Abstract
Aim: To develop a useful alternative approach to evaluate the gene function in hairy roots. Methods: Arabidopsis and tobacco (wild-type or mutant) were a host for Agrobacterium rhizogenes transformation. Results: The hairy roots formation efficiency ranged from 53 to 98% in tobacco and 53 to 66% in Arabidopsis. Hairy and intact roots showed similar gene expression pattern in response to salt and aluminum stress. Genomic polymerase chain reaction and fluorescent images showed high rate (>80%) of co-integration of T-DNAs and uniform cell transformation without use of any antibiotic selection. Whole processes of hairy roots were completed within 1 month after the infection of Agrobacterium. Conclusion: Aluminum-responsive orthologous gene function could be evaluated by NtSTOP1-KD and Atstop1 as a host for hairy roots transformation.
Lay abstract
Hairy roots have been used for various purposes over the last several years. In the present study we used Agrobacterium rhizogenes-mediated hairy roots as an alternative approach for Agrobacterium tumefaciens transformation. We developed a simple, effective and reproducible hairy root protocol in tobacco and Arabidopsis. Developed hairy roots were compared with intact roots to characterize the gene response to aluminum, NaCl stress and cellular localization of genes. Aluminum-responsive orthologous genes function could be evaluated by using NtSTOP1-KD and Atstop1 as a host for hairy roots transformation.
Recent molecular biological studies, in particular those using model plant systems such as Arabidopsis thaliana and rice (Oryza sativa), have identified various genes that regulate stress tolerance. Further studies of orthologs in crops provide valuable information for breeding of stress-tolerant varieties by molecular breeding. A number of the dehydration-responsive element binding protein (DREB) family, encoding transcription factors regulating drought, cold and salt tolerance, were originally identified in A. thaliana. Functional orthologs were identified from various major crops such as rice, soybean and from drought tolerance crop cowpea [Citation1–3]. Several Al tolerance genes, such as the genes encoding aluminum-activated transporter1 (ALMT1) and multidrug and toxic compound extrusion1 (MATE1) of malate and citrate transporters, respectively, were identified from various crop plants [Citation4–6]. These genes could be useful for molecular breeding in each crop species to improve stress tolerance.
Ectopic expression in different organisms is frequently used to characterize function of orthologs of crop species. Ectopic expression in oocytes or yeast is useful to study the molecular function of orthologs but in planta assay would have advantages to characterize its physiological functions. For example, in planta assay enables testing of orthologs if they regulate tolerance at a tissue or plant level. In fact, several transcription factors, including DREB, NAC and bZIP, were characterized by gene overexpression.
Agrobacterium-mediated transformation has been frequently used in planta assay. Agrobacterium species mostly are pathogenic, generating tumor and hairy roots in plant by integration of T-DNA containing pathogenic genes encoding phytohormone and opine synthesis enzymes [Citation7]. Agrobacterium tumefacience has been used in planta assays after the establishment of nonpathogenic and small-sized plasmids, the so called mini-Ti (tumor-inducing) plasmid, and nonpathogenic strains lacking native T-DNA [Citation8]. Although the bacterium infects mostly dicots [Citation7] in the natural environment, it can be used for transformation in various plants including monocots in laboratory conditions [Citation9]. This was established by modifications such as very high density infection (rice early protocol), and addition of the chemical activator (e.g, acetosyringone) of virulence reactions [Citation10]. An efficient and reliable transformation system was developed in the medically important planta Papaver bracteatum and Nepeta pogonosperma through optimization of several factors such as bacterial strain and nutrient composition that affect the rate of A. rhizogenes-mediated transformation and growth rate of hairy root [Citation11,Citation12]. In addition, an A. tumefacience method has been adapted in a transient assay that enables study of intracellular localization of proteins [Citation13]. However, there remain technical limitations for evaluating gene function. For example, it takes time to generate stable transgenic seed progenies. Also, it sometimes requires using uncommon antibiotic selection markers if subsequently transformed to transgenic plant materials such as T-DNA insertion mutants of Arabidopsis, which are used frequently as host of in planta complementation assays.
Mostly, the native strain of A. rhizogenes carrying pathogenicity (i.e., inducing hairy roots) could be used for evaluating function of genes in planta, if the phenotype can be observed in hairy roots. In fact, it has been used for studying synthesis of secondary metabolites in medicinal plants [Citation14–17], while it may also be applied to study functional genes that regulate tolerance of roots to rhizotoxic ions. Although the concept has been applied for evaluating Al tolerance genes (i.e., gene-encoding MATE-citrate transporter of eucalyptus using wild-type tobacco [Citation18]), it has not been evaluated whether it can apply to the complementation assays using transgenic mutant lines as host plant. By the complementation assay, it is better if the ortholog or other genes could be introduced without using any antibiotic selection marker, because the transgenic host plants usually carry marker genes for antibiotics. In the present study, we analyzed efficiency of transformation of target genes in the hairy roots, which were developed using transgenic host background. We also compared stress response of hairy and intact roots. Our analysis identified that hairy roots would be useful to study functions of transgenes that are responsible for regulation of stress tolerance in plant.
Materials & methods
Plant materials
Arabidopsis (Columbia [Col-0] and the stop1 mutant [Col-0 background; Iuchi et al., 2007]); tobacco (Nicotiana tabacum) (wild-type [cv. Xanthi] and homozygous T3 generation of NtSTOP1-KD [NtSTOP1-KD; RNA interference line of NtSTOP1], which were used in the previous study [19]), were used as the host of transformation in the present study. The NtSTOP1-KD was kanamycin (kan)- and hygromycin-resistant due to its caring of the selection marker of T-DNA. A hyper virulence strain ATCC15834 (Import permit No. 26N-406, MAFF, Japan) was used for A. rhizogenes-mediated transformation.
Plasmid DNA constructs & development of hairy roots
The overexpression constructs ([35S:AtSTOP1:sGFP] [synthetic green florescence protein] and 35S:CcMATE1:sGFP]) were similar from the previous studies [Citation19,Citation20]. Briefly, the cDNA sequence of AtSTOP1 and CcMATE1 were attached to the 5′ end of the open reading frame of the sGFP gene connected to the nopaline synthase terminator by overlapping extension polymerase chain reaction (PCR) [Citation21]. The amplicon of the respective gene was digested with SfiI and then introduced into the transfer DNA region of pBE2113 [Citation22], which contains a kan resistance gene as the selection marker. Similarly, the cDNAs of GUS and sGFP were digested with Sfi1 and then introduced into T-DNA region of pBE2113 under the control of cauliflower mosaic virus 35S promoter. These constructs were transformed into A. rhizogenes by electroporation and used for transgenic hairy roots (A. rhizogenes with Mini Ti plasmid) transformation or wild-type hairy roots (only A. rhizogenes infection) in Arabidopsis as well as tobacco (Supplementary Information). Similarly, the 35S:AtSTOP1:sGFP construct was used for A. tumefaciens-based plant transformation using the floral-dip method [Citation23].
Culture condition & RNA extraction
Arabidopsis and tobacco seedlings were grown hydroponically according to the method described [Citation19]; the culture solutions were renewed after every 2 days. The 1-month old transgenic hairy roots were precultured for 3 days in 1/50 MGRL (Modified Growth Regulators) solution with 1% sucrose. Arabidopsis (10 days old; 10 μM Al, 50 mM NaCl; pH 5.0 for transcript and 5 days old; 10 μM Al; 1% sucrose; pH 5.0 for malate; as described by [Citation19]), tobacco (14 days old; 30 μM Al, pH 5.0 for transcript and 7 days old 30 μM Al; 1% sucrose; pH 5.0 for citrate; as described by [Citation24]) plant and 3 days precultured (both Arabidopsis and tobacco) hairy roots were used. RNA isolation and cDNA synthesis were performed as described by [Citation25].
Collection of root exudates, & quantification of citrate & malate
Citrate and malate were quantified by the enzyme reaction (citrate lyase [EC 4.1.3.8] for citrate, malate dehydrogenase [EC 1.1.1.82] for malate)-coupled NADH/NAD+ cycling method developed by [Citation26] with minor modifications as described by [Citation27].
GUS staining & microscopic imaging
The GUS staining was performed as described by [Citation28]. Cellular localization and uniform transformation of 35S:AtSTOP1:sGFP and 35S:CcMATE1:sGFP were analyzed based on sGFP fluorescence. The samples were observed and photographed under an Olympus BX51 microscope 9 (Olympus, Tokyo, Japan) equipped with Olympus DP70 Camera System (Olympus) and fluorescence microscope (AXIO imager system, Carl-Zeiss-Japan, Tokyo, Japan). The transgenic hairy roots were examined for the integration and co-integration of the transformed gene by genomic PCR.
Results
Establishment of efficient hairy roots
We developed hairy root induction in Arabidopsis and tobacco by co-cultivating the explants with A. rhizogenes. In the preliminary experiment we used different explants such as leaf or stem of Arabidopsis or leaf or tobacco for hairy root development. The preliminary experimental results showed that stem of Arabidopsis and leaf of tobacco produced high efficiency of hairy roots compared with other explants (data not shown). We observed induction of tobacco and Arabidopsis hairy roots within <10 days, and the whole process of hairy roots was completed within 1 month from the day of infection with high efficiency and frequency of transformation (). We obtained a very high rate of hairy roots formation efficiency in tobacco that ranges from 53 to 98%, and 58 to 66% in Arabidopsis (). These results showed that hairy roots can be an approach for gene analysis.
Figure 1. Development of Agrobacterium rhizogenes-mediated transformation into Arabidopsis and tobacco hairy roots.
(A)Arabidopsis thaliana plants growing in containers. (B)Arabidopsis explants used for Agrobacterium rhizogenes infection. (C) Transgenic hairy root formation of Arabidopsis. (D, E & F) Transgenic hairy roots expressing the CaMV 35S-driven GUS and sGFP gene. (G) Hairy root formation of tobacco. (H & I) Excised tobacco hairy roots growth on solid and liquid medium. (J) Transgenic hairy roots of tobacco expressing the CaMV 35S-driven GUS gene. (K) Confocal laser scanning microscopy image of CaMV 35S-driven sGFP localization in tobacco transgenic hairy roots.
CaMV 35S: Cauliflower mosaic virus35S.
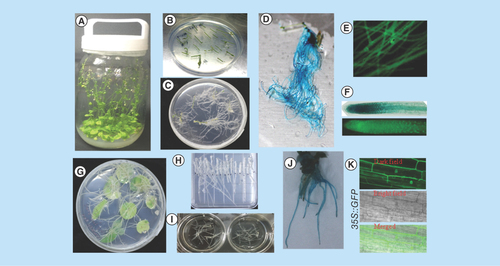
Table 1. Efficiency of hairy roots formation in different batch of transformation.
Transcript & organic acid analysis in A. rhizogenes-derived hairy roots & intact roots of wild-type or stop1 mutant of Arabidopsis
To explore the molecular function and functional similarity in response to stress physiology between A. rhizogenes-derived hairy roots and intact roots of Arabidopsis or tobacco, hairy and intact roots of Arabidopsis were exposed to Al or NaCl stress. Hairy and intact roots of Col-0 showed Al-induced ALMT1 and MATE1 expression, stop1 mutant hairy roots and intact roots showed the suppression of Al-induced ALMT1 and MATE1 expression (). The NaCl-exposed hairy and intact roots of Col-0 showed similar expression pattern of RD29A gene (). Similarly, the NtMATE expression was induced by Al treatment in wild-type tobacco intact roots and suppression in NtSTOP1-KD intact roots. The Al responsive NtMATE expression pattern in hairy roots was similar with intact roots (). These results revealed that roots derived from A. rhizogenes transformation or the intact roots showed similar expression level of compared genes when exposed to various stress condition (, B & C). Al-responsive malate secretion is regulated by ALMT1 in Arabidopsis and stop1 mutant could not excrete the malate when it is exposed to Al stress [Citation29]. Hairy or intact roots of col-0 showed that Al-responsive malate secretion is higher than compared with control, no Al stress (Supplementary Figure 1A), whereas stop1 mutant intact and hairy roots could not excrete Al-responsive malate (Supplementary Figure 1A). On the other hand, Al-inducible citrate secretion was not observed in both hairy and intact roots of Arabidopsis (Supplementary Figure 1B). Hairy and intact roots of tobacco showed the highest concentration of citrate secretion with Al inducible excretion pattern (Supplementary Figure 2A); however, NtSTOP1-KD hairy and intact roots showed suppressed Al-inducible citrate excretion (Supplementary Figure 2A). Suppression of Al responsive citrate secretion was similar to the previous report of [Citation24]. These results suggest that loss-of-function mutant (stop1 and NtSTOP1-KD) hairy roots can be used for gain-of-function analysis of genes related with Al tolerance from different plants, in particular those that are regulated by STOP1/MATE system.
Figure 2. Transcript analysis of hairy and intact roots of Arabidopsis and tobacco exposed to rhizotoxic stress.
Expression of AtALMT1(A), RD29(B), and NtMATE(C), in wild-type hairy roots (only Agrobacterium Rhizogenes infection) and intact roots exposed to various rhizotoxic stress with (pH 5.0; [10 μM- : Arabidopsis, 30 μM- : tobacco Al, and 50 mM NaCl)] or without (pH 5.0; Al and NaCl) for 24 h was detected by RT (real-time)-polymerase chain reaction. Hairy roots were precultured for 3 days in MGRL solution containing 1% sucrose. Ubq1 and ACT1 were used as the internal reference gene. Error bars indicate ± standard deviation (n = 3). The primers used for qRT-polymerase chain reaction were similar used from previous studies of Sawaki et al. and Ohayama et al.
![Figure 2. Transcript analysis of hairy and intact roots of Arabidopsis and tobacco exposed to rhizotoxic stress.Expression of AtALMT1(A), RD29(B), and NtMATE(C), in wild-type hairy roots (only Agrobacterium Rhizogenes infection) and intact roots exposed to various rhizotoxic stress with (pH 5.0; [10 μM- : Arabidopsis, 30 μM- : tobacco Al, and 50 mM NaCl)] or without (pH 5.0; Al and NaCl) for 24 h was detected by RT (real-time)-polymerase chain reaction. Hairy roots were precultured for 3 days in MGRL solution containing 1% sucrose. Ubq1 and ACT1 were used as the internal reference gene. Error bars indicate ± standard deviation (n = 3). The primers used for qRT-polymerase chain reaction were similar used from previous studies of Sawaki et al. and Ohayama et al.](/cms/asset/e31ca3ca-3d70-4e8f-bfa4-2e5d4a38475f/ifso_a_12364167_f0002.jpg)
Microscopic imaging & cellular localization pattern
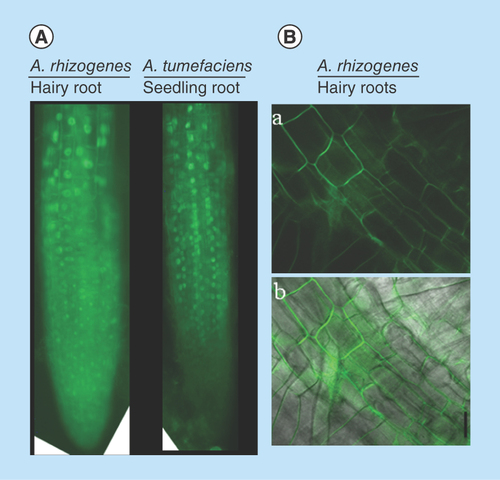
In the present study we found that there were no differences in the localization pattern of STOP1:sGFP in A. rhizogens-derived hairy roots and primary roots derived from A. tumefaciens (). Similarly, the tobacco hairy roots showed CcMATE1:sGFP localized in the plasma membrane (). Also, we observed a high rate of uniform cell transformation at cellular level based on sGFP fluorescence ().
Figure 3. Protein localization in Agrobacterium rhizogenes-transformed hairy roots and cellular transformation.
(A) Fluorescent microscopic image of 35S:AtSTOP1:sGFP localization in hairy and intact roots of Arabidopsis. (B)35S:CcMATE1:sGFP localization in tobacco hairy roots and detection of cellular transformation base on GFP fluorescence. The confocal laser scanning microscopic image of CcMATE1 localized in cell plasma membrane of tobacco hairy roots (A) (dark field) and (B) (merged). Uniform and single cell transformation of CcMATE1 was detected based on GFP fluorescence.
GFP: Green fluorescent protein.
Estimation of the ratio of co-integration of T-DNAs from Ti plasmid
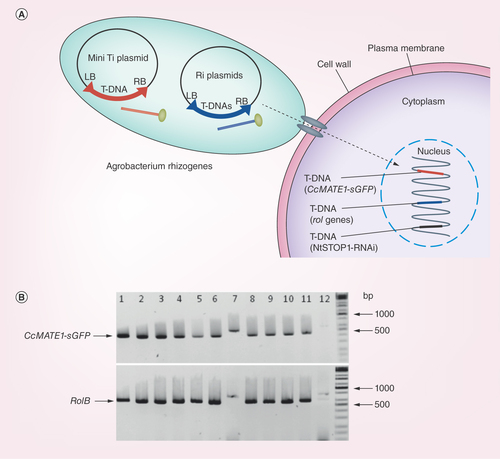
To confirm the transformation of hairy roots, we performed genomic PCR of tobacco hairy roots () and reporter assays of both tobacco and Arabidopsis (). We examined the integration of T-DNA of Ti and Ri (root-inducing) plasmids by genomic PCR. In this analysis, hairy roots were derived with the A. rhizogenes-carrying CcMATE1-sGFP in the T-DNA of Ti plasmid. Genomic PCR of hairy root gDNA amplified the expected size of amplicons; approximately 700 base pair with rol B primer, and 500 base pair with GFP primers (). The rate of co-integration was estimated as very high >80%. It indicated that a high rate of co-integration occurred in the hairy root generated by A. rhizogenes. These trends were confirmed by the GUS staining of hairy roots, which showed blue color in almost all of the hairy roots ().
Figure 4. Diagrammatic model and co-transformation efficiency of Ri and mini Ti plasmid in Agrobacterium rhizogenes developed tobacco hairy roots.
(A) Diagrammatic model illustrating co-integration of T-DNAs from mini Ti and Ri plasmid into the hairy roots of NtSTOP1-KD tobacco. Transgenic Agrobacterium rhizogenes carries two Ti plasmid; (1) T-DNA of mini Ti plasmid carries CcMATE1-sGFP, kanamycin as antibiotic resistance marker gene; (2) T-DNA of Ri plasmid carries root locus (rol; rolA, rolB and rolC) gene, responsible for opine production. The T-DNA of NtSTOP1-KD tobacco host plant carries sequence of STOP1-RNAi as well as kanamycin and hygromycin resistance antibiotic marker genes. (B) Genomic DNA was extracted from roots transformed with CcMATE1-sGFP, and genes were amplified using primers flanking the target sequence. The rolB gene present in the Ri plasmid of A. rhizogenes and CcMATE1-sGFP present in mini Ti plasmid. Presence of amplicon of hairy roots marker gene (rolB; 670 base pair) and mini Ti plasmid caring GUS (500 base pair) showed co-transformation. Lanes 1–11, tobacco hairy roots; lane 12, negative control (NtSTOP1-KD tobacco). The primer pairs of 5′-TAGCCGTGACTATAGCAAACCCCTCC-3′ and 5′-GGCTTCTTTCTTCAGGTTTACTGCAG-3′, and 5′-TACCCCGACCACATGAAGCAG-3′ and 5′- TACTTGTACAGCTCGTCCATGC-3′, were used for amplification of RolB and CcMATE1-sGFP genes, respectively.
Ri: Root-inducing; Ti: Tumor-inducing.
Discussion
In this study, we developed an efficient hairy root system using wild-type and mutant Arabidopsis or tobacco, by A. rhizogenes-mediated transformation, it allow us for rapid functional testing of genes (). The stop1 is sensitive to proton and Al rhizotoxicity [Citation29] and suppresses the expression of Al and H tolerance in Arabidopsis [Citation30]. The hairy roots of the stop1 mutant showed suppression of Al responsive genes (), and were not able to secret malate (Supplementary Figure 1A). This trend of gene expression and organic acid (OA) secretion of stop1 mutant hairy roots is very similar to intact roots of the stop1 mutant (Supplementary Figure 1A). Moreover, the stop1 mutant showed suppression of Al responsive ALMT1 and malate secretion [Citation29]. On the other hand, the salt responsive DR29 gene revealed very similar gene expression pattern with hairy and intact roots of Arabidopsis (). Moreover, NtSTOP1-KD was generated by A. tumefaciens-mediated transformation using a vector carrying an RNAi that targeted sequences of NtSTOP1 [Citation24]. Hairy roots of NtSTOP1-KD showed suppression of MATE, ALS3 gene and showed suppression of citrate excretion in response to Al stress. The suppression pattern of hairy roots of NtSTOP1-KD is very similar to that of intact roots of tobacco ( & Supplementary Figure 2A). However, the expression patterns of the genes relating to reactive oxygen species (ROS) are different between the intact and the hairy roots, possibly because they are hairy roots [Citation31]. Moreover, Al-tolerant cultured cells of tobacco showed Al tolerance and enhanced citrate excretion [Citation32]. Anatomy of tomato hairy roots is similar to that of primary roots; the only difference found was that hairy roots often contain one extra cortex layer [Citation33]. The TaNHX2 gene could enhance salt tolerance of soybean, and the A. rhizogenes-mediated transformation system could be used as a complementary tool of A. tumefaciens-mediated transformation to rapidly investigate candidate gene function in soybean [Citation34]. This suggests that hairy roots of mutants can be used as a background for further characterization of Al-responsive genes.
A. tumefaciens-mediated transient assay is a popular method to characterize gene localization. However, it needs a high level of skill to analyze results, and usually results in chimeric-transformed cell. A. rhizogenes-transformed hairy roots are an alternative approach to A. tumefaciens to study the gene localization (). A. tumefaciens and A. rhizogenes-transformed roots expressing a GFP-GUS transgene driven by the tomato SHR promoter (SlSHR) showed the same pattern of GUS and GFP fluorescence in the root stele of both primary and hairy roots [Citation33]. Multiple gene transformation can be achieved by sequential transformation. Such a method is time consuming, labor intensive and use of a different antibiotic resistance gene in each step of transformation or elimination of the antibiotic resistance gene from an earlier transgenic material is important. However, in the present study we used NtSTOP1-KD transgenic tobacco that carries kan and hygromycin antibiotic resistance genes, as a host for hairy roots production. Besides this, mini Ti plasmid carries CcMATE1-sGFP, also containing kan as an antibiotic selection marker. By using A. rhizogenes we successfully generated transgenic tobacco hairy roots caring CcMATE1-sGFP without using any antibiotic selection (). Moreover, co-transformation is an efficient approach that facilitates the delivery of more than one gene simultaneously. Co-transformation efficiency is very important in A. rhizogenes-mediated hairy roots transformation for generating transgenic hairy roots. Because of this, the gene of interest is present in the T-DNA region of mini Ti plasmid (transformed vector) and genes responsible for hairy roots production (i.e., rolA, rolB, rolC and rolD) were present in T-DNA of Ri plasmid. Some co-transformation strategies have been reported by using microprojectile bombardment [Citation35]. The CcMATE1-sGFP-transformed NtSTOP1-KD tobacco hairy roots revealed very high (>80%) co-transformation efficiency by PCR analysis (). Moreover, overexpressed GUS/GFP also showed high transformation efficiency (>95%) in both tobacco and Arabidopsis, using GUS/GFP fluorescence (). An average of 62% co-transformation efficiency using fluorescent markers in Eucalyptus grandis hairy roots by A. rhizogenes transformation was reported [Citation36]. Approximately 91% of hairy root transformation efficiency was obtained using the optimum conditions of A. rhizogenes K599 caring 35S::AhAREB1-GFP [Citation37]. A recent study showed that the hairy root induction efficiency of potato could depend on the cultivars [Citation38]. Similarly, we found higher efficiency of hairy root infection in tobacco camper with Arabidopsis.
Conclusion
We developed an efficient and simple A. rhizogenes-mediated hairy root development protocol in Arabidopsis and tobacco. The transgenic background can be used as host, without considering the antibiotic resistance gene in the transgenic background; the new gene can be efficiently introduced using A. rhizogenes transformation. The use of transgenic mutant background (i.e., NtSTOP1-KD or stop1 mutant) as a host for A. rhizogenes-mediated hairy roots transformation is a prominent approach for characterizing Al-responsive orthologous genes.
Future perspective
Hairy roots have been used for various purposes over the last several years; mostly for the production of secondary metabolite production. Recently, hairy root has been utilized as a biotechnological tool in different plant species to analyze gene function, promoters and cellular localization of proteins. Hairy root is a fast, simple and highly efficient system for utilization in biotechnology applications. However, maintenance and storage of hairy roots is still challenging and several previous reports revealed contradictions in transgene expression in intact and hairy roots. Further studies need to address issues such as storage, maintenance and effect of Rol genes that affect the expression of transgenes in diverse stress response.
Hairy roots have been used for various purposes over the last several years.
We used Agrobacterium rizogenes-mediated hairy roots as an alternative approach for Agrobacterium tumefaciens transformation.
We developed a simple, effective and reproducible hairy root protocol in tobacco and Arabidopsis.
Developed hairy roots were compared with intact roots to characterize the gene response to Al and NaCl stress.
Al and NaCl-responsive gene expression was found to be similar in hairy and intact roots of Arabidopsis and tobacco.
Similarly, organic acid excretion in response to Al stress was found to be similar in intact and hairy roots of Arabidopsis and tobacco.
Atstop1 mutant and NtSTOP1-KD were used as a host background for hairy root developments of Arabidopsis and tobacco, respectively.
Protein localization of STOP1 and CcMATE1 was found to be similar in intact and hairy roots of Arabidopsis and tobacco.
Genomic polymerase chain reaction and confocal imaging analysis revealed high co-transformation efficiency of T-DNA of mini Ti and Ri plasmids without use of any selection pressure.
Atstop1 mutant and NtSTOP1-KD can be used as a host background for hairy root transformation to evaluate the Al-responsive genes from orthologous plant species.
Author contributions
AA Daspute, M Gu, Y Kobayashi and H Koyama conceived the study and designed the experiments. AA Daspute, X Yunxuan and Y Kobayashi carried out the experiments and analyzed the data. AA Daspute, S Wagh, A Panche and H Koyama wrote the article and revised the text based on feedback from the co-authors. All authors have read and agreed to the submitted version of the manuscript.
supplementary figure 1
Download TIFF Image (177.4 KB)supplementary figure 2
Download TIFF Image (180.6 KB)supplementary material
Download MS Word (31.4 KB)Acknowledgments
We thank the RIKEN BioResource Center and the Arabidopsis Biological Resource Center for providing Arabidopsis seeds.
Supplementary data
See online at: https://www.tandfonline.com/doi/suppl/10.4155/fsoa-2018-0065
Financial & competing interests disclosure
This work was funded by a scientific research grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (15H04468). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- Karaba A, Dixit S, Greco R et al. Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. PNAS 104, 15270–15275 (2007).
- Preuss SB, Meister R, Xu Q et al. Expression of the Arabidopsis thaliana BBX32 gene in soybean increases grain yield. PLoS ONE 7, 1–12 (2012).
- Sadhukhan A, Kobayashi Y, Kobayashi Y et al. VuDREB2A, a novel DREB2-type transcription factor in the drought-tolerant legume cowpea, mediates DRE-dependent expression of stress responsive genes and confers enhanced drought resistance in transgenic Arabidopsis. Planta 240, 645–664 (2014).
- Sasaki T, Yamamoto Y, Ezaki Y et al. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37, 645–653 (2004).
- Furukawa J, Yamaji N, Wang H et al. An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 48, 1081–1091 (2007).
- Magalhaes JV, Liu J, Guimaraes CT et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genetics 39, 1156–1161 (2007).
- Zupan JR, Zambryski P. Transfer of T-DNA from Agrobacterium to the plant cell. Plant Physiol. 107, 1041–1047 (1995).
- Jen GC, Chilton MD. The right border region of pTiT37 T-DNA is intrinsically more active than the left border region in promoting T-DNA transformation. Genetics 83, 3895–3899 (1986).
- Hooykaas-Van Slogteren, Hooykaas PJJ, Schilperoort RA. Expression of Ti plasmid genes in monocotyledonous plants infected with Agrobacterium tumefaciens. Nature 311, 763–764 (1984).
- Gould J, Devey M, Hasegawa O, Ulian EC, Peterson G, Smith RH. Transformation of Zea mays L. using Agrobacterium tumefaciens and the shoot apex. Plant Physiol. 95, 426–434 (1991).
- Sharafi A, Sohi HH, Mousavi A, Azadi P, Razavi K, Ntui VO. A reliable and efficient protocol for inducing hairy roots in Papaver bracteatum. Plant Cell, Tissue and Organ Cult. 113, 1–9 (2013).
- Valimehr S, Sanjarian F, Sohi HH, Sharafi A, Sabouni F. A reliable and efficient protocol for inducing genetically transformed roots in medicinal plant Nepeta pogonosperma. Physiol. Mol. Biol. Plants 20, 351–356 (2014).
- Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1(4), 2019–2025 (2006).
- Ono NN, Tian L. The multiplicity of hairy root cultures: prolific possibilities. Plant Sci. 180, 439–446 (2011).
- Sharma P, Padh H, Shrivastava N. Hairy root cultures: a suitable biological system for studying secondary metabolic pathways in plants. Eng. Life Sci. 13, 62–75 (2013).
- Sharafi A, Sohi HH, Mousavi A, Azadi P, Dehsara B, Khalifani BH. Enhanced morphinan alkaloid production in hairy root cultures of Papaver bracteatum by over-expression of salutaridinol 7-o-acetyltransferase gene via Agrobacterium rhizogenes mediated transformation. World J. Micro. Biotech. 29, 2125–2131 (2013).
- Sharafi A, Sohi HH, Mousavi A, Azadi P, Khalifani BH, Razavi K. Metabolic engineering of morphinan alkaloids by over-expression of codeinone reductase in transgenic hairy roots of Papaver bracteatum, the Iranian poppy. Biotech. Lett. 35, 445–453 (2013).
- Sawaki Y, Kihara-Doi T, Kobayashi Y et al. Characterization of Al-responsive citrate excretion and citrate transporting MATEs in Eucalyptus camaldulensis. Planta 237, 979–989 (2013).
- Kobayashi Y, Hoekenga OA, Itoh H et al. Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol. 145, 843–852 (2007).
- Daspute AA, Kobayashi Y, Panda SK et al. Characterization of CcSTOP1; A C2h2-type transcription factor regulates AI tolerance gene in pigeonpea. Planta 247, 201–214 (2017).
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68 (1989).
- Mitsuhara I, Ugaki M, Hirochika H et al. Efficient promoter cassettes for enhanced expression of foreign genes in Dicotyledonous and Monocotyledonous plants. Plant Cell Physiol. 37, 49–59 (1996).
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
- Ohyama Y, Ito H, Kobayashi Y et al. Characterization of AtSTOP1 orthologous genes in tobacco and other plant species. Plant Physiol. 162, 1937–1946 (2013).
- Suzuki Y, Kawazu T, Koyama H. RNA isolation from siliques, dry seeds and other tissues of Arabidopsis thaliana. BioTechniques 37, 542–544 (2004).
- Hampp R, Goller M, Füllgraf H. Determination of compartmented metabolite pools by a combination of rapid fractionation of oat mesophyll protoplasts and enzymic cycling. Plant Physiol. 75, 1017–1021 (1984).
- Kihara T, Wada T, Suzuki Y, Hara T, Koyama H. Alteration of citrate metabolismin cluster roots of white lupin. Plant Cell Physiol. 44, 901–908 (2003).
- Kobayashi Y, Kobayashi Y, Sugimoto M et al. Characterization of the complex regulation of AtALMT1 expression in response to phytohormones and other inducers. Plant Physiol. 162, 732–740 (2013).
- Iuchi S, Koyama H, Iuchi A et al. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and co-regulates a key gene in aluminum tolerance. PNAS 104, 9900–9905 (2007).
- Sawaki Y, Iuchi S, Kobayashi Y et al. STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 150, 281–294 (2009).
- Wang JH, Lin HH, Liu CT, Lin TC, Liu LY, Lee KT. Transcriptomic analysis reveals that reactive oxygen species and genes encoding lipid transfer protein are associated with tobacco hairy root growth and branch development. Mol. Plant Microbe. Interact. 27, 678–687 (2014).
- Ojima K, Koyama H, Suzuki R, Yamaya T. Characterization of two tobacco cell lines selected to grow in the presence of either ionic Al or insoluble Al-phosphate. Soil Sci. Plant Nutr. 35, 545–551 (1989).
- Ron M, Kaisa K, Germain P et al. Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 166, 455–469 (2014).
- Cao D, Hou W, Liu W et al. Overexpression of TaNHX2 enhances salt tolerance of ‘composite’ and whole transgenic soybean plants. Plant Cell Tissue Organ Cult. 107, 541–552 (2011).
- Schmidt MA, LaFayette PR, Artelt BA, Parrott WA. A comparison of strategies for transformation with multiple genes via microprojectile-mediated bombardment. In Vitro Cell. Dev. Biol. 44, 162–168 (2008).
- Plasencia A, Soler MA, Dupas A et al. Eucalyptus hairy roots, a fast, efficient and versatile tool to explore function and expression of genes involved in wood formation. Plant Biotech. J. 14, 1381–1393 (2016).
- Liu J, Li Y, Wang W, Gai J, Li Y. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genomics 17, 223 (2016).
- Moehninsi, Navarre DA. Optimization of hairy root induction in Solanum tuberosum. Am. J. Potato Res. 95, 650–658 (2018).
