Abstract
Deregulation of STAT signaling has been implicated in the pathogenesis for a variety of cancers, including CTCL. Recent reports indicate that loss of STAT4 expression is an important prognostic marker for CTCL progression and is associated with the acquisition of T helper 2 cell phenotype by malignant cells. However, little is known about the molecular mechanism behind the downregulation of STAT4 in this cancer. In the current work we test the expression of STAT4 and STAT6 via RT-PCR and/or Western Blot in CTCL lesional skin samples and in immortalized patient-derived cell lines. In these malignant cell lines we correlate the expression of STAT4 and STAT6 with the T helper (Th) phenotype markers and test the effect of Histone Deacetylase (HDAC) inhibitors and siRNA-mediated knock down of miR-155 on STAT4 expression. Our findings demonstrate that STAT4 expression correlates with Th1 phenotype, while STAT6 is associated with the Th2 phenotype. Our results further document that STAT4 and STAT6 genes are inversely regulated in CTCL. Treatment with HDAC inhibitors upregulates STAT4 expression, while at the same time decreases STAT6 expression in MyLa cells. Also, siRNA-mediated knock down of miR-155 leads to upregulation in STAT4 expression in MyLa cells. In summary, our results suggest that loss of STAT4 expression and associated switch to Th2 phenotype during Mycosis Fungoides progression may be driven via aberrant histone acetylation and/or upregulation of oncogenic miR-155 microRNA.
Abbreviations:
| CTCL | = | cutaneous T-cell lymphoma |
| MF | = | mycosis fungoides |
| SS | = | sézary syndrome |
| HDAC | = | histone deacetylase |
| STAT | = | signal transducers and activators of transcription |
Introduction
Cutaneous T-Cell Lymphoma (CTCL) is a rare, but potentially devastating malignancy. Mycosis Fungoides (MF) and its leukemic form, Sézary Syndrome (SS), are the most common variants and account for ∼50% of all cutaneous lymphomas.Citation1 In Caucasians MF/SS primarily affects individuals over 55 years of age, while in African-Americans, Hispanics and Arabic individuals this disease presents at a significantly younger age (i.e. 20s and 30s).Citation2-4 Multiple epidemiologic studies documented that the incidence of CTCL has increased by ∼3 fold in the last 25–30 years.Citation4,5
In the early disease stages, which can last for several years, MF presents as flat erythematous skin patches resembling benign inflammatory diseases, whereas in the later stages, MF cells gradually form plaques or tumors and may disseminate to the lymph nodes and internal organs.Citation1 The early stages of CTCL are often difficult to distinguish clinically and even histologically from other benign entities including chronic eczema, psoriasis and pityriasis rubra pilaris.Citation6 In advanced disease, malignant cells may appear in the peripheral blood, leading to the leukemic stage of CTCL. A subset of leukemic CTCL, known as SS, is characterized by a triad of erythroderma, lymphadenopathy and detection of malignant T cells with convoluted/cerebriform nuclei on a peripheral blood smear.Citation1 The life expectancy of SS patients is usually less than 3 years.Citation1
The molecular pathogenesis of CTCL remains only partially understood. Recent reports elucidated the nature of cancer initiating cells for MF and SS.Citation7 Multiple studies attempted to clarify the genetic multistep carcinogenesis of CTCL.Citation8-11 Persistent activation of transcription factors of the signal transducers and activators of transcription (STAT) protein family has been implicated in the pathogenesis of a wide variety of human cancers, including CTCL. Expression and function of STAT3, STAT4 and STAT5 have been extensively studied in CTCL and these genes appear to play an important role in the disease pathogenesis and can be used as important prognostic markers.
Upregulation of STAT5 signaling occurs in the early stages of CTCL.Citation12,13 A growing body of experimental evidence suggests that this gene is important for expression of anti-apoptotic proteins (bcl-2 and bcl-x), cell cycle genes (Cyclin D and c-myc) and oncogenic miR-155 microRNA,Citation14-16 which work together to promote cancerogenesis. miR-155 was dubbed as the “bridge between inflammation and cancer”.Citation17
In the early stages of CTCL, STAT4 appears to be overexpressed, when compared to non-malignant skin samples.Citation18 The expression of STAT4, which is required for T helper (Th) 1 differentiation, is subsequently lost at the later stages, where the disease acquires predominantly Th2 phenotype.Citation19-23 In fact, in SS loss of STAT4 expression appears to be a robust and reliable diagnostic marker for this cancer.Citation21
Upregulation of STAT3 signaling has been consistently documented in advanced stages of CTCL.Citation24-28 During late stages autocrine IL-21 stimulation was shown to be responsible for constitutive activation of STAT3.Citation29 Furthermore, others have suggested that malignant T cells may acquire completely cytokine-independent constitutive activation of JAK3 signaling, which can drive a constitutive activation of STAT3.Citation24,25 Constitutively active STAT3 can increase survival and resistance to apoptosis in malignant T cells, promote sensitization to IL-2 stimulation,Citation24,29 promote Th2 phenotype in advanced disease and induce expression of miR-21 oncogenic microRNA.Citation30 STAT3 signaling in CTCL was also shown to upregulate VEGF,Citation31 the anti-inflammatory cytokine IL-10Citation32 and a suppressor of cytokine signaling-3 (SOCS).Citation33 The latter was demonstrated to confer resistance to IFN-α in malignant T cells.Citation32,33
Another important concept that has emerged in recent years is that the phenotype of malignant T cells is not fixed and this cancer exhibits a phenotypic plasticity that is highly sensitive to cytokine milieu in the tumor microenvironment.Citation32 Based on the above described evidence, STAT signaling appears to play a central role in this malignant phenotype switching.
Results
Correlation of STAT 4 and 6 expression with Th1, Th2, Treg, Th9, and Th17 phenotypes
Previous reports documented that many STAT members regulate Th differentiation phenotypes and determine cytokine expression in CTCL malignant cells.Citation34,35 Hence, we wanted to test the expression of Th1 (IFN-γ, IL-2, T-bet and IL-12A) vs. Th2 (IL-4 and IL-5) vs. Th9 (IL-9) vs. Th17 (IL-17A and IL-17F) vs. Treg (TGF-β1 and FOXP3) markers in these cells (Table S1). As demonstrated in and Fig. S1, most cell lines spontaneously do not express or only weakly express Th1 markers. However, T cell stimulation with PMA (phorbol 12-myristate 13-acetate) and ionomycin or with CD3/CD28 Dynabeads® in select cell lines (e.g. Hut78, HH, H9, SZ4, Sez4 and Hut102) led to an upregulation of a number of Th1 genes (; Fig. S1). In these cell lines the expression of Th1 markers correlated with the expression of STAT4 mRNA.
Figure 1. (A) Correlation of STAT4 and STAT6 expression with different T helper phenotype markers in CTCL cell lines under normal control vs. T cell stimulation culturing conditions (e.g., 10 ng/mL of PMA (phorbol 12-myristate 13-acetate) and 1 (M of ionomycin or with CD3/CD28 Dynabeads®). (B) Western blot analysis of STAT4 and STAT6 expression in patient-derived CTCL cell lines under normal control vs. T cell stimulation culturing conditions (i.e., PMA and ionomycin)
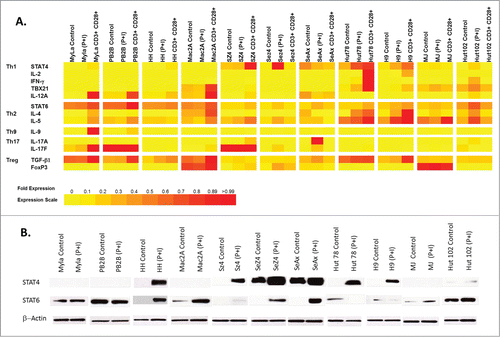
On the other hand, most cell lines heterogeneously express to some degree IL-4 and IL-5 Th2 markers and in a number of cases such expression corresponded to STAT6 expression (; Fig. S1).
Western blot analysis confirmed the expression of STAT4 and STAT6 genes on the protein level (). Specifically, cell lines that spontaneously express high levels of STAT6 (e.g., MyLa, PB2B, MaC2A, MJ and Hut102) did not express or only weakly expressed STAT4. On the other hand, cell lines that strongly express STAT4 (e.g., Sez4 and SeAx) did not express STAT6 under non-stimulated conditions.
IL-9 (Th9 marker) was only detected in MyLa cells upon stimulation with CD3/CD28 Dynabeads®, while most CTCL cell lines were IL-9-negative. Consistent with previous reports,Citation32,36,37 MyLa, PB2B, SZ4, Sez4 Hut102 and SeAx cells express IL-17F, IL-17A or both. Furthermore, these cytokines were further upregulated upon T cell stimulation (; Fig. S1). With respect to Treg phenotype, most malignant cells expressed TGF-β1 to a variable degree, but only MJ, Mac2A, SZ4 and SeAx SS-derived cells expressed detectable FoxP3.
Moreover, demonstrates that in immortalized malignant T cells the molecular definitions between different T helper subtypes are not strictly followed. Unlike, normal T cells, malignant T cell demonstrate molecular overlap across a number of Th phenotypes.
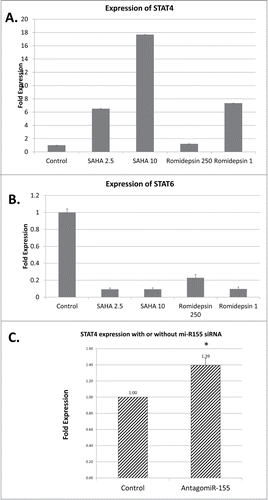
Effect of Histone Deacetylase (HDAC) inhibitors on STAT expression in MyLa patient-derived malignant cells
Recently, epigenetic changes became a significant focus of basic and clinical research in CTCL. A number of previous studies documented methylation/histone acetylation abnormalities in malignant CTCL cells.Citation38,39 In fact, 2 of the commonly used medications for advanced stages of this cancer are HDAC inhibitors (Romidepsin and Vorinostat).Citation40 Hence, we wanted to test whether histone acetylation mediates the expression of STAT4 and STAT6 thereby potentially affecting the balance between Th1 vs. Th2 phenotype in CTCL. To achieve that we treated MyLa cells for 24 hours with HDAC inhibitors, Romidepsin and Suberoylanilide Hydroxamic Acid (SAHA also known as Vorinostat). MyLa is a commonly used cell line derived from an MF patient skin biopsy.Citation41 As summarized in Table S2, out of 11 cell lines, this line is the only one derived from a skin biopsy of a typical MF patient. At baseline MyLa cells have very weak STAT4 expression and moderate STAT6 expression, which correlates with the advanced disease phenotype (). As demonstrated by RT-PCR, treatment with Vorinostat and Romidepsin upregulated the expression of STAT4 in these cells (), while concomitantly downregulating the expression of STAT6 (). Hence, this finding suggests that there is an inverse relationship in the expression of these genes, which is in part regulated by histone acetylation.
siRNA-mediated knock down of miR-155 leads to upregulation of STAT4 in MyLa cells
Oncogenic miR-155 microRNA was recently documented to play an important role in CTCL pathogenesis and was found to be upregulated in advanced stages of this cancer.Citation15,42 Using Bielefeld University Bioinformatic Server database we performed a screening analysis for matches in the STAT4 3'UTR, which identified a putative miR-155 binding site (position 2560 to 2690 of STAT4 mRNA; NM_003151.3). Hence, we wanted to test whether siRNA–mediated knock down of miR-155 expression will lead to the upregulation of STAT4 mRNA. As documented in , downregulation of miR-155 results in restoration of STAT4 expression in MyLa CTCL malignant cells. This suggests that the observed disease-associated upregulation of miR-155 may be responsible for downregulation of STAT4 and subsequent loss of Th1 phenotype in malignant T cells.
Comparison of STAT4 expression between CTCL lesional skin, normal skin from healthy volunteers and skin from patients with benign inflammatory dermatoses
Previous reports suggested that expression of certain STAT genes may have diagnostic and prognostic value in CTCL patients.Citation15,16,21 Hence, we wanted to compare the expression of STAT4 by RT-PCR between CTCL lesional skin, normal skin samples from healthy volunteers and biopsy specimens from patients affected by benign inflammatory dermatoses that often mimic CTCL (e.g., psoriasis, chronic eczema and pityriasis rubra pilaris or PRP). As demonstrated in , STAT4 was preferentially expressed in CTCL lesional skin, but not in normal skin or skin from patients with benign inflammatory dermatoses. It is of interest that upregulation of STAT4 was previously observed in CTCL lesional skin samples,Citation18 when compared to non-malignant inflammatory dermatoses.
Figure 3. (A) STAT4 is preferentially expressed in CTCL lesional skin compared to benign inflammatory dermatoses or normal skin from healthy volunteers. (B) Within the CTCL skin samples STAT4 low expression is observed in patients with progressive disease. (C) Kaplan-Meier analysis documented that loss of STAT4 expression is associated with poor disease prognosis.
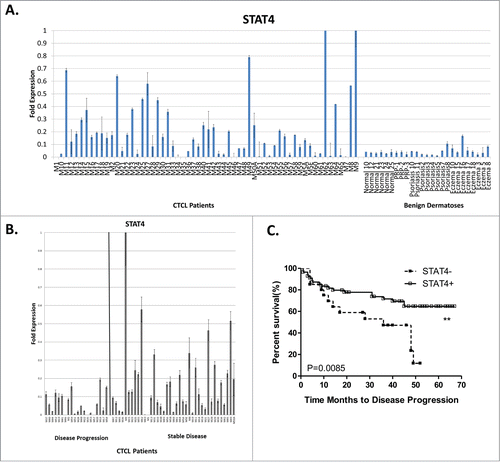
Further analysis of STAT4 expression in our patient samples documented that this gene was more strongly expressed in patients with stable disease and is downregulated in patients with progressive/advanced disease (). Using X-Tile softwareCitation43 we established a cut off <0.02 fold expression for low STAT4 expression and ≥0.02 cut off for high STAT4 expression and documented using Kaplan-Meier analysis () that loss of STAT4 expression is associated with progressive disease (Monte Carlo p value = 0.02; Miller-Seigmund p value = 0.0059). This finding is consistent with previous reports demonstrating that loss of STAT4 occurs as malignant cells shift from Th1 toward Th2 phenotype in advanced disease stages.Citation19-23
Discussion
In the current work we document that the expression of STAT4 correlates with the expression of Th1 markers in immortalized malignant CTCL cells, while STAT6 expression is associated with the Th2 phenotype. As summarized in Suppementary Table 2, most cell lines were derived from CTCL patients with advanced disease. Hence, our findings that the majority of tested cells (e.g., MyLa, PB2B, Mac2a, MJ and Hut102) strongly expressed STAT6 and corresponding Th2 markers, but not STAT4 and Th1 markers, are consistent with this observation. The expression of IL-17 in immortalized cells was addressed in previous experimental studiesCitation36,44 and may correlate with advanced disease in patients.Citation37,44,45
Our experimental work further suggests that aberrant histone acetylation in CTCL may result in a switch from STAT4-driven Th1 phenotype toward STAT6-associated Th2 phenotype in malignant cells and HDAC inhibitors may be able to restore this balance and upregulate STAT4 expression. More importantly our siRNA knock down experimental data demonstrates that STAT4 is one of the targets of miR-155 and inhibition of this oncogenic microRNA results in upregulation of STAT4. Hence, previously observed upregulation of miR-155 in advanced CTCL stagesCitation42 may be responsible for downregulation of STAT4 in CTCL lesional skin.
STAT4 was previously shown to be essential for Th1 differentiation and is typically upregulated via IL-12 signaling.Citation46 STAT4 binding to IFN-γ promoter/enhancer region is required for IFN-γ transcription.Citation47 As a result, STAT4 −/- CD4+ T cells produce low levels of IFN-γ47. Mice with a targeted disruption of the STAT4 gene have decreased responsiveness to IL-12, an impaired Th1 response and skewing toward markedly enhanced Th2 development.Citation48 Hence, it is not surprising that in advanced stages of CTCL downregulation of STAT4 appears to play a key role in a subsequent shift from Th1 toward Th2 disease phenotype.
In summary, STAT signaling appears to play a central role in the pathogenesis of this cancer where early in the disease STAT5 upregulation drives the expression of miR-155 oncogene, which targets STAT4 and contributes to a switch from the Th1 to Th2 phenotype, while during later stages activation of STAT3 leads to increased survival and resistance to apoptosis, expression of miR-21 oncogenic microRNA and upregulation of IL-5 and IL-10 signaling, all working in concert to promote carcinogenesis. Further research into the molecular pathogenesis of STAT signaling in this cancer may enable us to develop effective therapies for our patients.
Materials and Methods
Patients and Samples
All patients were enrolled in an IRB-approved study protocol with informed consent.Citation9 CTCL patients were recruited from the Cutaneous Lymphoma Clinic at the Dana Farber Cancer Institute (DFCI)/Brigham and Women's Hospital (BWH). All tissue samples were obtained and processed as previously described.Citation9 Briefly, punch biopsies from involved skin were collected from 60 CTCL patients between January 26, 2003 and June 1, 2005. Clinical stages of CTCL at the time of diagnosis for each patient are summarized in Supplementary Table 3. The obtained 6 mm biopsies were immediately snap-frozen in liquid nitrogen. Tissue was powdered in liquid nitrogen (Cryo-Press; Microtec Co, http://nition.com/en/products/cryopress.htm), and total RNA was extracted using Trizol (Invitrogen, https://www.lifetechnologies.com/order/catalog/product/15596026) and converted to cDNA using the iScript RT-PCR kit (Bio-Rad, http://www.bio-rad.com/en-us/sku/170-8893-iscript-one-step-rt-pcr-kit-with-sybr-green) according to the manufacturer's instructions. The biopsy samples analyzed in this report are the same samples that were analyzed in previous studies.Citation8,9,49,50 The diagnosis and clinical staging were established according to the diagnostic criteria of CTCL.Citation51 Similarly, volunteers with normal healthy skin (N = 6) and benign inflammatory dermatoses (N = 19) were recruited from the outpatient dermatology clinic of the University of British Columbia (Vancouver, Canada) with informed consent. Full-thickness lesional skin punch biopsies were obtained under local anesthesia as previously described.Citation50
Cell culture
HH, H9, Hut78, MJ and Hut102 patient-derived CTCL cell lines were previously describedCitation52,53 and were purchased from the American Tissue Culture Collection (ATCC). Detailed information on each cell line is provided in Suppementary Table 2. H9 is a clonal derivative of Hut78 cell line.Citation54 MyLa, PB2B, Mac2A, SZ4 and SeAx were a generous gift from professor K. Kaltoft (Copenhagen, Denmark) and were previously described elsewhere.Citation41,55-58 MJ, Hut78 cells were serially passaged in IMDM media (Invitrogen) containing 10% fetal bovine serum (FBS) (Invitrogen, http://www.lifetechnologies.com/ca/en/home/life-science/cell-culture.html). HH, H9, Hut102, MyLa, Mac2A and SZ4 cells were grown in RPMI media containing 10% FBS. Finally, SeAx cells were grown in RPMI media containing 10% FBS, 5 ng/mL of recombinant human IL-2 and IL-4 (R&D Systems, http://www.rndsystems.com/Products/202-IL). All cells were grown in 5% CO2, 95% air humidified incubator at 37°C. For stimulation, malignant T cells were treated with 10 ng/mL of PMA (phorbol 12-myristate 13-acetate) and 1 (M of ionomycin (Tocris Bioscience, http://www.tocris.com/dispprod.php?ItemId=65810#.U3AsrODD99A) or with CD3/CD28 Dynabeads® (Invitrogen, http://www.lifetechnologies.com/ca/en/home/references/protocols/proteins-expression-isolation-and-analysis/t-cell-activation-and-expansion/dynabeads-human-t-activator-cd3-cd28.html) according to manufacturer's instructions. To inhibit histone deacetylase (HDAC) activity cells were treated with 2.5-10 (M of Suberoylanilide Hydroxamic Acid (SAHA also known as Vorinostat, Santa Cruz Biotechnology, http://www.scbt.com/datasheet-220139-suberoylanilide-hydroxamic-acid.html) or 250nM-1(M Romidepsin (Adooq Bioscience, http://www.adooq.com/romidepsin-fk228-depsipeptide.html). mRNA from cell lines was isolated using Qiagen RNeasy kit (http://www.qiagen.com/products/catalog/sample-technologies/rna-sample-technologies/total-rna/rneasy-mini-kit) and was converted into cDNA using Bio-Rad iScript cDNA synthesis kit (http://www.bio-rad.com/en-us/product/reverse-transcription-reagents/iscript-cdna-synthesis-kit). Lysates for western blotting were obtained and quantitated as previously described.Citation59
Quantitative real-time reverse transcription-PCR gene expression analysis
STAT gene expression was tested in CTCL patients lesional skin, normal skin form healthy volunteers, lesional skin from patients with benign inflammatory dermatoses, and in patient-derived CTCL cell lines as previously described.Citation8,11,50 Primers for candidate human genes were designed using Primer 3 web softwareCitation60 and were purchased from Invitrogen Inc. Primer pair sequences for tested cytokines, STAT genes and control housekeeping genes are listed in Table S1. RT-PCR was performed utilizing the obtained cDNA from patients and iScript RT-PCR mix (Bio-Rad, http://www.bio-rad.com/en-us/sku/170-8893-iscript-one-step-rt-pcr-kit-with-sybr-green) on Bio-Rad iCycler as previously described.Citation8,49,50 The expression was standardized using genorm methodCitation61 utilizing ACTB, SDHA and GAPDH housekeeping genes. The obtained data was analyzed using XLSTAT 2009 software to obtain Kaplan-Meier curves.Citation62 We determined the optimal cut off point of STAT4 mRNA expression as 0.02 using the X-Tile software.Citation43 In our analysis, STAT4 high and STAT4 low groups were defined by the expression levels higher or lower than 0.02, respectively. p values < 0.05 were considered to be statistically significant.
Western blotting
Western blotting was performed as described previously.Citation11,49 STAT antibodies were purchased from Cell Signaling (http://www.cellsignal.com/products/primary-antibodies/9939?Ntt=%239939&fromPage=search) as part of Stat Antibody Sampler kit (Catalog #9939). STAT4 antibody was purchased separately (http://www.cellsignal.com/products/primary-antibodies/2653?Ntt=stat4&fromPage=search). β-Actin antibody was purchased from Santa Cruz Biotechnology (http://www.scbt.com/datasheet-130656-β-actin-n-21-antibody.html). Chemiluminescent detection reagents (ECL) were purchased from Amersham Biosciences (http://www.gelifesciences.com/webapp/wcs/stores/servlet/productById/en/GELifeSciences/28980926).
Bioinformatics sequence analysis for putative targets of miR-155
Following our report on the key function of miR-155 in CTCL,Citation15 we observed a reverse correlation between miR-155 and STAT4 expression in malignant T cells, which prompted us to use RNAhybrid analysis from Bielefeld University Bioinformatic Server database (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) to test our hypothesis that miR-155 targets STAT4.Citation63 Screening for matches in the STAT4 3'UTR, identified a putative miR-155 binding site (position 2560 to 2690 of STAT4 mRNA NM_003151.3).
Transient transfection with miRNA inhibitors (antagomiRs)
Cells were transfected with miR-155 antagomiR and negative control (mirVanaTM miRNA inhibitors, Ambion, http://www.lifetechnologies.com/ca/en/home/life-science/epigenetics-noncoding-rna-research/mirna-profiling-/ambion-anti-mir-inhibitors.html) using 0.625 nmol of the respective miR/control on 5.0 × 106 cells with an Amaxa Nucleofector as descriped previously.Citation15
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
947759_Supplementary_Materials.zip
Download Zip (543.1 KB)Acknowledgments
We thank Mr. Gregory Cormack for his technical assistance in performing molecular experiments.
Supplemental Materials
Supplemental data for this article can be accessed on the publisher's website.
Additional information
Funding
References
- Willemze R, Jaffe ES, Burg G, Cerroni L, Berti E, Swerdlow SH, Ralfkiaer E, Chimenti S, Diaz-Perez JL, Duncan LM, et al. WHO-EORTC classification for cutaneous lymphomas. Blood 2005; 105:3768-85; PMID:15692063; http://dx.doi.org/10.1182/blood-2004-09-3502
- Alsaleh QA, Nanda A, Al-Ajmi H, Al-Sabah H, Elkashlan M, Al-Shemmari S, Demierre MF. Clinicoepidemiological features of mycosis fungoides in Kuwait, 1991-2006. Int J Dermatol 2010; 49:1393-8; PMID:21155090; http://dx.doi.org/10.1111/j.1365-4632.2010.04567.x
- Sun G, Berthelot C, Li Y, Glass DA, 2nd, George D, Pandya A, Kurzrock R, Duvic M. Poor prognosis in non-Caucasian patients with early-onset mycosis fungoides. J Am Acad Dermatol 2009; 60:231-5; PMID: 19026464; http://dx.doi.org/10.1016/j.jaad.2008.09.063
- Wilson LD, Hinds GA, Yu JB. Age, race, sex, stage, and incidence of cutaneous lymphoma. Clin Lymphoma Myeloma Leuk 2012; 12:291-6; PMID: 23040434; http://dx.doi.org/10.1016/j.clml.2012.06.010
- Criscione VD, Weinstock MA. Incidence of cutaneous T-cell lymphoma in the United States, 1973-2002. Arch Dermatol 2007; 143:854-9; PMID: 17638728; http://dx.doi.org/10.1001/archderm.143.7.854
- Jawed SI, Myskowski PL, Horwitz S, Moskowitz A, Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sezary syndrome): part I. Diagnosis: Clinical and histopathologic features and new molecular and biologic markers. JAm Acad Dermatol 2014; 70:205 e1-16; quiz 21-2; PMID: 24355276; http://dx.doi.org/10.1016/j.jaad.2013.08.033
- Campbell JJ, Clark RA, Watanabe R, Kupper TS. Sezary syndrome and mycosis fungoides arise from distinct T-cell subsets: a biologic rationale for their distinct clinical behaviors. Blood 2010; 116:767-71; PMID:20484084; http://dx.doi.org/10.1182/blood-2009-11-251926
- Litvinov IV, Jones DA, Sasseville D, Kupper TS. Transcriptional profiles predict disease outcome in patients with cutaneous T-cell lymphoma. Clin cancer res : an official j Am Assoc Cancer Res 2010; 16:2106-14; PMID:20233883; http://dx.doi.org/10.1158/1078-0432.CCR-09-2879
- Shin J, Monti S, Aires DJ, Duvic M, Golub T, Jones DA, Kupper TS. Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome. Blood 2007; 110:3015-27; PMID:17638852; http://dx.doi.org/10.1182/blood-2006-12-061507
- van Kester MS, Borg MK, Zoutman WH, Out-Luiting JJ, Jansen PM, Dreef EJ, Vermeer MH, van Doorn R, Willemze R, Tensen CP. A meta-analysis of gene expression data identifies a molecular signature characteristic for tumor-stage mycosis fungoides. J invest dermatol 2012; 132:2050-9; PMID: 22513784; http://dx.doi.org/10.1038/jid.2012.117
- Litvinov IV, Cordeiro B, Huang Y, Zargham H, Pehr K, Dore MA, Gilbert M, Zhou Y, Kupper TS, Sasseville D. Ectopic expression of cancer testis antigens in Cutaneous T-Cell Lymphoma (CTCL) patients. Clin Cancer Res. 2014; 20(14):3799-808; PMID:24850846; http://dx.doi.org/10.1158/1078-0432.CCR-14-0307
- Dobbeling U. Transcription factor profiling shows new ways towards new treatment options of cutaneous T cell lymphomas. Curr drug discov technol 2007; 4:24-30; PMID:17630925; http://dx.doi.org/10.2174/157016307781115467
- Qin JZ, Kamarashev J, Zhang CL, Dummer R, Burg G, Dobbeling U. Constitutive and interleukin-7- and interleukin-15-stimulated DNA binding of STAT and novel factors in cutaneous T cell lymphoma cells. J invest dermatol 2001; 117:583-9; PMID: 11564163; http://dx.doi.org/10.1046/j.0022-202x.2001.01436.x
- Qin JZ, Zhang CL, Kamarashev J, Dummer R, Burg G, Dobbeling U. Interleukin-7 and interleukin-15 regulate the expression of the bcl-2 and c-myb genes in cutaneous T-cell lymphoma cells. Blood 2001; 98:2778-83; PMID:11675351; http://dx.doi.org/10.1182/blood.V98.9.2778
- Kopp KL, Ralfkiaer U, Gjerdrum LM, Helvad R, Pedersen IH, Litman T, Jønson L, Hagedorn PH, Krejsgaard T, Gniadecki R. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell cycle 2013; 12:1939-47; PMID: 23676217; http://dx.doi.org/10.4161/cc.24987
- Litvinov IV, Pehr K, Sasseville D. Connecting the dots in cutaneous T cell lymphoma (CTCL): STAT5 regulates malignant T cell proliferation via miR-155. Cell cycle 2013; 12:2172-3; PMID:23803726; http://dx.doi.org/10.4161/cc.25550
- Tili E, Michaille JJ, Wernicke D, Alder H, Costinean S, Volinia S, et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Nat Acad Sci U S A 2011; 108:4908-13; PMID:21383199; http://dx.doi.org/10.1073/pnas.1101795108
- Tracey L, Villuendas R, Dotor AM, Spiteri I, Ortiz P, Garcia JF, Peralto JL, Lawler M, Piris MA. Mycosis fungoides shows concurrent deregulation of multiple genes involved in the TNF signaling pathway: an expression profile study. Blood 2003; 102:1042-50; PMID:12689942; http://dx.doi.org/10.1182/blood-2002-11-3574
- Johnson VE, Vonderheid EC, Hess AD, Eischen CM, McGirt LY. Genetic markers associated with progression in early mycosis fungoides. J Eur Acad Dermatol Venereol 2013; PMID:24171863; http://dx.doi.org/10.1111/jdv.12299
- Kari L, Loboda A, Nebozhyn M, Rook AH, Vonderheid EC, Nichols C, Virok D, Chang C, Horng WH, Johnston J. et al. Classification and prediction of survival in patients with the leukemic phase of cutaneous T cell lymphoma. The J exp med 2003; 197:1477-88; PMID:12782714; http://dx.doi.org/10.1084/jem.20021726
- Nebozhyn M, Loboda A, Kari L, Rook AH, Vonderheid EC, Lessin S, Berger C, Edelson R, Nichols C, Yousef M. et al. Quantitative PCR on 5 genes reliably identifies CTCL patients with 5% to 99% circulating tumor cells with 90% accuracy. Blood 2006; 107:3189-96; PMID:16403914; http://dx.doi.org/10.1182/blood-2005-07-2813
- Showe LC, Fox FE, Williams D, Au K, Niu Z, Rook AH. Depressed IL-12-mediated signal transduction in T cells from patients with Sezary syndrome is associated with the absence of IL-12 receptor beta 2 mRNA and highly reduced levels of STAT4. J immunol 1999; 163:4073-9; PMID:10491012.
- van Doorn R, Dijkman R, Vermeer MH, Out-Luiting JJ, van der Raaij-Helmer EM, Willemze R, Tensen CP.et al. Aberrant expression of the tyrosine kinase receptor EphA4 and the transcription factor twist in Sezary syndrome identified by gene expression analysis. Cancer Res 2004; 64:5578-86; PMID:15313894; http://dx.doi.org/10.1158/0008-5472.CAN-04-1253
- Sommer VH, Clemmensen OJ, Nielsen O, Wasik M, Lovato P, Brender C, Eriksen KW, Woetmann A, Kaestel CG, Nissen MH. et al. In vivo activation of STAT3 in cutaneous T-cell lymphoma. Evidence for an antiapoptotic function of STAT3. Leukemia 2004; 18:1288-95; PMID:15141228; http://dx.doi.org/10.1038/sj.leu.2403385
- Eriksen KW, Kaltoft K, Mikkelsen G, Nielsen M, Zhang Q, Geisler C, Nissen MH, Röpke C, Wasik MA, Odum N. Constitutive STAT3-activation in Sezary syndrome: tyrphostin AG490 inhibits STAT3-activation, interleukin-2 receptor expression and growth of leukemic Sezary cells. Leukemia 2001; 15:787-93; PMID:11368440; http://dx.doi.org/10.1038/sj.leu.2402093
- Nielsen M, Kaestel CG, Eriksen KW, Woetmann A, Stokkedal T, Kaltoft K, Geisler C, Röpke C, Odum N. Inhibition of constitutively activated Stat3 correlates with altered Bcl-2/Bax expression and induction of apoptosis in mycosis fungoides tumor cells. Leukemia 1999; 13:735-8; PMID:10374878; http://dx.doi.org/10.1038/sj.leu.2401415
- van Kester MS, Out-Luiting JJ, von dem Borne PA, Willemze R, Tensen CP, Vermeer MH. Cucurbitacin I inhibits Stat3 and induces apoptosis in Sezary cells. J invest dermatol 2008; 128:1691-5; PMID:18200050; http://dx.doi.org/10.1038/sj.jid.5701246
- Zhang C, Li B, Gaikwad AS, Haridas V, Xu Z, Gutterman JU, Duvic M. Avicin D selectively induces apoptosis and downregulates p-STAT-3, bcl-2, and survivin in cutaneous T-cell lymphoma cells. J invest dermatol 2008; 128:2728-35; PMID:18496567; http://dx.doi.org/10.1038/jid.2008.138
- van der Fits L, Out-Luiting JJ, van Leeuwen MA, Samsom JN, Willemze R, Tensen CP, Vermeer MH. Autocrine IL-21 stimulation is involved in the maintenance of constitutive STAT3 activation in Sezary syndrome. J invest dermatol 2012; 132:440-7; PMID:21938013; http://dx.doi.org/10.1038/jid.2011.293
- van der Fits L, van Kester MS, Qin Y, Out-Luiting JJ, Smit F, Zoutman WH, Willemze R, Tensen CP, Vermeer MH. MicroRNA-21 expression in CD4+ T cells is regulated by STAT3 and is pathologically involved in Sezary syndrome. J invest dermatol 2011; 131:762-8; PMID:21085192; http://dx.doi.org/10.1038/jid.2010.349
- Krejsgaard T, Vetter-Kauczok CS, Woetmann A, Lovato P, Labuda T, Eriksen KW, Zhang Q, Becker JC, Ødum N. Jak3- and JNK-dependent vascular endothelial growth factor expression in cutaneous T-cell lymphoma. Leukemia 2006; 20:1759-66; PMID: 16932349; http://dx.doi.org/10.1038/sj.leu.2404350
- Abraham RM, Zhang Q, Odum N, Wasik MA. The role of cytokine signaling in the pathogenesis of cutaneous T-cell lymphoma. Cancer biol ther 2011; 12:1019-22; PMID:22236880; http://dx.doi.org/10.4161/cbt.12.12.18144
- Brender C, Lovato P, Sommer VH, Woetmann A, Mathiesen AM, Geisler C, Wasik M, Ødum N. Constitutive SOCS-3 expression protects T-cell lymphoma against growth inhibition by IFNalpha. Leukemia 2005; 19:209-13; PMID:15618960; http://dx.doi.org/10.1038/sj.leu.2403610
- Mathew A, MacLean JA, DeHaan E, Tager AM, Green FH, Luster AD. Signal transducer and activator of transcription 6 controls chemokine production and T helper cell type 2 cell trafficking in allergic pulmonary inflammation. J exp med 2001; 193:1087-96; PMID:11342593; http://dx.doi.org/10.1084/jem.193.9.1087
- Yang Z, Chen M, Ellett JD, Fialkow LB, Carter JD, McDuffie M, Nadler JL. Autoimmune diabetes is blocked in Stat4-deficient mice. J autoimmun 2004; 22:191-200; PMID:15041039; http://dx.doi.org/10.1016/j.jaut.2003.08.006
- Krejsgaard T, Ralfkiaer U, Clasen-Linde E, Eriksen KW, Kopp KL, Bonefeld CM, Geisler C, Dabelsteen S, Wasik MA, Ralfkiaer E. et al. Malignant cutaneous T-cell lymphoma cells express IL-17 utilizing the Jak3/Stat3 signaling pathway. J invest dermatol 2011; 131:1331-8; PMID:21346774; http://dx.doi.org/10.1038/jid.2011.27
- Krejsgaard T, Litvinov IV, Wang Y, Xia L, Willerslev-Olsen A, Koralov SB, Kopp KL, Bonefeld CM, Wasik MA, Geisler C. et al. Elucidating the role of interleukin-17F in cutaneous T-cell lymphoma. Blood 2013; 122:943-50; PMID:23801634; http://dx.doi.org/10.1182/blood-2013-01-480889
- Wong HK. Novel biomarkers, dysregulated epigenetics, and therapy in cutaneous T-cell lymphoma. Discov Med 2013; 16:71-8; PMID:23998443
- Zhang C, Richon V, Ni X, Talpur R, Duvic M. Selective induction of apoptosis by histone deacetylase inhibitor SAHA in cutaneous T-cell lymphoma cells: relevance to mechanism of therapeutic action. J invest dermatol 2005; 125:1045-52; PMID:16297208; http://dx.doi.org/10.1111/j.0022-202X.2005.23925.x
- Hymes KB. The role of histone deacetylase inhibitors in the treatment of patients with cutaneous T-cell lymphoma. Clin Lymphoma Myeloma Leuk 2010; 10:98-109; PMID:20371442; http://dx.doi.org/10.3816/CLML.2010.n.014
- Kaltoft K, Bisballe S, Dyrberg T, Boel E, Rasmussen PB, Thestrup-Pedersen K. Establishment of two continuous T-cell strains from a single plaque of a patient with mycosis fungoides. In vitro cell develop biol : j Tissue Culture Assoc 1992; 28A:161-7; PMID:1582990; http://dx.doi.org/10.1007/BF02631086
- Kopp KL, Ralfkiaer U, Nielsen BS, Gniadecki R, Woetmann A, Odum N, Ralfkiaer E. Expression of miR-155 and miR-126 in situ in cutaneous T-cell lymphoma. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica 2013; 121:1020-4; PMID:24033365; http://dx.doi.org/10.1111/apm.12162
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin cancer res : an official j Am Assoc Cancer Res 2004; 10:7252-9; PMID:15534099; http://dx.doi.org/10.1158/1078-0432.CCR-04-0713
- Ciree A, Michel L, Camilleri-Broet S, Jean Louis F, Oster M, Flageul B, Senet P, Fossiez F, Fridman WH, Bachelez H. et al. Expression and activity of IL-17 in cutaneous T-cell lymphomas (mycosis fungoides and Sezary syndrome). In j cancer J int du cancer 2004; 112:113-20; PMID:15305382; http://dx.doi.org/10.1002/ijc.20373
- Willerslev-Olsen A, Litvinov IV, Fredholm SM, Petersen DL, Sibbesen NA, Gniadecki R, Zhang Q, Bonefeld CM, Wasik MA, Geisler C. et al. IL-15 and IL-17F are differentially regulated and expressed in mycosis fungoides (MF). Cell cycle 2014; 13:1306-12; PMID:24621498; http://dx.doi.org/10.4161/cc.28256
- Nishikomori R, Usui T, Wu CY, Morinobu A, O'Shea JJ, Strober W. Activated STAT4 has an essential role in Th1 differentiation and proliferation that is independent of its role in the maintenance of IL-12R beta 2 chain expression and signaling. J immunol 2002; 169:4388-98; PMID:12370372; http://dx.doi.org/10.4049/jimmunol.169.8.4388
- Lawless VA, Zhang S, Ozes ON, Bruns HA, Oldham I, Hoey T, Grusby MJ, Kaplan MH. Stat4 regulates multiple components of IFN-gamma-inducing signaling pathways. J immunol 2000; PMID:11120802; 165:6803-8; http://dx.doi.org/10.4049/jimmunol.165.12.6803
- Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature 1996; 382:174-7; PMID:8700209; http://dx.doi.org/10.1038/382174a0
- Litvinov IV, Kupper TS, Sasseville D. The role of AHI1 and CDKN1C in cutaneous T-cell lymphoma progression. Exp dermatol 2012; 21:964-6; PMID:23171462; http://dx.doi.org/10.1111/exd.12039
- Litvinov IV, Zhou Y, Kupper TS, Sasseville D. Loss of BCL7A expression correlates with poor disease prognosis in patients with early-stage cutaneous T-cell lymphoma. Leuk lymphoma 2012; 54(3):653-4; PMID:22856870; http://dx.doi.org/10.3109/10428194.2012.717695
- Olsen E, Vonderheid E, Pimpinelli N, Willemze R, Kim Y, Knobler R, Zackheim H, Duvic M, Estrach T, Lamberg S. et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood 2007; 110:1713-22; PMID:17540844; http://dx.doi.org/10.1182/blood-2007-03-055749
- Zhang C, Hazarika P, Ni X, Weidner DA, Duvic M. Induction of apoptosis by bexarotene in cutaneous T-cell lymphoma cells: relevance to mechanism of therapeutic action. Clin cancer res : an official j Am Assoc Cancer Res 2002; 8:1234-40; PMID:12006543
- Bunn PA, Jr., Foss FM. T-cell lymphoma cell lines (HUT102 and HUT78) established at the National Cancer Institute: history and importance to understanding the biology, clinical features, and therapy of cutaneous T-cell lymphomas (CTCL) and adult T-cell leukemia-lymphomas (ATLL). J Cell Biochem Suppl 1996; 24:12-23; PMID:8806090; http://dx.doi.org/10.1002/jcb.240630503
- Chen TR. Karyotypic derivation of H9 cell line expressing human immunodeficiency virus susceptibility. J Natl Cancer Inst 1992; 84:1922-6; PMID:1460674; http://dx.doi.org/10.1093/jnci/84.24.1922
- Abrams JT, Lessin S, Ghosh SK, Ju W, Vonderheid EC, Nowell P, Murphy G, Elfenbein B, DeFreitas E. A clonal CD4-positive T-cell line established from the blood of a patient with Sezary syndrome. J invest dermatol 1991; 96:31-7; PMID:1987293; http://dx.doi.org/10.1111/1523-1747.ep12514693
- Kaltoft K, Bisballe S, Rasmussen HF, Thestrup-Pedersen K, Thomsen K, Sterry W. A continuous T-cell line from a patient with Sezary syndrome. Archdermatol res 1987; 279:293-8; PMID:3498444; http://dx.doi.org/10.1007/BF00431220
- Starkebaum G, Loughran TP, Jr., Waters CA, Ruscetti FW. Establishment of an IL-2 independent, human T-cell line possessing only the p70 IL-2 receptor. Int j cancer J int du cancer 1991; 49:246-53; PMID:1879969; http://dx.doi.org/10.1002/ijc.2910490218
- Wasik MA, Seldin DC, Butmarc JR, Gertz R, Marti R, Maslinski W, Kadin ME. Analysis of IL-2, IL-4 and their receptors in clonally-related cell lines derived from a patient with a progressive cutaneous T-cell lymphoproliferative disorder. Leukemia & lymphoma 1996; 23:125-36; PMID:9021695; http://dx.doi.org/10.3109/10428199609054811
- Litvinov IV, Vander Griend DJ, Xu Y, Antony L, Dalrymple SL, Isaacs JT. Low-calcium serum-free defined medium selects for growth of normal prostatic epithelial stem cells. Cancer Res 2006; 66:8598-607; PMID:16951173; http://dx.doi.org/10.1158/0008-5472.CAN-06-1228
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000; 132:365-86; PMID:10547847
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3(7): research0034.1-11; PMID:12184808; http://dx.doi.org/10.1186/gb-2002-3-7-research0034
- Kaplan ELaM, P. Nonparametric estimation from incomplete observations. J Am Stat Assoc 1958; 53:457-81; http://dx.doi.org/10.1080/01621459.1958.10501452
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA 2004; 10:1507-17; PMID:15383676; http://dx.doi.org/10.1261/rna.5248604