Abstract
Aim: Retrospective scaling of human biliary amounts has been performed using allometry. Methods: Human biliary excretory data for 14 drugs were predicted using simple, bile flow-rate corrected and uridine diphosphate glucuronosyltransferase (UDPGT) activity corrected allometry methods. Allometry was performed using Y = aXb relationship with correction factors. Statistical tests consisting of fold difference (predicted/observed) and root-mean-square error (RMSE) computation were carried out. Results: UDPGT activity corrected allometry predicted higher biliary amounts in humans as compared with other methods. The RMSE values were 38, 32 and 81 for simple, bile flow-rate corrected and UDPGT activity corrected allometry, respectively. Conclusion: Although data showed the usefulness of the approaches for human biliary predictions in the decision process for nominating drug candidate(s) based on % RMSE and fold differences, for some drugs prediction appeared not satisfactory using any of the methods.
Biliary excretion of the parent drug and metabolite(s) is an important contributor for the drug disposition/elimination along with the renal route of excretion, hepatic and nonhepatic drug metabolism of xenobiotics. Because of difficulty, complexity and ethical issues in the collection of bile samples in human subjects, scanty data are available with respect to biliary excretory amounts of drugs in humans. In this context, the fecal excretion data which are often reported in 14C mass balance studies in humans provide the measure of biliary excretory pathway for intravenously administered drugs. However, for drugs that are orally administered, the fecal excretory data may contain both biliary excretory component(s) of the absorbed drug and the portion of the orally administered dose, which escapes absorption.
Recently, the interspecies scaling of urinary excretory amounts of a large number of drugs with diversified structures has been performed and the resultant human predictions of the excretory amounts in urine were found to be excellent [Citation1,Citation2]. While relatively, few drugs were used for interspecies scaling of the fecal excretory amounts, the limited data suggested the possibility of a satisfactory human prediction of the fecal recovery of drugs such as rifapentine, cabotegravir and dolutegravir [Citation2]. Despite the complexities associated with the biliary excretion process across species such as biliary flow-rate differences, molecular weight cutoff for biliary excretion, occurrence of the enterohepatic recycling process, differences in the reabsorption of the parent drug during the enterohepatic recycling process, the fecal excretory amount predictions for a few drugs suggested that there may be a merit in the interspecies scaling of the biliary excretory amounts and prediction of the corresponding human biliary excretion data in a retrospective manner. The prediction of biliary excretion data in humans, a priori, would be beneficial in several ways: it would help in the design of clinical pharmacology studies with respect to the timing of food ingestion; anticipation of enterohepatic recirculation and appropriate pharmacokinetic sampling around the event; exclusion of drug(s) and/or flavonoid containing beverages or dietary supplements that may influence the biliary excretory process; design appropriate preclinical drug–drug interaction study to understand the complexity of pharmacokinetic interaction at the biliary excretion level with various associated transporters. For example, what would be the likely impact by the inhibition of sinusoidal biliary efflux transporter on biliary excretion of a drug?
We were interested to evaluate if the biliary excretory amounts in laboratory animals such as mice, rats, dogs and monkeys obtained from drugs with structural diversity (n = 14) would render itself amenable for interspecies scaling and possible human prediction in a retrospective manner. The scope of the present work is to report a practical approach that has a potential value in the assessment of drug(s) from a biliary excretory amount perspective without being bogged down by the detailed study of biliary excretory mechanisms, which may be premature to evaluate at the stage of drug discovery. Previously, the use of bile-related correction factor and incorporation of uridine diphosphate glucuronosyltransferase (UDPGT) activity have been considered in the human predictability of the total body clearance values but found to produce errors in the scaling of clearance values [Citation3,Citation4]. However, because we were interested in scaling of biliary excretory amounts, we explored the use of bile flow-rate and UDPGT activity, which may be relevant for interspecies scaling of biliary excretory amounts as the correction factors. It should be noted that we did not factor either pharmacology or excretory transporters in the selection of drugs.
Methods
Data compilation & parameters calculation
No new data were generated for this work – all data used were obtained from the scientific literature and the references used for this are detailed in . The drugs with biliary excretion data of parent drug or parent drug plus metabolite(s) were selected. Because of limitations in getting biliary excretory amounts across three species, even data from two animal species were included for allometry predictions. In the event, human biliary amounts were not readily available for the tested drugs, fecal excretory data in humans (of parent drug and/or metabolite[s]) were considered. The drugs that were included in this exercise were atorvastatin, brivanib alaninate, bromocriptine, carvedilol, cefotetan, cefpiramide, ceftriaxone, digoxin, doxorubicin, erythromycin, linagliptin, muraglitazar, napsagatran and odanacatib. The compiled data were tabulated to provide the following information: species, dose amount (mg/kg), route of drug administration and the percent biliary recovery (). Several physiological parameters used in the scaling are shown in . The following methods were used to predict human % biliary recovery from animal data.
Table 1. Physiological parameters used in the allometry scaling.
Table 2. Listing of species, doses, route, labeling, collection period and percent recovery in bile of various compounds/drugs that were subjected to interspecies scaling for allometry prediction.
Simple allometry
The % biliary recovery of each drug was plotted against the corresponding body weight on a log–log scale and the following allometric equation was used to predict the % biliary recovery in humans.
Where X is the body weight, and a and b are the coefficient and exponent of the allometric equation, respectively.
Bile flow-rate correction factors
A correction factor based on the bile flow rate normalized by per kg liver weight of the species was applied to predict human biliary-excreted amount [Citation3–5] more accurately than the simple allometry. In this approach, product of % biliary recovery and the bile flow rate was plotted against the corresponding body weight on a log–log scale and the following allometric equation was used:
Where Qbile = bile flow in ml/min. The correction factors used in the scaling are shown in .
UDPGT activity correction factors
A correction factor based on the UDPGT activity of the species was applied to predict human biliary-excreted amount more accurately than the simple allometry as previously demonstrated by other researchers [Citation3–5]. In this approach, product of % biliary recovery and the UDPGT activity was plotted against the corresponding body weight on a log–log scale and the following allometric equation was used:
Where the UDPGT activity is in nmol/min/mg protein. The correction factors used in the scaling are shown in .
Statistical analysis
The fold difference of the prediction was calculated as follows:
Predicted % biliary recovery was used to define the fold difference of the prediction. The analysis was done separately and reported for the biliary excretory data. For the purpose of the current exploratory analysis, a prediction of 0.5- to twofold was considered satisfactory. The fold-difference statistics has been successfully utilized in the evaluation of several drugs [Citation6–10].
The mean absolute error (MAE) was calculated by the following equation:
Root-mean-square error (RMSE) was calculated using the following equation:
The RMSE values were used only to judge which of the methods was more suitable to predict the biliary excretory amounts of the diversified drugs included in the present analysis.
Results
The biliary excretion amounts (expressed as % recovery) for the various drugs (n = 14) are provided in . The interspecies scaling via allometry methods was carried out successfully using either two species (n = 11) or three species (n = 3), as the case may be.
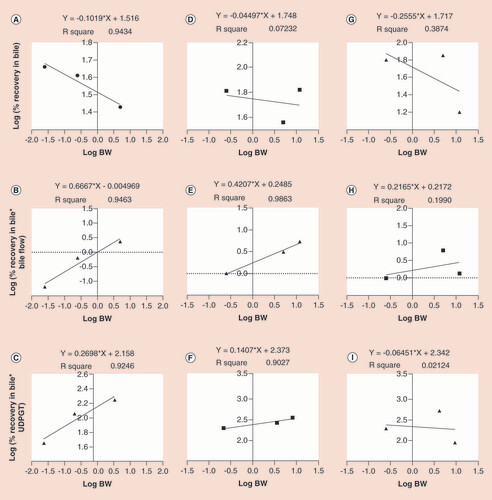
The allometry relationship between the biliary-excreted amounts of linagliptin, muraglitazar and odanacatib, which represented interspecies scaling of at least three species, is provided in . The correlation coefficient (r2) for the linagliptin and muraglitazar was more than 0.9, except for the simple allometry method of muraglitazar, suggesting there were no issues for the scalability of the biliary data across species. The correlation coefficient (r2) of odanacatib was weak for all three methods (range: 0.02–0.39), suggesting there was poor scalability of the biliary data across species for odanacatib using all three methods. The comparative data of the slopes and intercepts obtained for all 14 drugs are provided in Supplementary Table 1. Regardless of scaling in two or three species, the slope values were generally positive with the few instances where it was negative (18 out 42). The negative slope predominantly noticed for the simple allometry tended to indicate that the excretory amounts in the bile decreased as the function of body weight from lower to higher species. However, the application of bile flow-rate correction reversed the slope to a positive value in about 80% of the cases (Supplementary Table 1).
Using the simple allometry (A, D & G), bile flow-rate corrected (B, E & H) and UDPGT corrected (C, F & I) methods of linagliptine, muraglitazar and odanacatib, respectively.
UDPGT: Uridine diphosphate glucuronosyltransferase.
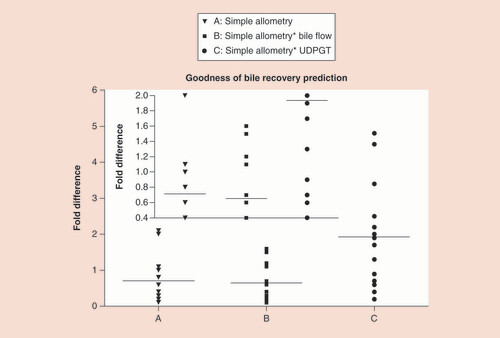
The comparison of human-predicted biliary excretory amounts from the three methods, namely the simple allometry, bile flow-rate corrected and UDPGT activity corrected methods, is provided in , and the spread of the values showing fold differences are displayed in . The mean fold differences (minimum - maximum range) for the simple allometry, bile flow-rate corrected and UDPGT activity corrected methods for the various drugs evaluated in the present analysis were 0.7 (0.1–2.1), 0.7 (0.1–1.6) and 1.9 (0.2–4.8), respectively. The UDPGT activity corrected approach predicted higher human biliary excretory amounts as compared with the other two methods. The quantitative predictions of the biliary excretory amounts appeared to be similar between the simple allometry versus bile flow-rate corrected methods ( & ). The predictions falling within a narrow boundary 0.5- to twofold difference were 5 out of 14 using the simple allometry method (36%), 8 out of 14 using the bile flow-rate corrected method (57%) and 7 out of 14 using the UDPGT activity corrected methods (50%). Using the % RMSE values, it appeared that bile flow-rate corrected allometry (32.0) had better error predictions as compared with the error predictions for either simple allometry (38.4) or the UDPGT activity corrected allometry (81.2).
Table 3. Fold difference in the predicted and observed percent recovery of drug/drug-related amount of various compounds/drugs using different allometry methods.
Using (A) simple allometry, (B) bile flow-rate corrected allometry and (C) UDPGT activity corrected allometry (inset: an expanded scale is provided for easy visualization of the fold differences for the three allometry methods).
UDPGT: Uridine diphosphate glucuronosyltransferase.
Discussion
During the selection of various drugs in this exercise, we ensured that the observed human biliary excretory amounts (direct bile or fecal excretion data) represented the four categories: ≤25%, >25–50%, >50–75% and >75%. This in turn would aid in the ease of predictability of human biliary excretory amounts and provide possible directions in the appropriateness of interspecies scaling and/or inclusion of other factors to improve the predictability. Although bile flow-rate and UDPGT activity corrections produced higher errors the predictions of human total clearance for biliary-excreted drugs [Citation4], we included both approaches in our analysis to compare and contrast with the predicted values obtained from a simple allometry scaling. The rationale for such inclusion stemmed from the fact that we wanted to introduce homogeneity in the biliary excretory amounts by including factors such as bile flow-rate and UDPGT activity, which are specific to biliary excretory pathways.
A recently published review has suggested that in the absence of active hepatic uptake mechanisms, the biliary clearances of drugs may be similar among rats, dogs and humans regardless of the efflux mechanisms such as P-glycoprotein, breast cancer-related protein, multiple drug resistance protein-2, etc. involved in the excretion of the drugs [Citation11]. This review critically examined the unbound biliary clearances for at least 22 drugs and concluded that in majority of the cases (19 out of 22), the biliary clearance in rats exceeded that of human biliary clearance by 9- to 2500-fold. Furthermore, if there was an involvement of a hepatic uptake transporter such as organic anion transporter protein-1B1, a discrepancy of at least tenfold should be expected between rat and human biliary clearance values [Citation11]. In a similar manner, the earlier work of Mahmood [Citation4] suggested a significant risk of only considering rat biliary excretion data for the prediction of human biliary clearance. However, collection of biliary excretion data from multiple species may aid in better prediction and reduce the risk of misleading and/or erroneous human prediction of the biliary excretion data [Citation11].
Therefore in the present work involving interspecies scaling, we did not opt to scale using only rat biliary excretory amount but included a minimum of two species (rat, dog or monkey as the case may be). There were a few instances where we used three species for interspecies scaling. The human data that were considered in this report for comparative purposes came from clinical studies where bile collection was possible through predefined collection procedures (T-tube and duodenal perfusion) or the fecal excretory amounts obtained from the 14C human mass balance excretory studies. Because the objective of the work was to simply examine the interspecies scalability of the biliary excretory amounts without regard to the uptake or efflux mechanisms that may govern the overall disposition of the biliary clearance of the drugs, it was reasoned that the use of total drug (parent plus metabolite or 14C data) may not introduce any bias in the data interpretation. More importantly, our goal was to seek an answer whether or not interspecies scaling of biliary excretory amounts was possible and therefore, it may be used as a tool in the late drug discovery stage for any potential risk assessment of biliary excretion in humans.
Although for orally administered drugs in humans there was a possibility that fecal excretory amounts may encompass the fraction of the administered dose-escaping absorption, no adjustments were done using the absolute bioavailability data. Also, it was not possible for us to obtain complete information on the fraction of the drug absorbed versus unabsorbed amount. Hence, in some instances, the observed fecal excretory amounts may be somewhat inflated value (i.e., containing both biliary-excreted amounts and unabsorbed dose of the drug).
The fold-difference evaluation clearly suggested that UDPGT activity corrected allometry had a tendency to predict higher biliary amounts as compared with either simple or bile flow-rate corrected allometry. Almost 36% of the UDPGT activity corrected predictions were greater than twofold; however, both simple and bile flow-rate corrected methods’ predictions were less than twofold, with the exception of a single value (2.1-fold for observed for ceftriaxone via simple allometry). Based on our assessment, although it appeared that either the simple allometry or bile flow-rate corrected allometry is applicable, the bile flow-rate corrected approach may render a lower error value. However, in some instances, UDPGT activity correction may render closer predictions.
There were four instances where simple or bile flow-rate corrected allometry showed poor predictions where the prediction was ≤0.3; this included drugs such as digoxin (0.1-fold), erythromycin (0.2-fold), atorvastatin (0.3-fold) and odanacatib (0.2-fold). Further scrutiny suggested that because both digoxin (MW: 780.94) and erythromycin (MW: 733.93) had higher molecular weights as compared with other drugs chosen in the exercise. Hence, it may be possible that biliary excretion in humans of either digoxin or erythromycin may have been much higher as compared with animal species where urinary excretion may have predominated for the two drugs. In case of other two drugs, a clear explanation for the lack of predictability could not be easily discerned. However, it may be speculated that for either atorvastatin or odanacatib, the representative human fecal data from the literature were inflated because of the use of total radioactivity measurements; and route-specific differences (oral vs intravenous) in the excretion may have contributed for the discrepancy. Interestingly, the application of UDPGT activity corrected allometry with the exception of digoxin improved the prediction for the remaining three drugs, namely atorvastatin, erythromycin and odanacatib.
We believe that the present allometry concept has been developed from a late drug discovery and early clinical candidate nomination perspective. Therefore, the notion of exact human prediction of the parent drug and/or differentiation of parent versus metabolites needs to be set aside given the exploratory nature of the predictions. The importance of the present work was to alert the drug discovery researchers if indeed human biliary excretion of drug (parent and metabolites) would be significant enough to warrant screening of transporters representing hepatobiliary excretory mechanism to be part of the clinical candidate nomination/decision process. Therefore, lack of specificity of parent + metabolites at this early stage of candidate nomination should have no bearing on the assessment and the decision rendered by this stage-gate process during the work leading to the nomination of clinical candidate.
We do not think that we can categorize that the allometry concept presented in the work failed because many predictions were outside of the 0.5- to two-fold difference criteria. For the purpose of early prediction and giving a sense of some direction, the suggested allometry tools may be useful. However, the values need to be interpreted with caution since the predictability of within twofold of the nominal values were not found in at least 50% of the cases for two out of the three allometry approaches.
Our work has several limitations that need to be highlighted. First, there were many data sets that had only two animals for interspecies scaling and the availability of a third species data perhaps may have rendered smoother predictions; Second, the bile collection periods in the various animal species varied across the chosen drugs – in some instances, the collection periods were shorter duration of time; and in other cases, it was longer duration of time. Third, the biliary excretory amounts in some cases were parent drugs only and in other cases were inclusive of parent + metabolites. Fourth, the inclusion of fecal data for oral drugs carried the risk of unabsorbed drug which may be erroneously included as part of the biliary-excreted component. Fifth, the dose(s) used in the individual animals studies from the collected literature data generally varied and were not based on human dose of the drug being considered. However, in spite of the above caveats, the present analysis has shown that there is a potential for using a prospective interspecies scaling approach for predicting biliary excretory amounts and this may be particularly helpful in drug discovery to make scaffold decisions to identify clinical candidate for development. Since it is important to make a decision on the allowable tolerance for biliary excretion during clinical candidate nomination process, the proposed tools in the work may find utility for consideration as a stage-gate process. We also want to point out that in the previous work reported by Mahmood [Citation4], the use of correction factors such as bile flow-rate and UDPGT activity resulted in prediction errors for interspecies scaling of total body clearance. However, we used the same correction factors in our analysis because our objective was to scale the excretory amounts of either parent and/or 14C radioactivity and therefore, use of both bile flow-rate and UDPGT activity was amply justified.
Conclusion
Interspecies scaling approaches based on the simple allometry, bile flow-rate corrected allometry and UDPGT activity corrected allometry were evaluated for retrospective prediction of biliary excretory amounts of 14 drugs representing diversity of chemical structures. Using the rigorous 0.5- to two-fold consideration for prediction purposes, the bile flow-rate corrected allometry was superior to either the simple or UDPGT corrected allometry. In terms of % RMSE values, the bile flow-rate corrected allometry showed a value of 32.0 marginally improved as compared with 38.4 for simple allometry; however, the UDPGT activity corrected allometry had the highest error rate of 81.2 of the observed biliary excretory amounts. In the drug discovery screening and identification of lead candidates, the bile flow-rate corrected allometry may be potentially used as a prospective tool to explore the quantum of biliary excretion in humans either using two or three animal species data. Regardless of the allometry method, the interpretation of the predicted data need to be made with caution for the decision-making.
Future perspective
The proposed methodology is simple to adopt inthe drug-discovery process
The adaptation of the proposed allometry approach in the drug-discovery process would enable to weed-off drug candidates that may have the potential of biliary excretion liability.
The availability of such biliary data may aid in the setting of transporters’ screen prior to clinical candidate nomination.
Allometry approach, despite its shortcomings, may be used as a useful during early drug discovery and drug development to assist in decision-making process.
Such predictions in early development would render the following benefits: designing clinical pharmacology studies, judging enterohepatic recirculation in humans, exclusion of drug(s) and/or flavonoid containing beverages or dietary supplements in early clinical trials that may influence the biliary excretory process, and designing appropriate preclinical drug–drug interaction studies.
An attempt was made to scale the biliary excretory amounts from animals to humans using simple, bile flow-rate corrected and uridine diphosphate glucuronosyltransferase (UDPGT) activity corrected allometric methods for 14 diversified chemical structures.
The UDPGT activity corrected allometry predicted higher biliary amounts in humans (1.94-fold difference) as compared with either simple or bile flow-rate corrected methods.
The predictions of biliary amounts were superior for bile flow-rate corrected allometry as compared with simple allometry.
The root-mean-square error values were 38, 32 and 81 for simple, bile flow-rate corrected and UDPGT activity corrected allometry, respectively.
Bile flow-rate corrected allometry showed satisfactory retrospective predictions for most of the drugs (8 out of 14 drugs within 0.5- to two-fold prediction) with root-mean-square error predictions of ≤32%.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles as outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Financial & competing interests disclosure
All the authors are employees of Cadila Healthcare Limited. The authors have no conflict of interest in the preparation of this manuscript (ZRC publication no. 484). No funding was available to support the contents of the work. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- Bhamidapati RK , MullangiR , SrinivasNR . Interspecies scaling of urinary excretory amounts of nine drugs belonging to different therapeutic areas with diverse chemical structures – accurate prediction of the human urinary excretory amounts . Xenobiotica47 ( 2 ), 112 – 118 ( 2016 ).
- Srinivas NR . Interspecies scaling of excretory amounts using allometry - retrospective analysis with rifapentine, aztreonam, carumonam, pefloxacin, miloxacin, trovafloxacin, doripenem, imipenem, cefozopran, ceftazidime, linezolid for urinary excretion and rifapentine, cabotegravir, and dolutegravir for fecal excretion . Xenobiotica46 ( 9 ), 784 – 792 ( 2016 ).
- Mahmood I , SahajwallaC . Interspecies scaling of biliary excreted drugs . J. Pharm. Sci.91 ( 8 ), 1908 – 1914 ( 2002 ).
- Mahmood I . Interspecies scaling of biliary excreted drugs: a comparison of several methods . J. Pharm. Sci.94 ( 4 ), 883 – 892 ( 2005 ).
- Ward KW , AzzaranoLM , BondinellWEet al. Preclinical pharmacokinetics and interspecies scaling of a novel vitronectin receptor antagonist . Drug Metab. Disp.27 ( 11 ), 1232 – 1241 ( 1999 ).
- Benjamin B , SahuM , BhatnagarU , AbhyankarD , SrinivasNR . The observed correlation between in vivo clinical pharmacokinetic parameters and in vitro potency of VEGFR-2 inhibitors. Can this be used as a prospective guide for the development of novel compounds?Arzneimittelforschung.62 ( 4 ), 194 – 201 ( 2012 ).
- Srinivas NR . Limited sampling strategy for the prediction of area under the curve (AUC) of statins: reliability of a single time point for AUC prediction for pravastatin and simvastatin . Drug Res. (Stuttg).66 ( 2 ), 82 – 93 ( 2016 ).
- Srinivas NR . Therapeutic drug monitoring of cyclosporine and area under the curve prediction using a single time point strategy: appraisal using peak concentration data . Biopharm. Drug Dispos.36 ( 9 ), 575 – 586 ( 2015 ).
- Srinivas NR . Differences in the prediction of area under the curve for a protease inhibitor using trough versus peak concentration: assessment using published pharmacokinetic data for indinavir . Am. J. Ther. ( 2015 ) ( Epub ahead of print ).
- Srinivas NR . Prediction of area under the curve for a P-glycoprotein, a CYP3A4 and a CYP2C9 substrate using a single time point strategy: assessment using fexofenadine, itraconazole and losartan and metabolites . Drug Dev. Ind. Pharm.42 ( 6 ), 945 – 957 ( 2016 ).
- Grime K , PaineSW . Species differences in biliary clearance and possible relevance of hepatic uptake and efflux transporters involvement . Drug Metab. Dispos.41 ( 2 ), 372 – 378 ( 2013 ).
- Davies B , MorrisT . Physiological parameters in laboratory animals and humans . Pharm. Res.10 ( 7 ), 1093 – 1096 ( 1993 ).
- Black AE , HayesRN , RothBD , WooP , WoolfTF . Metabolism and excretion of atorvastatin in rats and dogs . Drug Metab. Dispos.27 ( 8 ), 916 – 923 ( 1999 ).
- Zhang D , WangL , RaghavanNet al. Comparative metabolism of radiolabeled muraglitazar in animals and humans by quantitative and qualitative metabolite profiling . Drug Metab. Dispos.35 ( 1 ), 150 – 167 ( 2007 ).
- EMA/604444/2011, Trajenta® (Linagliptin): assessment report, (CHMP) . www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002110/WC500115748.pdf .
- Gong J , GanJ , CaceresCJet al. Metabolism and disposition of [14C]brivanib alaninate after oral administration to rats, monkeys, and humans . Drug Metab. Dispos.39 ( 5 ), 891 – 903 ( 2011 ).
- Kassahun K , BlackWC , NicollGDet al. Pharmacokinetics and metabolism in rats, dogs, and monkeys of the cathepsin k inhibitor odanacatib: demethylation of a methylsulfonyl moiety as a major metabolic pathway . Drug Metab. Dispos.39 ( 6 ), 1079 – 1087 ( 2011 ).
- Kassahun K , McIntoshI , KoeplingerKet al. Disposition and metabolism of the cathepsin k inhibitor odanacatib in humans . Drug Metab. Dispos.42 ( 5 ), 818 – 827 ( 2014 ).
- Maurer G , SchreierE , DelabordeS , NuferR , ShuklaAP . Fate and disposition of bromocriptine in animals and man. II: absorption, elimination and metabolism . Eur. J. Drug Metab. Pharmacokin.8 ( 1 ), 51 – 62 ( 1983 ).
- Roffey SJ , ObachRS , GedgeJI , SmithDA . What is the objective of the mass balance study? A retrospective analysis of data in animal and human excretion studies employing radiolabeled drugs . Drug Metab. Rev.39 ( 1 ), 17 – 43 ( 2007 ).
- Blech S , SchwellingerEL , Grafe ModyEU , WithopfB , WagnerK . The metabolism and disposition of the oral dipeptidyl peptidase-4 inhibitor, linagliptin, in humans . Drug Metab. Dispos.38 ( 4 ), 667 – 678 ( 2010 ).
- Schaefer WH , PolitowskiJ , HwangBet al. Metabolism of carvedilol in dogs, rats, and mice . Drug Metab. Dispos.26 ( 10 ), 958 – 969 ( 1998 ).
- Lau YY , OkochiH , HuangY , BenetLZ . Pharmacokinetics of atorvastatin and its hydroxy metabolites in rats and the effects of concomitant rifampicin single doses: relevance of first-pass effect from hepatic uptake transporters, and intestinal and hepatic metabolism . Drug Metab. Dispos.34 ( 7 ), 1175 – 1181 ( 2006 ).
