biogenesis of cellular components – 5% (6 proteins), Cell rescue – 4% (5 proteins), Energy – 14% (16 proteins), Unclassified – 13% (15 proteins), Cell-type differentiation – 1% (1 protein), Cell fate – 0% (0 protein).
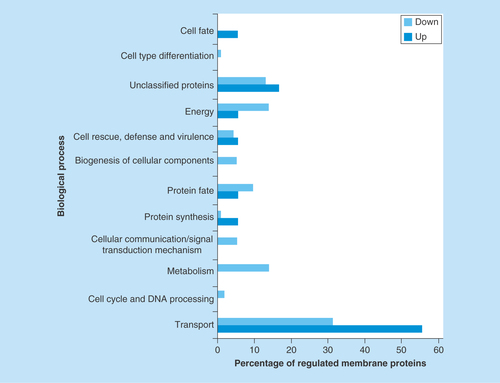
Eighteen proteins corresponding to 9.52% of those classified as belonging to membranes were upregulated upon zinc deprivation (Supplementary Table 5). Sixteen of those proteins were only identified upon zinc deprivation. A total of 56% (10 proteins) of the 18 proteins were involved in transport events and 6% (1 protein) was involved in protein fate ().
The classification of regulated proteins, according to the predicted membranes they belong to, is presented in Supplementary Figure 6. It was observed that 43% (49 proteins) of the downregulated membrane proteins are predicted to be mitochondrial, followed by endoplasmic reticulum 30% (34 proteins) and plasma membrane 15% (17 proteins). Among the upregulated proteins, 28% (5 proteins) were predicted as belonging mainly to mitochondrial membrane and 22% (4 proteins) to plasma membrane.
Modification of P. lutzii yeast cell wall upon zinc deprivation
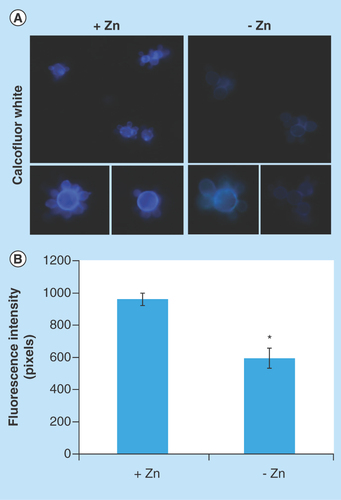
Chitin, glucans, lipids and proteins are the main constituents of the cell walls of mycelium and yeast forms of Paracoccidioides spp. [Citation59,Citation60]. Proteomic analysis revealed that proteins involved in the deposit of chitin in the cell wall, such as chitin synthase B (PAAG_03391) [Citation61] and phosphoinositide phosphatase Sac1 (PAAG_03162) [Citation62], were downregulated under zinc deprivation. Moreover, the enzyme 1-acyl-sn-glycerol-3-phosphate acyltransferase-β (1-AGPAT; PAAG_07503), involved in production of phospholipids, cell wall constituents of Paracoccidioides spp. [Citation63], was also downregulated at zinc-limited conditions. Therefore, fluorescence microscopy was performed in order to investigate changes in the cell wall using the fluorophore CFW that binds specifically the chitin present in the cell wall. As shown in , fluorescence of cell wall in zinc-deprived cells decreased when compared with the control. Quantitative analyzes of the fluorescence intensity (in pixels) of cells growing in the presence and absence of zinc, demonstrated a significant decrease of the fluorescence in cells under zinc deprivation (p < 0.05; ).
Figure 3. Effect of zinc deprivation in Paracoccidioides lutzii yeast cells wall.
Yeast cells were cultivated in MMcM medium depleted or supplemented with zinc for 24 h. (A) The cells were fixed and stained with CFW (increase of 40 times). (B) Fluorescence intensity (in pixels) of the cells under zinc deprivation. The AxioVision Software (Carl Zeiss) was used to obtain the values of fluorescence intensity. The values of fluorescence intensity and the standard error of each analysis were used to plot the graph. Data are expressed as mean ± standard error (represented using error bars).
*p ≤ 0.05.
CFW: Calcofluor white; MMcM: McVeigh/Morton’ liquid medium.
Zinc availability modulates glycosylation events
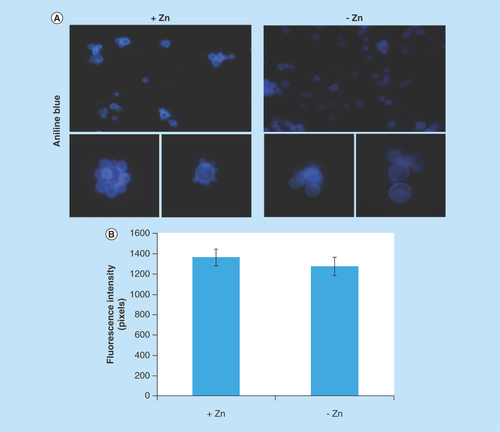
Several proteins of endoplasmic reticulum involved in events of glycosylation were downregulated during zinc deprivation. It includes dolichyl-phosphate-mannose-protein mannosyltransferase (PAAG_05910/PAAG_04725), α-1,2-mannosyltransferase KTR1 (PAAG_07238) and dolichyl-di-phosphooligosaccharide-protein glycotransferase (PAAG_04110). Staining with Schiff periodic acid allows the detection of glycoproteins, through the oxidation reaction between the periodic acid and the glycoproteins. Thus, the intensity of color depends on the number and the nature of sugar moieties bound to the glycoprotein [Citation64]. Under zinc deficiency, the intensity of Schiff staining was lower when compared with the control, as depicted in . The use of ConA conjugated to FITC revealed a significant decrease in the fluorescence of P. lutzii cells after zinc deprivation, as depicted in . ConA is a lectin that specifically binds glucan and mannan moieties [Citation65]. Additionally, the use of aniline blue which selectively stains cell β-(1,3) glucan [Citation52], did not show any difference on fluorescence in cells cultured in the presence or absence of zinc (). In this way, the reduction of protein glycosylation is probably a result of the stress caused by zinc deprivation and not a consequence of a reduced amount of glucans.
Figure 4. Zinc deprivation alters proteins glycosylation.
(A) Protein extracts of membranes were fractionated by electrophoresis and stained with periodic acid Schiff. The same extracts were stained with silver. (B) Fluorescence microscopy of P. lutzii cells that were cultured in the presence or absence of zinc for 24 h and subsequently incubated with ConA conjugated to FITC (increase of 40 times). (C) Fluorescence intensity graph. The data for fluorescence intensity evaluation were obtained through the AxioVision Software (Carl Zeiss). The values of fluorescence intensity (in pixels) and the standard error of each analysis were used to plot the graph. Data are expressed as mean ± standard error (represented using error bars), (*) represents p ≤ 0.05.
ConA: Concanavalin A; FITC: Fluorescein isothiocyanate.
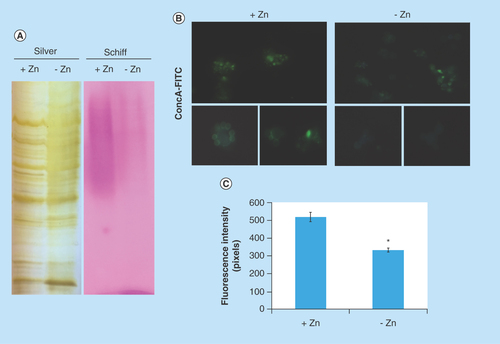
Figure 5. Evaluation of β-1,3 glucan quantities in the cell wall of Paracoccidioides lutzii.
(A) Aniline blue was used to evaluate, by fluorescence microscopy, the presence of β-1,3 glucan in the cell wall of P. lutzii after growth in the presence and absence of zinc (increase of 40 times). (B) Fluorescence intensity graph. The values of fluorescence intensity (in pixels) and the standard error of each analysis were used to plot the graph. Data are expressed as mean ± standard error (represented using error bars).
Discussion
In the whole membrane proteome of P. lutzii, obtained with the use of sodium carbonate pH 11 and ultracentrifugation, 188 proteins were described (Supplementary Table 2). Literature search and in silico analyses revealed proteins associated with different cell membranes. When cells are lysed in an aqueous environment, the cell-limiting membrane and membranes of organelles may fragment and form vesicles that can be separated from the cytosol by partitioning or sedimentation. However, cytoplasmic proteins may be entrapped in vesicles formed after cell lysis. Thus, in order to get a true membrane protein extract, steps employing solutions, such as sodium carbonate, are used to remove proteins that do not show a strong association with membranes [Citation25,Citation30,Citation66]. The use of sodium carbonate pH 11 has been described in the literature as a tool to obtain membrane proteins. Analysis of whole membrane system proteome of pathogenic and nonpathogenic microorganisms, such as Escherichia coli [Citation67], Bordetella pertussis [Citation30] and S. cerevisiae [Citation58], employing sodium carbonate pH 11 and steps of ultracentrifugation, allowed the identification of several membrane proteins. The success of the technique utilized for obtaining P. lutzii membrane proteins can be evaluated by comparing our data to those in the literature. In analyses of plasma membrane proteome of S. cerevisiae during salt stress, 24% of the identified proteins belong to the plasma membrane while 25 and 33% were represented by ribosomal proteins and proteins from the early secretory pathway belonging to endoplasmic reticulum and Golgi apparatus, respectively [Citation58]. At this work, 25.2% (188 proteins) of the total of identified proteins were predicted as belonging to membranes (Supplementary Figure 4).
In this study we identified 188 proteins of membranes, and from those, 63.3% (119 proteins) presented at least one transmembrane domain, 9.04% (17 proteins) presented post-translational modifications such as prenylation, myristoylation, palmitoylation, GPI anchor and 9.57% (18 proteins) presented signal peptide (Supplementary Table 2). In C. albicans, from 214 identified proteins in a preparation of plasma membrane, 47.66% (102 proteins) showed at least one transmembrane domain and 13.55% (29 proteins) presented signal peptide [Citation68]. Comparing the data, it is possible to see that the forms of association with cell membranes approximate to the number found in this work.
In this work, membrane proteins involved in synthesis or maintenance of cell wall, in glycosylation pathways and in the electron transport chain were identified (Supplementary Table 2), similarly to data described in C. albicans [Citation68]. Also, the whole membrane proteome of P. lutzii includes proteins related to vesicle traffic, such as ADP ribosylation factor (PAAG_07702), SNARE (PAAG_01588). In addition, some proteins were described only in this work compared with C. albicans [Citation68] and A. fumigatus [Citation57], such as zinc transporter (PAAG_00105), sideroflexin-1 (PAAG_09064) and peroxin (PAAG_08209; Supplementary Table 2). Bioinformatics analyses allowed the identification and classification of membrane proteins, not yet described in some proteomic works.
Some studies with Paracoccidioides spp. under metals deprivation allowed the characterization of metabolic and adaptive responses to iron [Citation69] and zinc [Citation19]. Due to the importance of zinc in pathogens survival and in the development of diseases, we evaluated membrane proteins in yeast cells of P. lutzii upon deprivation of this metal. Analysis of whole membrane system proteome during zinc deprivation allowed identification of a probable alteration on glycerophospholipid metabolism, as predicted by the regulation of enzymes located in endoplasmic reticulum membrane, such as the 1-AGPAT (PAAG_07503). This enzyme catalyzes the formation of phosphatidic acid (PA) from lysophosphatidic acid by incorporating an acyl moiety at sn-2 position. The PA produced by the 1-AGPAT activity is a substrate of the pathway for production of phospholipids, such as phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidylethanolamine (PE) and phosphatidylcholine (PC), which are present at the cell wall of S. cerevisiae [Citation70] and Paracoccidioides spp. [Citation63]. Similarly, in S. cerevisiae, the activities of the enzymes involved in PS, PE and PC pathways were decreased under zinc deficiency [Citation71]. As PA is the upstream substrate in the synthesis of all those phospholipids, we may suggest that the decreased production of PA by repression of the 1-AGPAT can lead to the reduction in PS, PE and PC levels and possible alterations in cell wall composition. Additionally to the descriptions above, another protein of the endoplasmic reticulum membrane, named Sac1, is involved in structure of cell wall and was downregulated in P. lutzii upon zinc deprivation, strongly suggesting modifications in the cell wall structure. Studies in C. albicans demonstrated that Sac1-p is related to the cell wall integrity. The deletion of Sac-1 increases the sensitivity to stress of the cell wall and alters the content and distribution of chitin in the mutant [Citation72]. Moreover, chitin synthase B (PAAG_03391), a plasma membrane protein, was also downregulated in this study in the absence of zinc. The reduced level of those proteins possibly alters the quantity of chitin at the cell wall, as depicted in .
Zinc deprivation repressed proteins present in membranes of organelles such as mitochondria and vacuole, as depicted in Supplementary Table 4. Studies performed with S. cerevisiae revealed that the transport of zinc into the vacuole has an ATP-dependent mechanism, requiring an H+ gradient generated by the V-ATPase. Thus, changes in the proton gradient generated by V-ATPase inhibit zinc uptake by the vacuole [Citation73]. In this work, a vacuolar ATP synthase of 98 kDa (PAAG_02679) was repressed during zinc deprivation. As the vacuole is the main organelle responsible for zinc storage [Citation14], the cells possibly inhibit the storage of this micronutrient in order to try to overcome zinc deprivation, allowing this metal available in the cytoplasm.
Protein glycosylation is the most common post-translational modification in eukaryotic cells. This process occurs by connecting a saccharide unit to a protein and is involved in the maintenance of protein conformation and activity, in protein protection from proteolytic degradation, and in protein intracellular trafficking and secretion [Citation74]. In studies performed with Candida albicans, it was observed that the process of glycosylation is important for cell wall integrity and for host–fungus interactions [Citation75,Citation76]. Also, in C. albicans, Hall et al. [Citation77] observed that the Mnn2 mannosyltransferase family is related in immune recognition, virulence and cell wall integrity. Based on these facts, the reduction in the level of glycosylated proteins may interfere in the success of infection. In this work, we described downregulated membrane proteins present at the endoplasmic reticulum and Golgi apparatus involved in the process of N-O-glycosylation [Citation57], such as α-1,2 mannosyltransferase KTR1 (PAAG_07238), dolichyl-di-phosphooligosaccharide-protein glycotransferase (PAAG_04110), calnexin (PAAG_07037), oligosaccharyl transferases (PAAG_04719/PAAG_01037). The dolichol-phosphate mannose Dpm1p protein, localized at the endoplasmic reticulum membrane, was downregulated under zinc-limited conditions. Dpm1p synthesizes Dol-P-Man, a compound that serves as mannosyl donor for glycosylation reactions in the endoplasmic reticulum lumen [Citation78]. We suggest that repression of those proteins upon zinc deprivation affects the glycosylation process in the endoplasmic reticulum in P. lutzii and, consequently, the profile of glycosylated proteins is less evident under zinc deprivation (). Furthermore, the aniline blue staining confirmed that the reduction of protein glycosylation is not a consequence of a reduced amount of glucans (), but probably a result of the stress caused by zinc deprivation. Furthermore, the deletion of genes-encoding proteins of the N-glycosylation pathway, such as α-glucosidase-I and O-Mannosyltransferases, in A. fumigatus and S. pombe resulted in abnormalities in the cell wall [Citation79–81]. We hypothesized that the repression of proteins involved in the glycosylation pathway in P. lutzii during zinc deprivation contributes to changes in the cell wall organization in this fungus.
Another response to zinc deprivation identified in this work was at the mitochondrial membrane level. Part of the energy used by cells is obtained during the process of oxidative phosphorylation that occurs in the inner mitochondrial membrane. Electrons generated in glycolysis and tricarboxylic acid cycle are used during energy achievement. A study with S. cerevisiae revealed that zinc deficiency leads to increased oxidative stress [Citation82]. Aspergillus niger submitted to long-term oxidative stress reduces glucose uptake [Citation83] and induces enzymes involved in the gluconeogenesis pathway. Parente et al. [Citation19] observed that P. lutzii yeast cells cultivated for 24 h under zinc deprivation also induce gluconeogenesis. Furthermore, Gupta et al. [Citation84] found that the tricarboxylic acid cycle had decreased activity when Aspergillus parasiticus is cultured in medium with zinc deficiency. In the present study, we observed that proteins of oxidative phosphorylation were downregulated upon zinc deficiency (Supplementary Table 4). Additionally, the mitochondrial inner membrane Sco1 protein (PAAG_06668) [Citation85], which is essential for cytochrome oxidase assembly [Citation86], was also repressed during zinc deficiency in P. lutzii, corroborating previous data that pointed to a metabolism shift in P. lutzii upon zinc deficiency.
Proteins associated with traffic of vesicles were regulated by zinc, as well. GTP-binding proteins play a role in vesicle traffic through a cycle of GTP-binding and hydrolysis. GTP-binding proteins present at the surface of carrier vesicles are responsible for the delivery of these structures to the appropriate acceptor compartment. The presence of Rab-GTPases GTP-bound (active) and GDP-bound (inactive) form is mediated by interaction with regulatory proteins. The active form of Rabp proteins interacts with Rab effectors and GTPase-activating proteins and in inactive form, these proteins are recognized by guanine nucleotide exchange factors [Citation87]. The Rab GDP-dissociation inhibitor protein (PAAG_06344) of P. lutzii was upregulated upon zinc deprivation (Supplementary Table 5). Its homolog Gdi1p, from S. cerevisiae induces the inhibition of GDP dissociation from Sec4p. A reversible modification of either Sec4p or Gdilp would cause a dissociation of this complex, allowing attachment of Sec4p to a new vesicle [Citation88]. The upregulation of Rab GDP-dissociation inhibitor in membrane proteome during zinc deprivation, here described, may induce the formation of a soluble inactive complex between Rabp and Sec4p proteins and, consequently, the presence of this complex could decrease the binding of Sec4 to new vesicles during zinc deprivation. The protein homolog to Sec4p, the GTP-binding protein SAS1 (PAAG_01500) was downregulated. The repression of those proteins may be associated with induction of Rab GDP-dissociation inhibitor protein and formation of complex Rabp/Sec4p, and consequently suppression on the traffic of vesicles under zinc limitation.
Upon microbial infection, the host sequesters zinc from either extra- and intracellular compartments [Citation89,Citation90] in order to limit microorganisms growth and proliferation. Considering this perspective, the use of zinc chelators during infection treatment may be suggested. Laskaris et al. [Citation91] showed that mice infected with A. fumigatus had survival improved with administration of zinc-chelating agents in monotherapy and in combination with the antifungal caspofungin. Also, the use of metal chelators has been well documented in respect of iron [Citation92–94]. As demonstrated here and also by Parente et al. [Citation19], zinc plays a role in essential processes that contribute to the maintenance of P. lutzii physiology. Taking this fact into account, the use of agents able to sequester this metal, alone or together with antifungals, may be an alternative in paracoccidioidomycosis treatment.
Conclusion & future perspective
The development and advance of strategies of treatment in the field of infectious diseases are based on the knowledge of a microorganism’s physiology and metabolism. Information of pathogen behavior in situations that mimic conditions found in the host are valuable, since they can be used as guides in the definition of targets for treatment. To our knowledge, this is the first report that describes whole membrane system proteome and the response of P. lutzii to micronutrient starvation at the membrane proteome level. Identification of total membrane proteome of a member of the genus Paracoccidioides provides a starting point for future studies on functional analysis. Protein glycosylation and chitin content at the cell wall, both processes important in host–fungus interaction, were decreased under zinc deprivation, a condition found in the host. These and other metabolic changes described in this study may contribute to the arsenal of targets used in the development of new drugs in the future. Additionally, the establishment of a protocol for Paracoccidioides spp. membrane protein extraction opens new possibilities for the understanding of fungus biology.
This was the first work describing the whole membrane proteins of the pathogenic fungus Paracoccidioides luztii and its regulation in a condition that mimics that found in the host.
In this study, employing a methodology for extracting membrane proteins and nanoUPLC-MSE, 188 proteins from the membrane system were identified.
Under conditions of zinc deprivation, membrane proteins are regulated. In general, this stress alters several cellular processes carried out by proteins such as glycosylation, energy production, storage of zinc in vacuoles and vesicle traffic.
Proteomic data revealed that membrane proteins involved in cell wall synthesis were repressed by zinc deprivation. This finding was confirmed by fluorescence microscopy. The cell wall is essential for the contact of the pathogen with the host.
The data of this article allowed the characterization of the membrane proteins of P. lutzii in response to stress mediated by zinc deprivation.
Author contributions
CMA Soares, JS de Curcio and MG Silva conceived and designed the experiments. JS de Curcio, MG Silva and L Casaletti performed the experiments. JS de Curcio, MG Silva, MGS Bailão, AM Bailão and CMA Soares analyzed and/or interpreted the data. SN Báo and CMA Soares contributed to reagents and materials. JS de Curcio, MG Silva and CMA Soares wrote the manuscript.
Supplementary docx
Download TIFF Image (2.3 MB)Supplementary docx
Download TIFF Image (1.7 MB)Supplementary docx
Download TIFF Image (2.5 MB)Supplementary docx
Download TIFF Image (919.6 KB)Supplementary docx
Download TIFF Image (1.3 MB)Supplementary docx
Download TIFF Image (967.1 KB)Supplementary Material For: Supplementary Table 1. Functional classification of proteins identified in the whole membrane system in P. lutzii
Download MS Word (232.3 KB)Supplementary Material For: Supplementary Table 2.Membranes proteins of Paracoccidioides lutzii.
Download MS Word (85.2 KB)Supplementary Material For: Supplementary Table 3.Subcellular localization of membrane proteins in P. lutzii
Download MS Word (64.9 KB)Supplementary Material For: Supplementary Table 4. Membranes proteins of P. lutzii down-regulated upon zinc deprivation, their biological functions and cellular localization (organelle) provided by FunCat2 and Loctree, respectively.
Download MS Word (56.6 KB)Supplementary Material For: Supplementary Table5. Membranes proteins of P. lutziiup regulated upon zinc deprivation, their biological functions and cellular localization (organelle) provided for FunCat2 and Loctree, respectively.
Download MS Word (24.1 KB)Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.tandfonline/doi/full/10.4155/fsoa-2017-0044
Financial & competing interests disclosure
This work was supported by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado de Goiás (FAPEG) – Instituto Nacional de Ciência e Tecnologia (INCT) de Estratégias de Interação Patógeno Hospedeiro (IPH) and Fundo Newton. JS de Curcio and MG Silva are fellows from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and CNPq, respectively. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- Restrepo AM. The ecology of Paracoccidioides brasiliensis: a puzzle still unsolved. Sabouraudia J. Med. Vet. Mycol. 23(5), 323–334 (1985).
- San-Blas G, Niño-Vega G, Iturriaga T. Paracoccidioides brasiliensis and paracoccidioidomycosis: molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med. Mycol. 40(3), 225–242 (2002).
- McEwen JG, Bedoya V, Patino MM, Salazar ME, Restrepo A. Experimental murine paracoccidiodomycosis induced by the inhalation of conidia. J. Med. Vet. Mycol. 25(3), 165–175 (1987).
- Shikanai-Yasuda MA, Telles FD, Mendes RP, Colombo AL, Moretti ML, Paracoccidioidomycose G. Guideliness in paracoccidioidomycosis. Rev. Soc. Bras. Med. Trop. 39(3), 297–310 (2006).
- Van Ho A, Ward DM, Kaplan J. Transition metal transport in yeast. Annu. Rev. Microbiol. 56, 237–261 (2002).
- Eide DJ. Multiple regulatory mechanisms maintain zinc homeostasis in Saccharomyces cerevisiae. J. Nutr. 133(5 Suppl. 1), S1532–S1535 (2003).
- Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl Acad. Sci. USA 93(6), 2454–2458 (1996).
- Zhao H, Eide D. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271(38), 23203–23210 (1996).
- Zhao H, Eide DJ. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 17(9), 5044–5052 (1997).
- Gitan RS, Luo H, Rodgers J, Broderius M, Eide D. Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem. 273(44), 28617–28624 (1998).
- Rutherford JC, Bird AJ. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eurkaryot. Cell 3(1), 1–13 (2004).
- Wu YH, Frey AG, Eide DJ. Regulation of the Zrg17 zinc transporter in the yeast secretory pathway. Biochem. J. 435(1), 259–266 (2011).
- Ellis CD, Wang F, MacDiarmid CW, Clark S, Lyons T, Eide DJ. Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J. Cell Biol. 166(3), 325–335 (2004).
- Ramsay LM, Gadd GM. Mutants of Saccharomyces cerevisiae defective in vacuolar function confirm a role for the vacuole in toxic metal ion detoxification. FEMS Microbiol. Lett. 152, 293–298 (1997).
- MacDiarmid CW, Gaither LA, Eide D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19(12), 2845–2855 (2000).
- Eide DJ. Zinc transporters and the cellular trafficking of zinc. Biochim. Biophys. Acta 1763(7), 711–722 (2006).
- Silva MG, Schrank A, Bailão EFLC et al. The homeostasis of iron, copper, and zinc in Paracoccidioides brasiliensis, Cryptococcus neoformans var. Grubii, and Cryptococcus gattii: a comparative analysis. Front. Microbiol. 2, 1–19 (2011).
- Bailão EFLC, Parente AFA, Parente JA et al. Metal acquisition and homeostasis in fungi. Curr. Fungal Infect. Rep. 6(4), 257–266 (2012).
- Parente AFA, de Rezende TCV, de Castro KP et al. A proteomic view of the response of Paracoccidioides yeast cells to zinc deprivation. Fungal Biol. 117(6), 399–410 (2013).
- Bailão AM, Schrank A, Luiz C et al. Differential gene expression by Paracoccidioides brasiliensis in host interaction conditions: representational difference analysis identifies candidate genes associated with fungal pathogenesis. Microbes Infect. 8(12-13), 2686–2697 (2006).
- Bailão AM, Shrank A, Borges CL et al. The transcriptional profile of Paracoccidioides brasiliensis yeast cells is in influenced by human plasma. FEMS Immunol. Med. Microbiol. 51(1), 43–57 (2007).
- Ephritikhine G, Ferro M, Rolland N. Plant membrane proteomics. Plant Physiol. Biochem. 42(12), 943–962 (2004).
- Tan S, Hwee TT, Chung MCM. Membrane proteins and membrane proteomics. Proteomics 8(19), 3924–3932 (2008).
- Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 7(4), 1029–1038 (1998).
- Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis 21(6), 1054–1070 (2000).
- Hopff D, Wienkoop S, Lüthje S. The plasma membrane proteome of maize roots grown under low and high iron conditions. J. Proteomics 91, 605–618 (2013).
- Rucevic M, Hixson D, Josic D. Mammalian plasma membrane proteins as potential biomarkers and drug targets. Electrophoresis 32(13), 1549–1564 (2011).
- Fava-Netto C. Estudos quantitativos sobre fixação de complemento na blastomicose sul americana, com antígeno polissacarídico. Arq. Cir. Clin. Exp. 18, 197–254 (1955).
- Restrepo A, Jimenez BE. Growth of Paracoccidioides brasiliensis yeast phase in a chemically defined culture medium. J. Clin. Microbiol. 12(2), 279–281 (1980).
- Vidakovics MLP, Paba J, Lamberti Y, Andre C, De Sousa MV, Rodriguez ME. Profiling the Bordetella pertussis proteome during iron starvation research. J. Proteome Res. 6, 2518–2528 (2006).
- Da Fonseca CA, Jesuino RSA, Felipe MS, Cunha DA, Brito WA, Soares CMA. Two-dimensional electrophoresis and characterization of antigens from Paracoccidioides brasiliensis. Microbes Infect. 3(7), 535–542 (2001).
- Barbosa SM, Báo SN, Andreotti PF et al. Glyceraldehyde-3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect. Immun. 74(1), 382–389 (2006).
- Bradford M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
- Murad AM, Souza GHMF, Garcia JS, Rech EL. Detection and expression analysis of recombinant proteins in plant-derived complex mixtures using nanoUPLC-MSE. J. Sep. Sci. 34(19), 2618–2630 (2011).
- Geromanos SJ, Vissers JPC, Silva JC et al. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9(6), 1683–1695 (2009).
- Curty N, Kubitschek-Barreira PH, Neves GW et al. Discovering the infectome of human endothelial cells challenged with Aspergillus fumigatus applying a mass spectrometry label-free approach. J. Proteomics 97, 126–140 (2014).
- Silva JC. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5(1), 144–156 (2006).
- Silva JC, Denny R, Dorschel CA et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 77(7), 2187–2200 (2005).
- Murad AM, Rech EL. NanoUPLC-MSE proteomic data assessment of soybean seeds using the Uniprot database. BMC Biotechnol. 12(1), 82 (2012).
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet. Epidemiol. 19(Suppl. 1), S36–S42 (2000).
- Horton P, Park KJ, Obayashi T et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35(Suppl. 2), 585–587 (2007).
- Frishman D, Mokrejs M, Kosykh D et al. The PEDANT genome database. Nucleic Acids Res. 31(1), 207–211 (2003).
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305(3), 567–580 (2001).
- Eisenhaber B, Schneider G, Wildpaner M, Eisenhaber F. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyce. J. Mol. Biol. 337(2), 243–253 (2004).
- Bologna G, Yvon C, Duvaud S, Veuthey AL. N-terminal myristoylation predictions by ensembles of neural networks. Proteomics 4(6), 1626–1632 (2004).
- Maurer-Stroh S, Eisenhaber F. Refinement and prediction of protein prenylation motifs. Genome Biol. 6(6), R55 (2005).
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8(10), 785–786 (2011).
- Goldberg T, Hecht M, Hamp T et al. LocTree3 prediction of localization. Nucleic Acids Res. 42(W1), 1–6 (2014).
- Zambuzzi-Carvalho PF, Tomazett PK, Santos SC et al. Transcriptional profile of Paracoccidioides induced by oenothein B, a potential antifungal agent from the Brazilian Cerrado plant Eugenia uniflora. BMC Microbiol. 13, 227 (2013).
- Maizels RM, Blaxter ML, Robertson BD, Selkirk ME. Parasite Antigens, Parasite Genes: A Laboratory Manual for Molecular Parasitology. Cambridge University Press, Cambridge, UK (1992).
- Sagaram US, Shaw BD, Shim WB. Fusarium verticillioides GAP1, a gene encoding a putative glycolipid-anchored surface protein, participates in conidiation and cell wall structure but not virulence. Microbiology 153(9), 2850–2861 (2007).
- Renshaw H, Vargas-Muñiz JM, Richards AD, Asfaw YG, Juvvadi PR, Steinbach WJ. Distinct roles of myosins in Aspergillus fumigatus hyphal growth and pathogenesis. Infect. Immun. 84, IAI.01190–IAI.01115 (2016).
- Knauer R, Lehle L. The oligosaccharyltransferase complex from yeast. Biochim. Biophys. Acta 1426(2), 259–273 (1999).
- Ruiz-Canada C, Kelleher DJ, Gilmore R. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Clin. Lymphoma. 9(1), 19–22 (2009).
- Wang N, Seko A, Takeda Y, Kikuma T, Ito Y. Cooperative role of calnexin and TigA in Aspergillus oryzae glycoprotein folding. Glycobiology 25(10), 1090–1099 (2015).
- Goto M. Protein O-glycosylation in fungi: diverse structures and multiple functions. Biosci. Biotechnol. Biochem. 71(6), 1415–1427 (2007).
- Ouyang H, Luo Y, Zhang L, Li Y, Jin C. Proteome analysis of Aspergillus fumigatus total membrane proteins identifies proteins associated with the glycoconjugates and cell wall biosynthesis using 2D LC-MS/MS. Mol. Biotechnol. 44(3), 177–189 (2010).
- Szopinska A, Degand H, Hochstenbach JF, Nader J, Morsomme P. Rapid response of the yeast plasma membrane proteome to salt stress. Mol. Cell. Proteomics 10(11), M111.009589 (2011).
- Kanetsuna F, Carbonell LM. Cell wall glucans of the yeast and mycelial forms of Paracoccidioides brasiliensis. J. Bacteriol. 101(3), 675–680 (1970).
- San-blas G. The cell wall of fungal human pathogens: its possible role in host-parasite relationships dermatophytes. Young 184(Ivic), 159–184 (1982).
- Preechasuth K, Anderson JC, Peck SC, Brown AJP, Gow NAR, Lenardon MD. Cell wall protection by the Candida albicans class I chitin synthases. Fungal Genet. Biol. 82, 264–276 (2015).
- Whitters EA, Cleves AE, McGee TP, Skinner HB, Bankaitis VA. SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J. Cell Biol. 122(1), 79–94 (1993).
- Longo LVG, Nakayasu ES, Gazos-Lopes F et al. Characterization of cell wall lipids from the pathogenic phase of Paracoccidioides brasiliensis cultivated in the presence or absence of human plasma. PLoS ONE 8(5), e63372 (2013).
- Matthieu JM, Quarles RH. Quantitative scanning of glycoproteins on polyacrylamide gels stained with periodic acid-schiff reagent (PAS). Anal. Biochem. 55(1), 313–316 (1973).
- Shaw BD, Hoch HC. The pycnidiospore of Phyllosticta ampelicida: surface properties involved in substratum attachment and germination. Mycol. Res. 103(7), 915–924 (1999).
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93(1), 97–102 (1982).
- Molloy M, Herbert B, Slade M. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. 2881, 1–11 (2000).
- Cabezón V, Llama-Palacios A, Nombela C, Monteoliva L, Gil C. Analysis of Candida albicans plasma membrane proteome. Proteomics 9(20), 4770–4786 (2009).
- Parente AFA, Bailão AM, Borges CL et al. Proteomic analysis reveals that iron availability alters the metabolic status of the pathogenic fungus Paracoccidioides brasiliensis. PLoS ONE 6(7), e22810 (2011).
- Carman GM, Han G-S. Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc depletion. Biochim. Biophys. Acta 1771(3), 322–330 (2007).
- Iwanyshyn WM, Han G-S, Carman GM. Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc. J. Biol. Chem. 279(21), 21976–21983 (2004).
- Zhang B, Yu Q, Jia C et al. The actin-related protein Sac1 is required for morphogenesis and cell wall integrity in Candida albicans. Fungal Genet. Biol. 81, 261–270 (2015).
- MacDiarmid CW, Milanick MA, Eide DJ. Biochemical properties of vacuolar zinc transport systems of Saccharomyces cerevisiae. J. Biol. Chem. 277(42), 39187–39194 (2002).
- Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3(2), 97–130 (1993).
- Albrecht A, Felk A, Pichova I et al. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281(2), 688–694 (2006).
- Mora-Montes HM, Bates S, Netea MG et al. Endoplasmic reticulum alpha-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction. Eukaryot. Cell 6(12), 2184–2193 (2007).
- Hall RA, Bates S, Lenardon MD et al. The Mnn2 mannosyltransferase family modulates mannoprotein fibril length, immune recognition and virulence of Candida albicans. PLoS Pathog. 9(4), 13–17 (2013).
- Burda P, Aebi M. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426(2), 239–257 (1999).
- Willer T, Brandl M, Sipiczki M, Strahl S. Protein O-mannosylation is crucial for cell wall integrity, septation and viability in fission yeast. Mol. Microbiol. 57(1), 156–170 (2005).
- Zhou H, Hu H, Zhang L et al. O-mannosyltransferase 1 in Aspergillus fumigatus (AfPmt1p) is crucial for cell wall integrity and conidium morphology, especially at an elevated temperature. Eukaryot. Cell 6(12), 2260–2268 (2007).
- Zhang L, Feng D, Fang W et al. Comparative proteomic analysis of an Aspergillus fumigatus mutant deficient in glucosidase I (AfCwh41). Microbiology 155(7), 2157–2167 (2009).
- Wu C, Bird AJ, Winge DR, Eide DJ. Regulation of the yeast TSA1 peroxiredoxin by ZAP1 is an adaptive response to the oxidative stress of zinc deficiency. J. Biol. Chem. 282(4), 2184–2195 (2007).
- Li Q, Abrashev R, Harvey LM, McNeil B. Oxidative stress-associated impairment of glucose and ammonia metabolism in the filamentous fungus, Aspergillus niger B1-D. Mycol. Res. 112(9), 1049–1055 (2008).
- Gupta SK, Maggon KK, Venkitasubramanian TA. Effect of Zinc on tricarboxylic acid cycle intermediates and enzymes in relation to aflatoxin biosynthesis. J. Gen. Microbiol. 99(1), 43–48 (1977).
- Buchwald P, Krummeck G, Rödel G. Immunological identification of yeast SCO1 protein as a component of the inner mitochondrial membrane. Mol. Gen. Genet. 229(3), 413–420 (1991).
- Glerum DM, Shtanko A, Tzagoloff A. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 271(34), 20531–20535 (1996).
- Ignatev A, Kravchenko S, Rak A, Goody RS, Pylypenko O. A structural model of the GDP dissociation inhibitor Rab membrane extraction mechanism. J. Biol. Chem. 283(26), 18377–18384 (2008).
- Garrett MD, Zahner JE, Cheney CM, Novick PJ. GDI1 encodes a GDP dissociation inhibitor that plays an essential role in the yeast secretory pathway. EMBO J. 13(7), 1718–1728 (1994).
- Corbin BD, Seeley EH, Raab A et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319(5865), 962–965 (2008).
- Vignesh KS, Figueroa JAL, Porollo A, Caruso JA, Deepe GS Jr. Granulocyte macrophage-colony stimulating factor-induced Zn sequestration enhances macrophage superoxide and limits intracellular pathogen survival. Immunity 39(4), 697–710 (2013).
- Laskaris P, Atrouni A, Calera JA, Enfert C. Administration of zinc chelators improves survival of mice infected with Aspergillus fumigatus both in monotherapy and in combination with caspofungin. Antimicrob. Agents Chemother. 60(10), 5631–5639 (2016).
- Ibrahim AS, Gebremariam T, French SW, Edwards JE Jr, Spellberg B. The iron chelator deferasirox enhances liposomal amphotericin B efficacy in treating murine invasive pulmonary aspergillosis. J. Antimicrob. Chemother. 65(2), 289–292 (2009).
- Leal SM, Roy S, Vareechon C et al. Targeting iron acquisition blocks infection with the fungal pathogens Aspergillus fumigatus and Fusarium oxysporum. PLoS Pathog. 9(7), 1–16 (2013).
- Zarember KA, Cruz AR, Yuang C-Y, Gallin JI. Antifungal activities of natural and synthetic iron chelators alone and in combination with azole and polyene antibiotics against Aspergillus fumigatus. Antimicrob. Agents Chemother. 53(6), 2654–2656 (2009).