Abstract
Tumor-specific expression of antitumor drugs can be achieved using attenuated Salmonella typhimurium harboring the PBAD promoter, which is induced by L-arabinose. However, L-arabinose does not accumulate because it is metabolized to D-xylulose-5-P by enzymes encoded by the ara operon in Salmonellae. To address this problem, we developed an engineered strain of S. typhimurium in which the ara operon is deleted. Linear DNA transformation was performed using λ red recombinase to exchange the ara operon with linear DNA carrying an antibiotic-resistance gene with homology to regions adjacent to the ara operon. The ara operon-deleted strain and its parental strain were transformed with a plasmid encoding Renilla luciferase variant 8 (RLuc8) or cytolysin A (clyA) under the control of the PBAD promoter. Luciferase assays demonstrated that RLuc8 expression was 49-fold higher in the ara operon-deleted S. typhimurium than in the parental strain after the addition of L-arabinose. In vivo bioluminescence imaging showed that the tumor tissue targeted by the ara operon-deleted Salmonella had a stronger imaging signal (∼30-fold) than that targeted by the parental strain. Mice with murine colon cancer (CT26) that had been injected with the ara operon-deleted S. typhimurium expressing clyA showed significant tumor suppression. The present report demonstrates that deletion of the ara operon of S. typhimurium enhances L-arabinose accumulation and thereby drives PBAD-promoted expression of cytotoxic agents and imaging agents. This is a promising approach for tumor therapy and imaging.
Introduction
Specific strains of Escherichia coli and Salmonella typhimurium spontaneously accumulate and proliferate in tumors, and this eventually leads to cancer cell death.Citation1-8 In addition, bacteria have been used as vehicles to deliver therapeutic agents against cancer. Treatment with such vehicles results in remarkable tumor regression.Citation9-13
S. typhimurium has several advantages over other strains: (1) it can grow in aerobic or anaerobic conditions, both of which are found in solid tumors; (2) it has specialized systems that mediate the invasion of epithelial cells and macrophages; and (3) there is a vast body of information on its genetics, which facilitates the genetic engineering of this bacterium for cancer therapies.Citation1,4,7,13 Genetic engineering of Salmonella has become extremely important to lower its toxicity and maximize its therapeutic efficacy.Citation12,14 We previously reported that treatment with attenuated S. typhimurium defective in ppGpp synthesis (ΔppGpp S. typhimurium)Citation15 suppresses tumor growth by monotherapy and eradicates tumors when used to deliver cytotoxic molecules.Citation10
A major problem preventing cytotoxin-carrying Salmonella from being fully exploited in cancer therapy is its toxicity in non-tumor reticuloendothelial organs, mainly the liver and spleen, due to the initial localization of the bacteria in these organs following tail vein injection.Citation3,6 Thus, constitutive cytotoxin expression would inevitably result in hepatic and splenic injury. Therefore, for tumor-specific drug production with reduced toxicity in normal organs, inducible promoter systems have been employed such as the PBAD promoter, which can be activated by the sugar L-arabinose.Citation16,17
The PBAD promoter is positively and negatively regulated by the product of the araC gene,Citation18 a transcriptional regulator that forms a complex with L-arabinose. In the absence of arabinose, a single AraC dimer contacts the 2 widely separated O2 and I1 half-sites of the araBAD operon, forming a 210 bp DNA loop and repressing transcription from the PBAD promoter. Binding of arabinose to araC causes the protein to cease DNA looping and favor binding to the adjacent I1 and I2 half-sites, resulting in activation of transcription from the PBAD promoter.Citation19
L-arabinose, the inducer of the PBAD system, is metabolized to L-ribulose, L-ribulose-5-P, and D-xylulose-5-P, which are intermediates of the pentose phosphate pathway, by enzymes encoded by genes of the ara operon (ara A, B, and D). Therefore, in the bacterial cytosol, some L-arabinose is metabolized by these enzymes ().Citation20-23 In the current study, we hypothesized that deletion of the ara operon may allow the level of L-arabinose in the bacterial cytosol to increase since L-arabinose would not be metabolized and would thereby be available to bind araC, eventually leading to enhanced transcriptional activity.
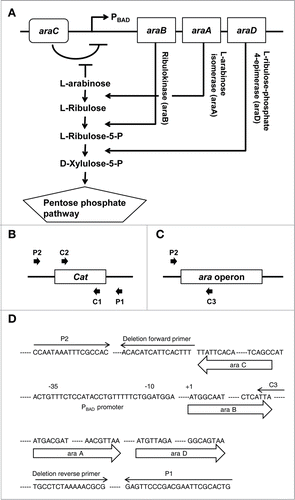
Figure 1. Deletion of the ara operon. (A) Schematic representation of the ara operon in S. typhimurium and the metabolic pathway by which enzymes encoded by the ara operon metabolize L-arabinose. (B) Diagram of PCR to confirm recombination between the ara operon and an antibiotic-resistance gene using pairs of primers (P1 and C2 or C1 and P2). (C) Diagram of PCR to confirm deletion of the ara operon using the C3 and P2 primers. Cat, chloramphenicol acetyltransferase. (D) Sequence of the ara operon showing the precise positions of primers used for targeted deletion of ara operon and for PCR verification.
In the present report, using an attenuated strain of ΔppGpp S. typhimurium, we engineered an inducible bacterial gene expression system in which expression of a bioluminescence reporter protein or a cytotoxic protein was significantly enhanced in response to administration of a small amount of the exogenous inducer L-arabinose. This system improved imaging and therapeutic efficacy of Salmonellae-mediated anti-cancer therapy, respectively.
Results and Discussion
Construction of an ara operon-deleted S. typhimurium strain
To demonstrate the feasibility of using ara operon-deleted bacteria to enhance gene expression, we first developed an engineered strain of ΔppGpp S. typhimurium in which the ara operon was deleted (ΔaraΔppGpp). This was achieved by transforming linear DNA using λ red recombinase to exchange the ara operon with linear DNA carrying an antibiotic-resistance gene (). The growth of the ΔaraΔppGpp strain was compared with that of its parental strain (ΔppGpp). The ΔaraΔppGpp strain showed no growth retardation in log phase (), indicating that the growth of S. typhimurium was not affected by deletion of the ara operon.
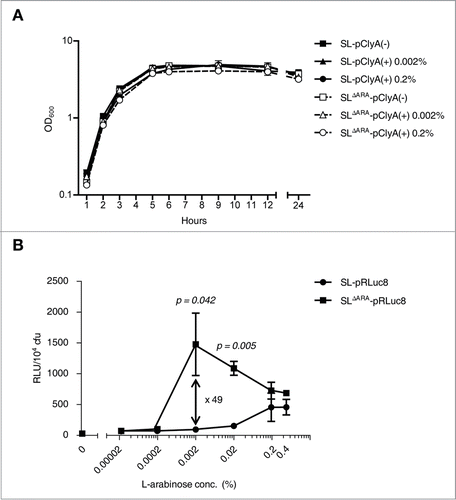
Figure 2. In vitro characteristics of ara operon-deleted S. typhimurium. (A) Bacterial growth curves. Growth was assessed for ΔppGpp S. typhimurium (SL) and ara operon-deleted ΔppGpp S. typhimurium (SLΔARA). SL and SLΔARA was transformed with pClyA (SL-pClyA and SLΔARA-pClyA). SL-pClyA (-) or SLΔARA-pClyA (-) indicates without induction, while SL-pClyA (+) or SLΔARA-pClyA (+) indicates with induction by 0.002% or 0.2% L-arabinose. L-arabinose was added to the growth medium at 1.5 h. (B) SL and SLΔARA were transformed with pRLuc8. Various concentrations (0–0.4%) of L-arabinose were added to fresh bacterial cultures (after 1.5 h of culture) when the OD600 was 0.5–0.7. After 4.5 h incubation, ∼8 × 107 bacteria were collected for the luciferase assay (n = 5 per group). Bioluminescence, indicative of RLuc8 expression, was detected using a luminometer.
Enhanced protein expression induced by L-arabinose in ara operon-deleted S. typhimurium
To assess the transcriptional activity of the PBAD promoter in the ΔaraΔppGpp and ΔppGpp strains, both strains were electroporated with a bacterial expression plasmid harboring Renilla luciferase 8 (RLuc8) under the control of the PBAD promoter (pBAD-RLuc8 (pRLuc8), Fig. S1A), which is only active in the presence of the inducer L-arabinose.Citation10
The ΔaraΔppGpp strain exhibited significantly increased expression of RLuc8 and increased sensitivity to the inducer L-arabinose (). The level of bioluminescence per bacterium was highest after induction with 0.002% L-arabinose, which increased bioluminescence 49.2 ± 21.4-fold compared with basal bioluminescence in the ΔppGpp strain. When treated with 0.02% L-arabinose, bioluminescence was 11.5 ± 0.8-fold higher in the ΔaraΔppGpp strain than in the ΔppGpp strain. In the control ΔppGpp strain, bioluminescence was highest after induction with 0.2% L-arabinose. Bioluminescence was 3-fold higher in the ΔaraΔppGpp strain after induction with 0.002% L-arabinose than in the control ΔppGpp strain with 0.2% L-arabinose, indicating a greater than 300-fold increase in sensitivity by ara operon deletion. These results indicate that in comparison to the ΔppGpp strain, gene expression driven by the PBAD promoter was saturated with a significantly lower concentration of L-arabinose in the ΔaraΔppGpp strain since L-arabinose was not metabolized to D-xylulose-5-P by enzymes encoded by the ara operon in bacterial chromosomes and was therefore stable.
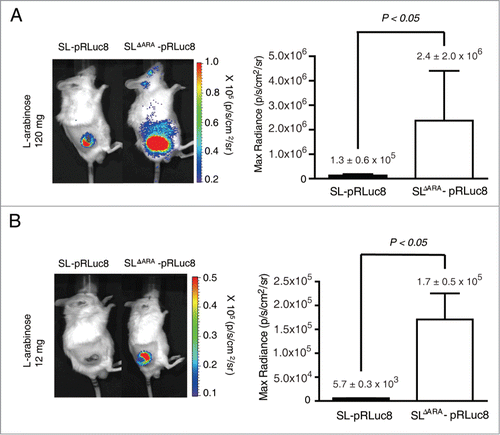
To evaluate whether gene expression in the ΔaraΔppGpp strain could be enhanced in vivo, the ΔaraΔppGpp or ΔppGpp strain was intravenously injected into immunocompetent BALB/c mice bearing syngeneic tumors derived from CT26 colon carcinoma cells. When induced with 120 mg L-arabinose, bioluminescence was higher (18.5-fold) in tumors of mice that received the ΔaraΔppGpp strain (2.4 ± 2.0 × 106 p/s/cm2/sr) than in tumors in mice that received the ΔppGpp strain (1.3 ± 0.5 × 105 p/s/cm2/sr) (). When induced with a 10-fold lower dose of L-arabinose (12 mg), bioluminescence was strongly detected in tumors colonized by the ΔaraΔppGpp strain, but not in those colonized by the ΔppGpp strain (30.2-fold higher in the former than in the latter) (). Bioluminescence in tumors following injection of 12 mg of arabinose for the ΔaraΔppGpp strain (1.7 ± 0.5 × 105 p/s/cm2/sr) was comparable to that following injection of 120 mg of arabinose for the ΔppGpp strain (1.3 ± 0.5 × 105 p/s/cm2/sr). These in vitro and in vivo results indicate that gene expression could be enhanced by the deletion of the ara operon of ΔppGpp strain.
Figure 3. Bacterial bioluminescence and RLuc8 expression in colonized tumors. (A and B) In vivo expression of RLuc8 in tumors colonized by bacteria at 6 d post inoculum (dpi). BALB/c mice were inoculated subcutaneously with CT26 cells (1 × 106) and then injected with SL or SLΔARA carrying pBAD-RLuc8 (SL- pRLuc8 and SLΔARA- pRLuc8). Mice received an intraperitoneal injection of 120 mg (A) or 12 mg (B). L-arabinose at 3 dpi. Bioluminescence imaging was performed following intravenous injection of coelenterazine (0.7 mg/kg). A region of interest was selected manually to quantify photon flux. The region of interest area was kept constant, and the maximum intensity within this area was recorded (photons·s−1·cm−2·sr−1).
Enhanced expression of a cytotoxic protein by ara operon-deleted S. typhimurium
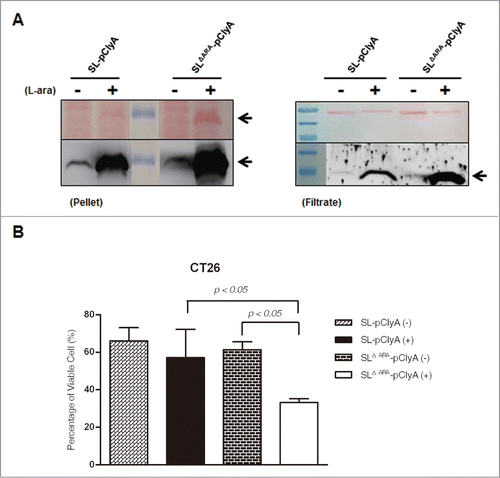
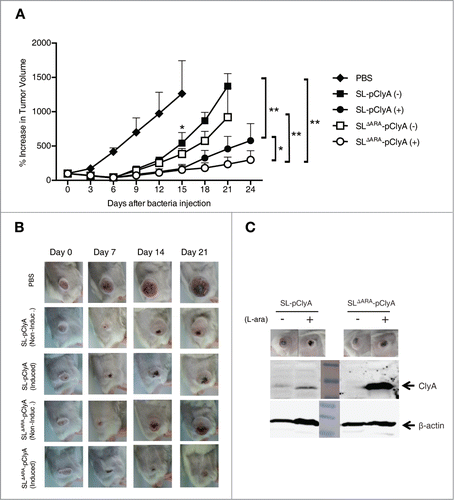
To explore whether the ara operon-deleted S. typhimurium has enhanced antitumor efficacy, we electroporated the ΔaraΔppGpp and ΔppGpp strains with a bacterial expression plasmid harboring the cytotoxic gene cytolysin A (ClyA) under the control of the PBAD promoter (pBAD-ClyA (pClyA), Fig. S1B).Citation10 Western blot analysis demonstrated that the ClyA protein (34 kDa) was present in the bacterial cell pellet and culture medium of both strains carrying pClyA, and these levels were significantly increased after L-arabinose administration (). In particular, ClyA expression was significantly enhanced in the bacterial cell pellet and culture medium of the ΔaraΔppGpp strain following L-arabinose administration. This was further verified by examining the cytotoxicity associated with the conditioned bacterial medium. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay demonstrated that cultured CT26 tumor cells were significantly killed by ClyA present in the spent media of the ΔaraΔppGpp strain carrying pClyA after induction with L-arabinose (). This result indicates that ClyA protein expression was specifically induced by L-arabinose and significantly increased in the ara operon-deleted strain. A larger amount of ClyA was secreted from the ΔaraΔppGpp strain, which led to enhanced cytotoxic efficacy against tumor cells (Fig. S2).
Figure 4. L-arabinose-induced expression and secretion of ClyA in ara operon-deleted S. typhimurium. SL and SLΔara were transformed with pClyA. (A) Expression and secretion of ClyA (34 kDa) from both transformed strains (SL- pClyA and SLΔARA- pClyA) was verified by immunoblotting using an anti-ClyA antibody. Bacterial pellets (Pellet) and filtered culture medium (Filtrate) were collected at 4.5 hour after induction at 1.5 hours with or without 0.2% L-arabinose. Arrows indicate ClyA. Upper panel is Ponseau S staining. (B) Cytotoxicity due to secretion of ClyA from transformed bacteria. SL- pClyA and SLΔARA- pClyA were grown in the presence (+) or absence (-) of L-arabinose. The bacterial spent medium was filtered (0.2 μm filter), centrifuged (3,000 × g for 5 min), and concentrated to 20 mg/ml using a concentrator. The cytotoxic effects of the concentrated bacterial supernatant (0.2 mg protein) were tested using cultured CT26 cells (1 × 104). Cell death was determined using an MTT assay kit (n = 5 per group).
A previous study investigated the secretion mechanism of ClyA in bacteriaCitation24 and demonstrated that ClyA protein is exported from the bacterial cell via an outer membrane vesicle (OMV) mechanism that is independent of previously described type I - V secretion systems. The ClyA protein, which is present in monomeric form in the periplasm, undergoes oligomerization and forms active pore assemblies in the OMVs, which export ClyA in its active form to the external environment.
Enhanced tumor suppression by the ara operon-deleted strain in a mouse tumor model
To further explore the antitumor activity of ara operon-deleted Salmonella, BALB/c mice transplanted with CT26 tumors were treated with engineered S. typhimurium (ΔaraΔppGpp or ΔppGpp strains) carrying pClyA in the presence or absence of induction with L-arabinose, or with PBS. Tumor-bearing mice tolerated all treatments. No fatalities were observed when either strain was used. The body weight of treated mice was not significantly different from that of the control group, suggesting that expression of ClyA via the intravenous administration of engineered Salmonella did not elicit explicit toxicity (Fig. S3). Similar numbers of each strain colonized tumors at various time-points after bacterial injection (Fig. S4), indicating that the ΔaraΔppGpp and ΔppGpp strains were targeted to tumors with a similar efficiency.
The maximum anti-tumor effect was observed with the ΔaraΔppGpp strain carrying pClyA induced with L-arabinose, followed by the ΔppGpp strain induced with L-arabinose (). No statistically significant difference in the antitumor efficacies of the 2 strains was observed in the absence of L-arabinose induction, although the antitumor efficacies of the 2 strains were significantly higher than that of the PBS control mice. At the end of the experiment (24 days), mean tumor volume was 300 mm3 and 579 mm3 in the groups treated with the ΔaraΔppGpp or ΔppGpp strain, respectively, and induced with L-arabinose. By contrast, in mice treated with either strain in the absence of L-arabinose induction or those that received only PBS, tumor volume reached > 1,000 mm3 (). Immunoblot analysis of excised tumor tissue demonstrated that ClyA was expressed in tumors colonized by bacteria in response to L-arabinose administration, and that the level of ClyA was higher in tumor tissue colonized by the ΔaraΔppGpp strain than in tumors colonized by the ΔppGpp strain (). These results indicated that the ΔaraΔppGpp strain carrying pClyA significantly suppressed tumor growth.
Figure 5. Effect of ara operon-deleted S. typhimurium on tumor growth. BALB/c mice were inoculated subcutaneously with CT26 cells (1 × 106). When the tumor volume reached ∼120 mm3, mice were injected with PBS (n = 6), SL carrying pClyA (SL- pClyA, n = 6), or SLΔARA carrying pClyA (SLΔARA- pClyA, n = 7), but were not treated with L-arabinose (-). In separate groups (n = 8 per group), mice were injected with SL- pClyA or SLΔARA- pClyA, and then received a daily intraperitoneal injection of 120 mg L-arabinose (+), starting 3 d after they had received the bacteria. (A) Changes in tumor volume. * P = 0.04 (PBS vs. SL-pClyA (-) or SLΔARA- pClyA (-) at day 15, SL- pClyA (+) vs. SLΔARA- pClyA (+) at day 24), ** P = 0.03 (SL-pClyA (-) vs. SL-pClyA (+) at day 21, SLΔARA- pClyA (-) vs. SLΔARA- pClyA (+) at day 21, SL-pClyA (-) vs. SLΔARA- pClyA (+) at day 21) (B) Images of subcutaneous tumors in representative mice. (C) Western blot analysis of the 34 kDa ClyA protein from CT26 tumor tissues of mice injected with SL- pClyA or SLΔARA- pClyA, and with or without L-arabinose induction.
In summary, these results provide proof of principle that deletion of the ara operon in highly attenuated S. typhimurium can potentiate the therapeutic efficacy of L-arabinose-induced bacteriotherapy. In a previous study, an inducible cell communication system was introduced into Salmonella to amplify the L-arabinose-induced gene expression in tumor tissue.Citation25 This system uses two genes: LuxI and LuxR. The LuxI protein induced by L-arabinose catalyzes formation of the autoinducer acylhomoserine lactone (AI-1), which freely diffuses across bacterial membranes. The LuxR protein is active in the presence of AI-1 and induces transcription by binding to the PLuxI promoter.Citation26,27 Self-induction creates a feedback amplification loop that increases production of any gene incorporated into the operon. While the cell communication system focused on designing a gene circuit for multi-step amplification, the present study focused on designing a bacterial strain in which the ara operon is deleted, thereby allowing L-arabinose to elicit the maximal possible expression of a target gene. Therefore, many conventional pBAD vectors can be applied to this engineered bacterial system to amplify gene expression. The amplification effects of these 2 systems seem to be comparable. Most importantly, the present bacterial strain transformed with a conventional pBAD plasmid encoding the cytotoxic protein ClyA significantly suppressed tumor growth in tumor-bearing mice. Further studies should be directed toward improving the anti-tumor efficacy of this system, such as testing it in a more diverse range of mouse tumor models and determining the effects of variations in the doses of the engineered bacteria administered and differences in cargo molecules.
Materials and Methods
Cell lines, bacterial strains, and plasmids
Murine CT26 colon carcinoma cells were grown in high-glucose Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 1% penicillin-streptomycin. The bacterial strains and plasmids used in this study are described in . The Δara operon::cat ΔppGpp S. typhimurium (SLΔara) strain was constructed from the ΔppGpp S. typhimurium (SMR2130) (SL) strain by linear DNA transformation, as described previously.Citation28 The expression plasmids pRLuc8 and pClyA have been previously described.Citation10,29
Table 1. Bacterial strains and plasmids used in this study
Deletion of the ara operon
The ara operon was deleted to generate the SLΔara strain by linear DNA transformation, as described previously.Citation28 SL transformants carrying the λ red helper plasmid (pKD-46) were grown in LB medium containing ampicillin (5 μg/ml) and L-arabinose (10 mM) at 30°C to an OD600 of ∼0.6. Thereafter, cells were made electrocompetent by concentrating 100-fold and washing 3 times with ice-cold 10% glycerol. A linear DNA fragment was generated by PCR from the template plasmid. This fragment included an antibiotic-resistance gene and sequences homologous to the regions adjacent to the ara operon that was to be deleted. The primers used are shown in (A and B). The antibiotic-resistance gene was flanked by a FRT (FLP recognition target) site. Electroporation was performed using a Cell-porator with a voltage booster and 0.15 cm chambers, according to the manufacturer's instructions (GIBCO/BRL) using 25 μl of cells and 10-100 ng of PCR product. Shocked cells were added to 1 ml SOC medium and incubated at 37°C for 1 h, and then one-half of the mixture was spread onto agar to select chloramphenicol-resistant transformants. If no colonies grew within 24 h, the remainder of the mixture was spread onto agar after standing overnight at room temperature. After primary selection, mutants were maintained on medium lacking an antibiotic. They were colony-purified once in the absence of antibiotics at 37°C, and then tested for ampicillin sensitivity to evaluate the loss of the helper plasmid. If ampicillin sensitivity was not detected, a few colonies were purified one time at 43°C and similarly tested.
Table 2. Primer sequences used in this study
PCR verification
Four PCRs were used to demonstrate that all mutants had the correct structure. A freshly isolated colony was suspended in 20 μl distilled H2O using a plastic tip. From this solution, a volume of 5 μl was used in 4 separate 20 μl PCRs following preincubation (“hot start”) at 95°C for 2 min. The test primers are shown in (c, d, e, f, and g). Two reactions were performed using primers specific for a nearby locus with the respective common junction primer (P1 and C2 or C1 and P2) to test for both junction fragments (; Fig. S1A). A third reaction was carried out with the flanking locus-specific primers (C3 and P2) to verify simultaneous loss of the parental (non-mutant) fragment (; Fig. S5B). In all cases, control colonies were tested side-by-side. The sequence of the ara operon sowing the precise positions of primers is depicted in .
Luciferase assay
To compare gene expression among the parental strain (SL) and SLΔara, bacteria were transformed with a plasmid harboring RLuc8 under the control of the PBAD promoter (pRLuc8). The transformants (SL- pRLuc8 and SLΔara- pRLuc8) were grown in LB medium containing ampicillin (5 μg/ml) and various concentrations of L-arabinose (0.00002-0.4%). Samples were induced after 1.5 hours of culture and harvested after 4.5 hours of culture. Harvested bacteria were resuspended in 1 ml PBS, diluted to an OD600 of 0.1, and then diluted further. Luminescence was measured using a luminometer (Berthold Technologies, DE/LB 96V) after the substrate coelenterazine (400 ng) was mixed with the bacterial sample. The number of relative light units (RLUs) per 104 colony-forming units (CFU) of bacteria was calculated.
Animal models
Male BALB/c mice (5-6-weeks-old) were purchased from the Orient Company (Seongnam, Korea). All animal experiments were performed in accordance with protocols approved by Chonnam National University Animal Research Committee (CNU IACUC-H-2013-3). Mice were anesthetized using isoflurane (2%) for imaging and measurement of tumors. To generate BALB/c mice with subcutaneous colon adenocarcinoma, CT26 cells were harvested and the number of viable cells was determined using trypan blue. Thereafter, 50 μl of PBS containing 1 × 106 viable CT26 cells was injected subcutaneously into the right lateral thigh of each mouse.
Bioluminescence imaging
Bacteria (3 × 107 CFU) suspended in 100 μl PBS were injected intravenously using a 1-ml, 29 gauge, insulin syringe through the lateral tail vein. To induce Renilla luciferase variant 8, mice were injected daily and intraperitoneally with L-arabinose (either 120 mg or 12 mg in 300 μl PBS) starting 3 d after injection with the bacteria. Bioluminescence imaging was performed on mice at day 7 after injection of the bacteria. To obtain bacterial bioluminescence images, anesthetized animals were placed in a light-tight chamber of the IVIS100 imaging system (Caliper), equipped with a cooled charge-coupled device camera. Photons emitted from luciferase-expressing bacteria were collected and integrated over 1 min periods. Pseudo color images indicating photon counts were overlaid onto photographs of the mice using Living Image software v. 2.25 (Caliper). A region of interest was manually selected based on the signal intensity. The area of the region of interest was kept constant, and maximum intensity was recorded within each region of interest.
Western blot analysis
Bacterial cell lysates and culture medium were quantified using the Bradford protein assay.
Total proteins (40 μg) were separated by electrophoresis and blotted as described previously.Citation15 Protein samples were boiled for 5 min and separated by SDS-PAGE. The separated proteins were transferred electrophoretically to a nitrocellulose membrane. The membrane was stained with Ponseau S staining solution (0.1% (w/v) Ponseau S in 5% (v/v) acetic acid) for 5 min to locate protein bands on Western blots. After washing, the membrane was blocked with 5% skimmed milk and probed with a rabbit anti-ClyA antibody (1:500 dilution) at 4°C overnight. Membranes were then incubated with anti-rabbit IgG antibody (1:2000 dilution) linked to horseradish peroxidase (Sigma-Aldrich, UK) for 1 hour, and bound proteins were visualized using the LAS-3000 imaging system (FUJIFILM).
MTT assay to assess cell viability and proliferation
The MTT assay was performed. Bacteria carrying a plasmid harboring a cytotoxic gene (ClyA) were cultured in 100 ml LB broth and then induced with 0.2% L-arabinose. The culture medium was separated from the bacterial cell pellet by centrifugation at 1,073 g for 4 min. The culture medium was filtered through a 0.2 μm filter, centrifuged (3,000 × g for 5 min), and the 34 kDa ClyA protein was concentrated using a 10 kDa concentrator (Amicon Ultra-15 Centrifugal Filter Unit with Ultracel-10 membrane, Millipore). The amount of concentrated extracellular protein was 12 mg.
CT26 cells (10,000 cells/well) were cultured in a 96-well plate for 18 h. Cells were treated with various dilutions (0.05 mg ∼ 0.2 mg) of the culture media for 6 h, washed, and further incubated in DMEM for 20 h. MTT (10 μl of a 10 mg/ml solution) was added to 200 μl DMEM for 20 min. After washing each well with PBS, cells were incubated with DMSO for 20 min on a shaker. Absorbance was measured at 570 nm (US/SpectraMax 190).
Intravenous injection of bacteria expressing ClyA into tumor-bearing mice
Bacteria were administered (3 × 107 CFU suspended in 100 μl PBS) via the lateral tail vein using a 1-ml, 29 gauge, insulin syringe (Becton Dickinson). PBS or bacteria harboring plasmid pBAD-ClyA was injected intravenously at 10 d (100 mm3 ∼ 150 mm3) after subcutaneous implantation of CT26 cells. To induce ClyA, mice were administered a daily intraperitoneal injection of L-arabinose (120 mg) starting 3 d after injection with the bacteria. Tumor growth was monitored by measuring tumors with a digital caliper. All animals were feed with L- arabinose-free mouse food (AIN-76, Damool Science, Korea) to prevent induction of ClyA from the PBAD promoter.
Statistical analysis
Statistical significance was determined using the Mann-Whitney U-test. The 2-tailed Student's t-test was used to determine statistical significant differences in primary tumor growth between control and treatment groups. A P-value < 0.05 was considered significant for all analyses. All data are expressed as means ± standard deviation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
949527_Supplementary_Materials.zip
Download Zip (1,004.1 KB)Acknowledgments
We thank Misun Yun and Dong-Yeon Kim for technical supports.
Additional information
Funding
References
- Kishimoto H, Aki R, Urata Y, Bouvet M, Momiyama M, Tanaka N, Fujiwara T, Hoffman RM. Tumor-selective, adenoviral-mediated GFP genetic labeling of human cancer in the live mouse reports future recurrence after resection. Cell Cycle 2011; 10:2737-41; PMID:21785265; http://dx.doi.org/10.4161/cc.10.16.16756
- Zhao M, Yang M, Li XM, Jiang P, Baranov E, Li S, Xu M, Penman S, Hoffman RM. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc Natl Acad Sci U S A 2005; 102:755-60; PMID:15644448; http://dx.doi.org/10.1073/pnas.0408422102
- Yu YA, Shabahang S, Timiryasova TM, Zhang Q, Beltz R, Gentschev I, Goebel W, Szalay AA. Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light-emitting proteins. Nat Biotechnol 2004; 22(3):313-20; PMID:14990953; http://dx.doi.org/10.1038/nbt937
- Pawelek JM, Low KB, Bermudes D. Bacteria as tumour-targeting vectors. Lancet Oncol 2003; 4(11):658; http://dx.doi.org/10.1016/S1470-2045(03)01194-X
- Min JJ, Nguyen VH, Kim HJ, Hong Y, Choy HE. Quantitative bioluminescence imaging of tumor-targeting bacteria in living animals. Nat Protoc 2008; 3(4):629-36; PMID:18388945; http://dx.doi.org/10.1038/nprot.2008.32
- Min JJ, Kim HJ, Park JH, Moon S, Jeong JH, Hong YJ, Cho KO, Nam JH, Kim N, Park YK, et al.. Noninvasive real-time imaging of tumors and metastases using tumor-targeting light-emitting Escherichia coli. Mol Imaging Biol 2008; 10(1):54-61; PMID:17994265; http://dx.doi.org/10.1007/s11307-007-0120-5
- Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer 2010; 10(11):785-94; PMID:20944664; http://dx.doi.org/10.1038/nrc2934
- Ganai S, Arenas RB, Sauer JP, Bentley B, Forbes NS. In tumors Salmonella migrate away from vasculature toward the transition zone and induce apoptosis. Cancer Gene Ther 2011; 18(7):457-66; PMID:21436868; http://dx.doi.org/10.1038/cgt.2011.10
- Seow Y, Wood MJ. Biological gene delivery vehicles: beyond viral vectors. Mol Ther 2009; 17(5):767-77; PMID:19277019; http://dx.doi.org/10.1038/mt.2009.41
- Nguyen VH, Kim HS, Ha JM, Hong Y, Choy HE, Min JJ. Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Cancer Res 2010; 70(1):18-23; PMID:20028866; http://dx.doi.org/10.1158/0008-5472.CAN-09-3453
- Jiang SN, Phan TX, Nam TK, Nguyen VH, Kim HS, Bom HS, Choy HE, Hong Y, Min JJ. Inhibition of tumor growth and metastasis by a combination of Escherichia coli-mediated cytolytic therapy and radiotherapy. Mol Ther 2010; 18(3):635-42; PMID:20051939; http://dx.doi.org/10.1038/mt.2009.295
- Zhao M, Suetsugu A, Ma H, Zhang L, Liu F, Zhang Y, Tran B, Hoffman RM. Efficacy against lung metastasis with a tumor-targeting mutant of Salmonella typhimurium in immunocompetent mice. Cell Cycle 2012; 11:187-93; PMID:22186786; http://dx.doi.org/10.4161/cc.11.1.18667
- Momiyama M, Zhao M, Kimura H, Tran B, Chishima T, Bouvet M, Endo I, Hoffman RM. Inhibition and eradication of human glioma with tumor-targeting Salmonella typhimurium in an orthotopic nude-mouse model. Cell Cycle 2012; 11:628-32; PMID:22274398; http://dx.doi.org/10.4161/cc.11.3.19116
- Chorobik P, Marcinkiewicz J. Therapeutic vaccines based on genetically modified Salmonella: a novel strategy in cancer immunotherapy. Pol Arch Med Wewn 2011; 121(12):461-6; PMID:22129784
- Song M, Kim HJ, Kim EY, Shin M, Lee HC, Hong Y, Rhee JH, Yoon H, Ryu S, Lim S, et al. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J Biol Chem 2004; 279(33):34183-90; PMID:15161921; http://dx.doi.org/10.1074/jbc.M313491200
- L Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 1995; 177(14):4121-30; PMID:7608087
- Loessner H, Endmann A, Leschner S, Westphal K, Rohde M, Miloud T, Hämmerling G, Neuhaus K, Weiss S. Remote control of tumour-targeted Salmonella enterica serovar Typhimurium by the use of L-arabinose as inducer of bacterial gene expression in vivo. Cell Microbiol 2007; 9(6):1529-37; PMID:17298393; http://dx.doi.org/10.1111/j.1462-5822.2007.00890.x
- Ogden S, Haggerty D, Stoner CM, Kolodrubetz D, Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A 1980; 77(6):3346-50; PMID:6251457; http://dx.doi.org/10.1073/pnas.77.6.3346
- Soisson SM, MacDougall-Shackleton B, Schleif R, Wolberger C. Structural basis for ligand-regulated oligomerization of AraC. Science 1997; 276(5311):421-5; PMID:9103202; http://dx.doi.org/10.1126/science.276.5311.421
- Schleif R. AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol Rev 2010; 34(5):779-96; PMID:20491933
- Lee N, Francklyn C, Hamilton EP. Arabinose-induced binding of AraC protein to araI2 activates the araBAD operon promoter. Proc Natl Acad Sci U S A 1987; 84(24):8814-8; PMID:2962192; http://dx.doi.org/10.1073/pnas.84.24.8814
- Miyada CG, Stoltzfus L, Wilcox G. Regulation of the araC gene of Escherichia coli: catabolite repression, autoregulation, and effect on araBAD expression. Proc Natl Acad Sci U S A 1984; 81(13):4120-4; PMID:6377308; http://dx.doi.org/10.1073/pnas.81.13.4120
- Doyle ME, Brown C, Hogg RW, Helling RB. Induction of the ara operon of Escherichia coli B-r. J Bacteriol 1972; 110(1):56-65; PMID:4553005
- Wai SN, Lindmark B, Soderblom T, Takade A, Westermark M, Oscarsson J, Jass J, Richter-Dahlfors A, Mizunoe Y, Uhlin BE. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 2003; 115:25-35; PMID:14532000; http://dx.doi.org/10.1016/S0092-8674(03)00754-2
- Dai Y, Toley BJ, Swofford CA, Forbes NS. Construction of an inducible cell-communication system that amplifies Salmonella gene expression in tumor tissue. Biotechnol Bioeng 2013; 110(6):1769-81; PMID:23280328; http://dx.doi.org/10.1002/bit.24816
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol 2001; 55:165-99; PMID:11544353; http://dx.doi.org/10.1146/annurev.micro.55.1.165
- Li L, Hooi D, Chhabra SR, Pritchard D, Shaw PE. Bacterial N-acylhomoserine lactone-induced apoptosis in breast carcinoma cells correlated with down-modulation of STAT3. Oncogene 2004; 23:4894-902; PMID:15064716; http://dx.doi.org/10.1038/sj.onc.1207612
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 2000; 97(12):6640-5; PMID:10829079; http://dx.doi.org/10.1073/pnas.120163297
- Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng Des Sel 2006; 19(9):391-400; PMID:16857694; http://dx.doi.org/10.1093/protein/gzl023
