Abstract
Defective DNA damage response (DDR) is frequently associated with carcinogenesis. Abrogation of DDR leads to chromosomal instability, a most common characteristic of tumors. However, the molecular mechanisms underlying regulation of DDR are still elusive. The ubiquitin ligase RNF8 mediates the ubiquitination of γH2AX and recruits 53BP1 and BRCA1 to DNA damage sites which promotes DDR and inhibits chromosomal instability. Though RNF8 is a key player involved in DDR, regulation of its expression is still poorly understood. Here, we show that miR-214 could abrogate DDR by repressing RNF8 expression through direct binding to 3′-untranslated region (3′ UTR) of RNF8 mRNA in human ovarian cancer cells. Antagonizing miR-214 by expressing its inhibitors in A2780 cells significantly increased RNF8 expression and thus promoted DNA damage repair. Consistent with the role of miR-214 in regulating RNF8 expression, the impaired DNA repair induced by miR-214 overexpression can be rescued by overexpressing RNF8 mRNA lacking the 3′ UTR. Together, our results indicate that down-regulation of RNF8 mediated by miR-214 impedes DNA damage response to induce chromosomal instability in ovarian cancers, which may facilitate the understanding of mechanisms underlying chromosomal instability.
Abbreviations
| DDR | = | DNA damage response |
| CIN | = | Chromosomal instability |
| 3′ UTR | = | 3′untranslated region |
| miRNAs | = | microRNAs |
Introduction
Chromosomal instability (CIN) are gross chromosomal abnormalities involving gain or loss of whole or fractions of chromosomes that can lead to tumor formation.Citation1 To ensure genomic integrity, cells apply several mechanisms including the DDR pathway.Citation2 RNF8, a RING-finger E3 ubiquitin ligase, ubiquitylate histone H2A and H2AX, which mediates the recruitments of 53BP1 and BRCA1 at sites of DNA damage to promote the transduction of DDR.Citation4,5 Decreased expression of RNF8 can significantly block DDR and induce CIN. Many types of cancers with specific defects in the DNA damage response (DDR) have the characteristic of genomic instability.Citation6 RNF8−/− mice exhibit increased genomic instability and have an elevated risk of tumorigenesis, indicating that RNF8 is a tumor suppressor gene.Citation7 Consistent with this finding, it has been observed that RNF8 expression is lower in many types of cancer cell lines than in normal cell lines.Citation8 Although the regulations and functions of many genes in DDR have been studied, the mechanism by which RNF8 expression is regulated remains unknown.
MicroRNAs (miRNAs) are small noncoding RNAs (20–22 nucleotides), which can cause mRNA degradation or a post-transcriptional repression of target genes by complementary binding to their 3′ UTR.Citation9 Several genes involved in DDR are regulated by specific miRNAs, such as BRCA1,Citation10 BRCA2,Citation11 Rad51,Citation12,13 NBS1,Citation14 H2AX,Citation15,16 SNF2H,Citation17 ATMCitation18 and DNA-PKcs.Citation18 RNF8 mRNA has less than 2 K bp of coding sequence, with about 4 K bp of 3′ UTR sequence. This suggests a high possibility that specific miRNAs could directly suppress RNF8 expression by targeting its long 3′ UTR leading to chromosomal instability.
In ovarian cancer, gross chromosomal instability such as structural rearrangements are frequently reported.Citation19,20 Germline-associated BRCA1 or BRCA2 mutations can only be found in a small proportion of ovarian cancers.Citation21-24 Even loss of BRCA1 or BRCA2 function due to genetic or epigenetic processes only accounted for a small proportion of ovarian cancers.Citation25 Somatic mutations or epigenetic alterations in genes for chromosomal instability need to be further discovered. Over-expression of miR-214 has been reported in many types of cancers, such as melanoma,Citation26 pancreatic cancer,Citation27 lung cancer,Citation28 oral carcinoma,Citation29 gastric cancerCitation30 and especially in ovarian cancer.Citation31-34 It has been documented that both miR-214 overexpressionCitation34,35 and chromosomal instabilityCitation36-39 are individually associated with the development of ovarian cancer. Here, for the first time we investigated the relationship between miR-214 overexpression and chromosomal instability in ovarian cell lines and are reporting that miR-214 negatively regulates RNF8 expression and can induce chromosomal instability leading to ovarian cancer.
Results
MiR-214 overexpression increases CIN after ionizing radiation (IR) treatment
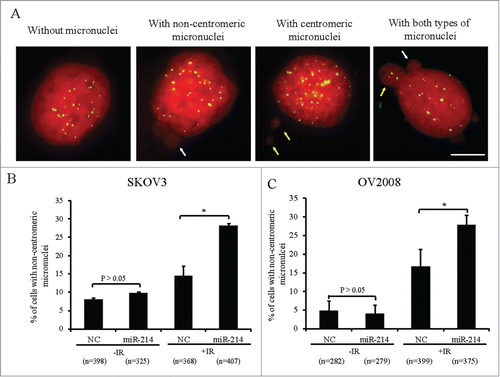
To find potential miRNAs that can directly target human RNF8, 3 online computational softwares: Targetscan (http://www.targetscan.org), Miranda (http://www.microrna.org) and miRDB (http://mirdb.org/miRDB) were used to perform an “in silico” search for putative miRNA binding-sites in the 3′ UTR of RNF8. Only miR-214 was predicted by all 3 software packages (Fig. S1A) and it was found to be evolutionarily conserved throughout the vertebrates (Fig. S1B). The Cancer Genome Atlas (TCGA) database was used to explore further, the relationship between miR-214 and the expression of RNF8 mRNA. It was observed that miR-214 expression was inversely correlated with RNF8 mRNA expression in grade 1 and 2 ovarian cancer (Fig. S2). Next, we examined the relative miR-214 and RNF8 mRNA levels in several ovarian cancer cell lines. Cell lines with relatively lower endogenous miR-214 (SKOV3, OV2008) showed higher concentrations of RNF8 mRNA and those with higher endogenous miR-214 (ES-2, A2780) had lower levels of RNF8 mRNA (Fig. S3). To study whether miR-214 overexpression can disturb DDR to induce chromosomal instability, miR-214 mimics were transfected into SKOV3 and OV2008 cell lines. About 66 h after transfection, cells were exposed to IR and then the micronucleus frequency was analyzed 20 h post IR treatment. The results showed that miR-214 overexpression in OV2008 and SKOV3 cells did not significantly increase the percentage of cells with non-centromeric micronuclei as compared with control cells without IR treatment, but this frequency was significantly increased after IR treatment in both cell lines (). The other types of cells with micronuclei in OV2008 and SKOV3 cell lines did not showed a significant increase either with or without IR treatment (Supplementary Table). These results indicated that miR-214 overexpression can increase CIN after IR treatment, suggesting that miR-214 may disturb some genes involved in DDR. As RNF8 was a predicted target for miR-214, we focused on this interaction.
Figure 1. MiR-214 increases DNA damage-induced chromosomal instability. Cells were treated with 5 Gy IR 66 h after transfection and micronuclei were detected 20 h later. (A) Four types of SKOV3 cells were observed on the basis of micronuclei: without micronucleus, with centromeric micronucleus, with non-centromeric micronucleus and with both types of micronuclei. Green, Human centromeres; Red, DNA; Bars = 10 μm. Non-centromeric (white arrows) or centromeric (yellow arrows) micronucleus. (B and C) The percentage of cells with non-centromeric micronuclei in SKOV3 (B) or OV2008 (C) cells. n = the number of cells counted. Mean ± SD, from two independent experiments. *P < 0.05, chi-square test.
MiR-214 directly down regulates RNF8 expression by binding to 3′ UTR of its mRNA
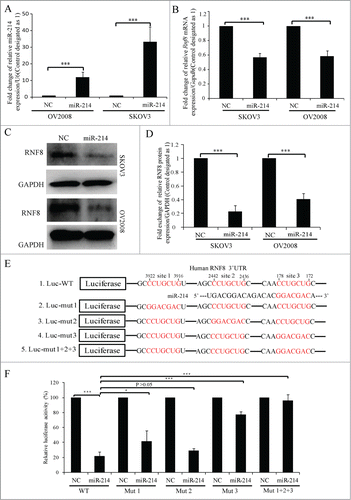
To test whether miR-214 can regulate RNF8 expression, miR-214 mimics were overexpressed in SKOV3 and OV2008 cells, then Western blotting or real-time PCR were performed at 48 or 72 h post transfection respectively. The results indicated that miR-214 had highest expression 48 h post transfection, while the mRNA and protein levels of RNF8 were significantly reduced (). Computational analysis suggested that there are 3 putative binding sites for miR-214 in the 3′ UTR of human RNF8 mRNA. Every site is the 7-mer-m8 seed CCAGCAG, which means Watson-Crick base-pairing from positions 2 to 8 at positions 172–178, 2436–2442 and 3916–3922 from the start of the 3′ UTR (). To examine whether miR-214 directly binds to the RNF8 3′ UTR at these putative binding sites, a dual-luciferase reporter vector was constructed by inserting the wild-type 3′ UTR of human RNF8 gene into the 3′ end of the renilla luciferase gene. Human kidney 293T cells were transfected with this vector together with either miR-214 or negative control (NC) oligonucleotides. These results showed that the luciferase activity was greatly repressed by miR-214 compared to NC at 24 h post transfection (). We prepared equivalent luciferase constructs with mutations in individual putative binding sites as well as in all 3 binding sites. Mutation in each putative binding site had a different influence on luciferase activity. Mutation in site 2 had no significant effect on luciferase activity, while mutation in sites 1 or 3 significantly reduced the suppression by miR-214. Mutation in all 3 putative sites further decreased this suppression (). Taken together, these results confirm that RNF8 is the target of miR-214.
Figure 2. MiR-214 directly regulates RNF8 expression. Relative expression levels of miR-214 (A), RNF8 mRNAs (B) or protein (C and D) were detected at 48 h or 72 h post transfection by real-time PCR or Western blotting, respectively. (E) Mature miR-214 sequence and its putative binding sites in the 3′ UTR of human RNF8 mRNA. Dual luciferase reporter constructs contain a DNA sequence encoding wild-type (E1) or mutant RNF8 mRNA 3′ UTR for each individual (E2-4) or all (E5) putative binding sites, respectively. Mean ± SD, from 3 independent experiments. (F) Relative luciferase activity was measured 24 h after co-transfection of each luciferase vectors (100 ng) together with miR-214 mimics (50 nM) or controls in 293T cells. *P < 0.05, ***P < 0.001, 2-tailed t-test (B and D), chi-square test (F).
MiR-214 disturbs DNA damage response via downregulation of RNF8
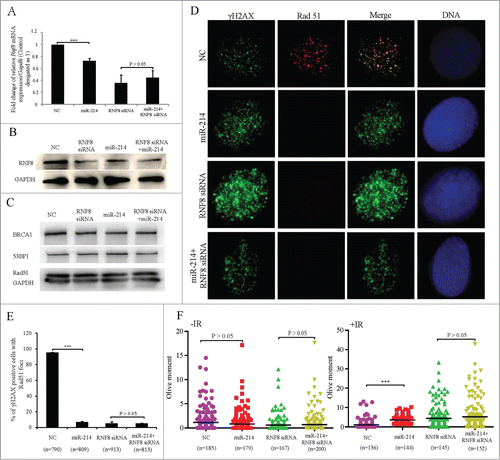
To study whether miR-214 can increase the IR-induced chromosomal instability by down regulation of RNF8, SKOV3 cells were transfected with miR-214 mimics or RNF8 siRNA individually or together in a combination. We found that miR-214 did not change the protein level of 53BP1, BRCA1 and Rad51, while RNF8 expression at mRNA and protein levels was significantly decreased (). Furthermore, co-transfection of miR-214 mimics and RNF8 siRNA did not further alter the expression of 53BP1, BRCA1, Rad51 and RNF8 compared with transfection of RNF8 siRNA in SKOV3 cells. However, miR-214 inhibited the recruitment of 53BP1, BRCA1 and Rad51 to DNA damage sites after IR treatment (, Fig. S4). Co-transfection of miR-214 mimics together with RNF8 siRNA did not further disturb the recruitment of these proteins (). A neutral comet assay showed that miR-214 mimic-transfected cells displayed significantly higher residual level of DNA damage as compared to the control cells 2 h after IR-treatment, and there was no significant difference in the presence of DNA damage in cells between co-transfection of miR-214 with RNF8 siRNA and RNF8 siRNA alone ().
Figure 3. MiR-214 disturbs DNA damage response by downregulating RNF8 expression. A hundred nM MiR-214 mimics or RNF8 siRNAs were transfected individually or together into SKOV3 cells. Forty 8 or 72 h later, relative RNF8 mRNA (A) or protein (B) expression was detected by real-time PCR or Western blotting. The expression of downstream proteins 53BP1, BRCA1 or Rad51 in DNA damage response was also detected by Western blotting (C). Cells were treated with 5 Gy IR 72 h after transfection and then fixed for immunofluorescent staining for RAD51 and γH2AX. Representative images (D) and quantitative results (E) of RAD51 and γH2AX staining. Green,γH2AX; RAD51, Red; Blue, DNA; Bars = 10 μm. One-2 hours following treatment of cells with 5 Gy IR 72 h after transfection, neutral comet assay was performed to detect DNA damage. (F) Quantitative results were calculated by olive moment. n = the number of cells counted. ***P < 0.001, chi-square test (D), two-tailed t-test (A and F).
Inhibition of miR-214 facilitates IR-induced DNA damage repair
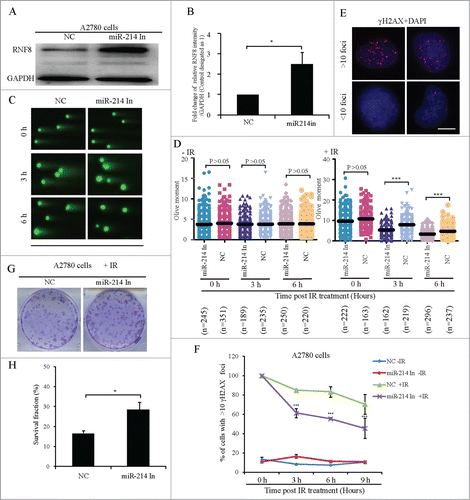
Our next focus was whether inhibition of miR-214 in A2780 cells, which had the high level of endogenous miR-214 expression as compared to the other ovarian cancer cell lines (Fig. S3), can rescue RNF8 expression and facilitate DNA damage repair or not. Our results revealed that overexpression of miR-214 inhibitor significantly increased the expression of RNF8 protein (). To determine whether inhibition of endogenous miR-214 can affect DNA repair or not, the persistence of DNA damage was measured by neutral comet assay 2 h after 5 Gy IR treatment. Results indicated that inhibition of endogenous miR-214 significantly facilitated DNA repair after IR treatment (). We also analyzed the percentage of cells with >10 γH2AX foci in a time course after 5 Gy IR treatment in both control and miR-214-inhibitor overexpressing cells. We observed that the percentage of cells with >10 γH2AX foci deceased more rapidly in miR-214-inhibitor overexpressing cells than controls (). Inhibition of endogenous miR-214 also facilitated clone formation after 5 Gy IR treatment (), while in treatment without IR there was no significant change clone formation (Fig. S5). These results indicated that inhibition of endogenous miR-214 results in increased RNF8 expression which facilitates DNA damage repair.
Figure 4. Inhibition of endogenous miR-214 increases RNF8-mediated DNA damage response. Representative image (A) and quantitation (B) of RNF8 expression at 72 h after miR-214 inhibitor transfection detected by Western blotting. (C) At 72 h after transfection, neutral comet assay was performed to detect DNA damage and representative images were shown, Bars = 20 μm. (D) Quantitative results were calculated by olive moment. n = the number of cells counted. Seventy-2 hours post transfection, the percentage of cells with >10 γH2AX foci were analyzed at different time points after IR treatment. (E) Representative images of γH2AX staining, Bars = 10 μm and (F) quantitative results of γH2AX staining. More than 100 cells were analyzed for each category. Seventy-2 hours post transfection, 1 × 103 A2780 cells transfected with miR-214 inhibitors or controls were treated with 5 Gy IR and followed by clone formation assay, respectively. Representative image (G) and quantitative analysis (H) of clone formation assay 7 days after IR. A2780 cells were transfected with or without miR-214 inhibitors for 72 h and followed by 5 Gy IR treatment. *P < 0.05, ***P < 0.001, chi-square test (F and H), two-tailed t-test (B and D). Mean ± SD, from two independent experiments.
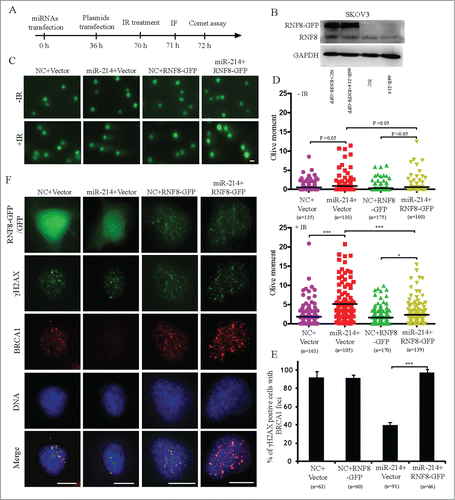
Overexpression of RNF8 mRNA lacking 3′ UTR rescues the disturbance of DNA repair induced by miR-214
To further validate whether miR-214 disturbs the ability of DNA damage repair through downregulation of RNF8, we tested whether overexpression of RNF8 without 3′ UTR can rescue the ability of DNA repair after miR-214 overexpression. miR-214 mimics or negative control were transfected respectively; at 36 h after the first transfection, RNF8-GFP cDNA (lacking the 3′ UTR with putative miR-214 binding sites) or backbone vector were subsequently transfected. Cells were irradiated 34 hours after the final transfection and comet assays or immunofluorescence staining was performed one or two hours after irradiation to measure DNA damage (). The results indicated that overexpression of RNF8 without 3′ UTR significantly reduced IR-induced DNA damage (). The results of immunofluorescent staining revealed that after IR treatment, the 53BP1, BRCA1 and Rad51 foci formation was significantly rescued in RNF8-GFP expressing cells (, Fig. S6). Taken together, these results indicated that miR-214-mediated RNF8 downregulation disturbs DNA damage response by binding at 3′ UTR of RNF8 mRNA.
Figure 5. Overexpression of RNF8 mRNAs without 3′ UTR can antagonize the effects of miR-214 on DNA damage response. (A) Experimental scheme for cell transfection. Thirty-6 hours after transfection either with negative control or miR-214 mimics, SKOV3 cells were transfected with plasmids with or without RNF8-GFP cDNA lacking the 3′ UTR. Western blotting was performed 36 h later after plasmids transfection to detect RNF8 expression (B). Cells were then treated with 5 Gy IR and followed by recovery for about 1 or 2 h followed by immunofluorescent staining or neutral comet assay to detect DNA damage. Representative images (C) and Quantitative analysis (D) of comet assay. n = the number of cells counted. Bars = 20 μm. (E) Representative images and (F) quantitative results of RAD51 and γH2AX staining in various experimental treatments. Bars = 10 μm, Mean ± SD, from two independent experiments. *P < 0.05, ***P < 0.001, 2-tailed t-test (D) and chi-square test (F).
Discussion
Many cancers are characterized by CIN.Citation1 The DNA damage response acts as a barrier to inhibit CIN by sensing DNA lesions, signal transduction and promoting damage repair which is commonly defective in many cancers.Citation2,6 In this process, mutations in several genes such as ATM,Citation40,41 BRCA1,Citation21,22,42 BRCA2,Citation21,42 or NBS1Citation43 predispose to carcinogenesis. Apart from mutations, many recent studies have suggested that repair is also impaired by overexpression of several specific miRNAs in different cancers; for example, in breast cancer, low BRCA2 expression is induced by overexpressed miR-1245.Citation11 Similarly down regulation of BRCA1 by overexpression of miR-182 is observed in ovarian cancerCitation10 and decreased NBS1 expression by miR-629 overexpression has been documented in lung cancer.Citation14 miR-214 was also found up-regulated in human pancreatic cancerCitation27 or gastric cancer,Citation30 melanoma,Citation26 lung cancer,Citation28 pediatric osteosarcomaCitation48 and oral carcinoma,Citation29 and therefore has been considered to be an oncogene. In the present study, we have found that miR-214 overexpression can lead to CIN by disturbing RNF8-mediated DNA damage response in ovarian cancer.
Gross chromosomal instability such as structural rearrangements are frequently identified in ovarian cancer.Citation19,20 However, the underlying mechanism leading to CIN in ovarian cancer is still poorly understood except that germ line associated BRCA1 or BRCA2 mutations can cause a variety of cancer-prone chromosomal instability syndromes.Citation21,22 Recently, miR-214 overexpression has been reported in many ovarian cancer tissues.Citation31-33 The magnitude of miR-214 overexpression correlates with ovarian cancer progressionCitation34,35 and CIN is considered to be an early event that drives this progression.Citation1,44,45 Here for the first time we have studied the role of miR-214 in CIN of ovarian cancer and found that chromosomal integrity is significantly disturbed by miR-214 overexpression after IR treatment, suggesting that miR-214 is associated with DDR in ovarian cancer cell lines ().
RNF8 is a RING-finger E3 ubiquitin ligase and it is a key protein in DDR that mediates ubiquitin conjugation to help the recruitment of 53BP1 and BRCA1 to the sites of DNA lesions. Here for the first time we have reported that miR-214 inhibited RNF8 expression and disturbed DDR by impeding recruitment of 53BP1, Rad51 and BRCA1 at DNA damage sites ( and ). MiR-214 reduces RNF8 levels by directly binding to 3 seeds in 3′ UTR of human RNF8 (). Our results indicated that miR-214 overexpression causes CIN by disturbing DNA damage response in ovarian cancer cell lines. Additionally, miR-214 overexpression also facilitated the stemness of ovarian cancer stem cells,Citation46 suggesting that miR-214 contributes to ovarian carcinogenesis through more than one mechanism.
Most prevalent radiotherapy or chemotherapy for anticancer relies on the generation of DNA damage, because most cancer cells are DDR-impaired and rapidly proliferating as compared to normal cells.Citation2,6 The inhibition of DDR could increase the effectiveness of radiotherapy or chemotherapy.Citation6 In previous reports, miR-214 has been reported to induce sensitivity against DNA-damage agent cisplatin in cervical cancer cellsCitation49 and inhibit cancer cells growth in hepatocellular carcinoma,Citation50 breast cancerCitation51 and oesophageal squamous cell carcinoma.Citation52 Furthermore, miR-214 also inhibits cancer metastasis in intrahepatic cholangiocarcinoma.Citation53 These analyses implied that overexpression miR-214 in cancer cells might benefit cancer therapy. However, other reports showed that miR-214 has the negative effects on cancer therapy. MiR-214 can increase its ability of radio-resistance in non-small-cell lung cancerCitation54 and facilitate cell proliferation in nasopharyngeal carcinoma.Citation55 Several other studies have shown that miR-214 also contributes to metastasis in melanomaCitation26 or gastric cancer.Citation56 Furthermore, in cisplatin-resistant ovarian cancers, miR-214 can increase cell survival after cisplatin treatment. The contradicting effects of miR-214 on cancer therapy need more detailed studies to find exact relationships to cancer development. In the current study, we found that miR-214 disturbs DNA damage response through RNF8 down regulation, which can induce gross CIN. CIN can randomly activate large amounts of oncogenes and ensue DNA replication stress leading to cancer heterogeneityCitation6,57 which may be the underlying reason for these differing miR-214 phenotypes in different types of cancers. Resistance to DDR-associated cancer therapy can be due to many factors and miR-214 is one of them. Our results may broaden the knowledge of cancer detection and therapy. Detection of carcinogenesis can be improved by analysis of miR-214 expression, while the management of radio- or drug- resistant cancers needs more clinical studies on DNA damage response in different cancers.
Materials and Methods
Cell culture
Human SKOV3 cell line was gifted by Professor Dr. Jing Liu (University of Science and Technology of China; City Hefei), human OV2008, ES-2 and A2780 cell lines were gifted by Professor Dr. HengYu Fan (Zhe Jiang University, City Hangzhou, China) and 293T cell line was purchased from ATCC. These cell lines were cultured in Dulbecco´s modified Eagle´s medium (Gibco 12800 -017) supplemented with 10% fetal bovine serum (HyClone SV30087), 100 U/ml penicillin and 100 U/ml streptomycin (Gibco 15140-122).
Cell transfection
RNF8 specific siRNA (5′-AGAAUGAGCUCCAAUGUAUUUTT -3′) was used as previously reported.Citation58 Transfection of RNF8 siRNA, miRNA mimics or inhibitors and miRNA mimic negative control or siRNA negative control (100 nM, Genepharma, Shanghai, China) were carried out using Metafectene (Biontex) according to the manufacturers´ protocols. MiR-214 inhibitors are chemically modified, single stranded nucleic acids designed to specifically bind and to inhibit endogenous miR-214 (Genepharma, Shanghai, China). SiRNA and miRNA mimics negative control (5′-TTCTCCGAACGTGTCACG-3′, Genepharma) or miR-214 inhibitor negative control (5′-CAGUACUUUUGUGUAGUACAA-3′, Genepharma) are random double- or single-stranded sequence molecules which can mimic and were validated not to produce identifiable effects on the known RNAs expression. 400 ng/ml RNF8-GFP or backbone plasmids was transfected by lipofectamine 2000 transfection reagent, according to the manufacturers´ protocols.
Construction of luciferase reporter vectors
The human wild type (WT) RNF8 3′ UTR sequences (1–3980 nt from the start of 3′ UTR) containing the 3 putative miR-214 binding sites were cloned in psiCHECKTM-2 luciferase reporter vector (Promega) using the following set of primers: forward primer (F) 5′-AATCTCGAGAGACCGTGCTCTAAGGGCATT TGAA-3′ and reverse primer (R) 5′-AGTGCGGCCGCAACTATTGCTGAATTGAA TTTATT-3′. These two primers contain Xho I and Not I recognition sites at the 5′end of the primers, respectively. The sequence encoding the 3′ UTR of mutant RNF8 mRNA that lacks the putative miR-214 binding site, was synthesized by PCR. The detail of these methods can be found elsewhere.Citation59 For mutation of each putative binding site, the following pairs of primers were employed with the pairs of primers used for amplification of wild-type 3′ UTR: mut1, 5′-GTGCCACCGGTCGCCTC TCGGTAGTAACCGAC-3′; mut2, 5′-AAACTTAGAACCGCCTGCTCAATCAT TGACACA-3′; mut3, 5′-CACTTATGAAGTTCCTGCTCACTAAACCCCGGC-3′. The enzymes for cDNA amplification were purchased by PrimeSTAR (Takara, R044A). The sequences of inserted DNA fragments were verified by DNA sequencing.
Luciferase reporter transfection and dual luciferase assay
5 × 104 HEK 293T cells were plated in a 24-well cell culture plate a day before transfection. In 3′ UTR luciferase reporter assay, the luciferase reporter constructs (100 ng), together with miR-214 mimics (50 nM) (GenePharma) were incubated with 1.5 μl of Lipofectamine 2000. Cells were then transfected using Lipofectamine 2000 (Invitrogen, 11668) according to the manufacturer´s instructions. Twenty-four hours later, cell lysates were harvested and each reporter activity was measured by Dual Luciferase Assay (Promega E1910) as previously described.Citation60 The relative Renilla luciferase activity was normalized to that of the firefly luciferase.
Real-time PCR
Total RNAs were extracted using Trizol reagent (Takara) and reverse-transcribed using the the prime Script™ 1st Strand cDNA Synthesis Kit (Tiangen, Beijing, China). To detect the relative level of RNF8 mRNA, GAPDH mRNA was used as internal normalization control. The following primers were used: RNF8 Forward (F), CAGG CTCTGCAGGAGCATTGGG; Reverse (R) GTGGGCACAGTTCAAGGTGACA, GCGAPDH (F) GTCAAGGCTGAGAACGGGAA; and (R) AAATGAGCCCCA GCCTTCTC. To detect the relative level of miR-214 expression, U6 was used as internal normalization control. microRNAs were reverse transcribed to cDNAs using miR-214 specific reversal primer (F) GTCGTATCCAGTGCGTGTCGTGGAG TCGGCAATTGCACTGGATACGACACTGCCT and U6 specific reversal primer CGCTTCACGAATTTGCGTGTCAT. The real-time PCR for miRNA was performed using the follow primers. miR-214 (F) AGGACAGCAGGCACAGAC; (R) CAGT GCGTGTCGTGGAGT; U6 (F) GCTTCGGCAGCACATATACTAAAAT and (R) CGCTTCACGAATTTGCGTGTCAT. All real-time PCRs were performed by Applied Biosystems StepOne™ real-time PCR System (Applied Bio systems, USA) using Faststart universal SYBR Green Master (Roche), according to our previous report.Citation61
Clone formation assay
5 × 104 A2780 cells were seeded in 24-well plate and 24 h after transfection, 1000 or 150 cells were seeded in each 6-well plate and treated with or without 5 Gy IR at 72 h after transfection, respectively. The detail protocol for clone formation assay was performed following previous report.Citation62
Western blotting analysis
SDS-PAGE was performed with whole-cell extracts as our previously described.Citation37 Primary antibodies included mouse anti-GAPDH (MAB374, 1:1000; Millipore) and BRCA1 (D-9,1:200; Santa Cruz), and rabbit anti-53BP1 (NB100–926, 1:500; Novus Biologicals), RNF8 (H-300, 1:100; Santa Cruz), RAD51 (H-92, 1:200; Santa Cruz). Secondary primary antibodies were alkaline phosphatase (AP)-conjugated anti-mouse IgG (H&L) or anti-Rabbit IgG(Fc) (S3721 or S3731, 1:500; Promega). The protein levels were visualized with a Lumi-Phos kit (Thermo, 34150), and band intensity on Western blotting was quantified by Image-Pro Plus software 6.0 and normalized to GAPDH.
Immunofluorescence staining
Immunofluorescence staining was performed as our previous described.Citation63 Cells grown on coverslips were permeabilized with PBS containing 0.25% Triton X-100 for 5 min. The primary antibodies included rabbit anti-53BP1 (NB100–305, 1:200; Novus Biologicals), Mouse anti-γ-H2AX (05–636,1:250; Millipore), rabbit anti-Rad51 (H-92, 1:200; Santa Cruz), goat anti-Brca1 (M20, 1:200; Santa Cruz) and human auto-antibody against centromere antibody (HCT-0100, 1:100; Immunovision). Secondary antibodies as goat anti-rabbit IgG or anti-mouse IgG conjugated with Alexa Fluor 647 or Alexa Fluor 568 (Invitrogen), donkey anti-goat IgG conjugated with Dylight 649 (Jackson ImmunoResearch) or with Alexa Fluor 555 (Invitrogen),or donkey anti-rabbit IgG or anti-human IgG or sheep anti-mouse conjugated with AMCA (Jakson ImmunoResearch) were diluted 1:100 for immunofluorescence staining assay. DNA was stained by Propidium Iodine (537059, 20ug/ml; calbiochem), and all slides were examined by Olympus BX-61 fluorescence microscope.
Statistical Analysis
Differences among various treatments were analyzed for statistical significance using chi-square or two-tailed t-test by using Microsoft Office Excel 2007 or Graphpad 5 software, respectively. All quantitative data presented are the Mean ± SD from at least 2 independent experiments. The correlation between miR-214 expression and RNF8 mRNA was analyzed by The Pearson's product-moment correlation coefficient (Pearson's r) using SPSS version 12.0 software. Significance level was set at P < 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
958413_Supplementary_Materials.zip
Download Zip (1.3 MB)Acknowledgments
We thank Professor Dr. Jing Liu (University of Science and Technology of China, China) for the gift of human SKOV3 cell line, and Professor Dr. Hengyu Fan (ZheJiang University, China) for the gift of human OV2008, ES-2 and A2780 cell lines. We also thank Professor Dr. Stephen P. Jackson (University of Cambridge, UK) for the gift of RNF8-GFP plasmid.
Funding
This work was supported by grants from National Basic Research Program of China (2013CB947900 and 2012CB944402), National Natural Science Foundation of China (Grant 30900794), and Doctoral Fund of Ministry of Education of China (20123402130004).
Supplemental Materials
Supplemental data for this article can be found on the publisher's website.
References
- Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining 'chromosomal instability'. Trends Genet 2008; 24:64-9; PMID:18192061; http://dx.doi.org/10.1016/j.tig.2007.11.006
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009; 461:1071-8; PMID:19847258; http://dx.doi.org/10.1038/nature08467
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007; 131:887-900; PMID:18001824; http://dx.doi.org/10.1016/j.cell.2007.09.040
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 2007; 318:1637-40; PMID:18006705; http://dx.doi.org/10.1126/science.1150034
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 2007; 131:901-14; PMID:18001825; http://dx.doi.org/10.1016/j.cell.2007.09.041
- Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nature Rev Cancer 2012; 12:587-98; PMID:22918414; http://dx.doi.org/10.1038/nrc3342
- Li L, Halaby MJ, Hakem A, Cardoso R, El Ghamrasni S, Harding S, Chan N, Bristow R, Sanchez O, Durocher D, et al. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J Exp Med 2010; 207:983-97; PMID:20385750; http://dx.doi.org/10.1084/jem.20092437
- Yoshioka T, Kimura M, Saio M, Era S, Okano Y. Plk1 is negatively regulated by RNF8. Biochem Biophys Res Commun 2011; 410:57-61; PMID:21635870; http://dx.doi.org/10.1016/j.bbrc.2011.05.104
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 2013; 14:475-88; PMID:23800994; http://dx.doi.org/10.1038/nrm3611
- Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell 2011; 41:210-20; PMID:21195000; http://dx.doi.org/10.1016/j.molcel.2010.12.005
- Song L, Dai T, Xie Y, Wang C, Lin C, Wu Z, Ying Z, Wu J, Li M, Li J. Up-regulation of miR-1245 by c-myc targets BRCA2 and impairs DNA repair. J Mol Cell Biol 2012; 4:108-17; PMID:22158906; http://dx.doi.org/10.1093/jmcb/mjr046
- Wang Y, Huang JW, Calses P, Kemp CJ, Taniguchi T. MiR-96 downregulates REV1 and RAD51 to promote cellular sensitivity to cisplatin and PARP inhibition. Cancer Res 2012; 72:4037-46; PMID:22761336; http://dx.doi.org/10.1158/0008-5472.CAN-12-0103
- Huang JW, Wang Y, Dhillon KK, Calses P, Villegas E, Mitchell PS, Tewari M, Kemp CJ, Taniguchi T. Systematic screen identifies miRNAs that target RAD51 and RAD51D to enhance chemosensitivity. Mol Cancer Res : MCR 2013; 11:1564-73; PMID:24088786; http://dx.doi.org/10.1158/1541-7786.MCR-13-0292
- Yang L, Li Y, Cheng M, Huang D, Zheng J, Liu B, Ling X, Li Q, Zhang X, Ji W, et al. A functional polymorphism at microRNA-629-binding site in the 3'-untranslated region of NBS1 gene confers an increased risk of lung cancer in Southern and Eastern Chinese population. Carcinogenesis 2012; 33:338-47; PMID:22114071; http://dx.doi.org/10.1093/carcin/bgr272
- Wang Y, Huang JW, Li M, Cavenee WK, Mitchell PS, Zhou X, Tewari M, Furnari FB, Taniguchi T. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol Cancer Res : MCR 2011; 9:1100-11; PMID:21693595; http://dx.doi.org/10.1158/1541-7786.MCR-11-0007
- Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, Bentwich Z, Lieberman J, Chowdhury D. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol 2009; 16:492-8; PMID:19377482; http://dx.doi.org/10.1038/nsmb.1589
- Mueller AC, Sun D, Dutta A. The miR-99 family regulates the DNA damage response through its target SNF2H. Oncogene 2013; 32:1164-72; PMID:22525276; http://dx.doi.org/10.1038/onc.2012.131
- Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo YY, Mao H, Hao C, Olson JJ, Curran WJ, et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PloS One 2010; 5:e11397; http://dx.doi.org/10.1371/journal.pone.0011397
- Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S, Scudiero DA, Weinstein JN, Kirsch IR. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res 2003; 63:8634-47; PMID:14695175
- Iwabuchi H, Sakamoto M, Sakunaga H, Ma YY, Carcangiu ML, Pinkel D, Yang-Feng TL, Gray JW. Genetic analysis of benign, low-grade, and high-grade ovarian tumors. Cancer Res 1995; 55:6172-80; PMID:8521410
- Maxwell KN, Domchek SM. Cancer treatment according to BRCA1 and BRCA2 mutations. Nat Rev Clin Oncol 2012; 9:520-8; PMID:22825375; http://dx.doi.org/10.1038/nrclinonc.2012.123
- George SH, Shaw P. BRCA and Early Events in the Development of Serous Ovarian Cancer. Frontiers Oncol 2014; 4:5; PMID:24478985; http://dx.doi.org/10.3389/fonc.2014.00005
- Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474:609-15; PMID:21720365; http://dx.doi.org/10.1038/nature10166
- Alsop K, Fereday S, Meldrum C, deFazio A, Emmanuel C, George J, Dobrovic A, Birrer MJ, Webb PM, Stewart C, et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian ovarian cancer study group. J Clin Oncol : official J Am Soc Clin Oncol 2012; 30:2654-63; PMID:22711857; http://dx.doi.org/10.1200/JCO.2011.39.8545
- Press JZ, De Luca A, Boyd N, Young S, Troussard A, Ridge Y, Kaurah P, Kalloger SE, Blood KA, Smith M, et al. Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC Cancer 2008; 8:17; PMID:18208621; http://dx.doi.org/10.1186/1471-2407-8-17
- Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De Pitta C, et al. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J 2011; 30:1990-2007; PMID:21468029; http://dx.doi.org/10.1038/emboj.2011.102
- Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol 2010; 3:46; PMID:21106054; http://dx.doi.org/10.1186/1756-8722-3-46
- Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009; 10:42-6; PMID:19289371; http://dx.doi.org/10.3816/CLC.2009.n.006
- Scapoli L, Palmieri A, Lo Muzio L, Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M, Carinci F. MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int J Immunopathol Pharmacol 2010; 23:1229-34; PMID:21244772
- Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol 2010; 11:136-46; PMID:20022810; http://dx.doi.org/10.1016/S1470-2045(09)70343-2
- Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res 2008; 68:425-33; PMID:18199536; http://dx.doi.org/10.1158/0008-5472.CAN-07-2488
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res 2007; 67:8699-707; PMID:17875710; http://dx.doi.org/10.1158/0008-5472.CAN-07-1936
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008; 110:13-21; PMID:18589210; http://dx.doi.org/10.1016/j.ygyno.2008.04.033
- Marchini S, Cavalieri D, Fruscio R, Calura E, Garavaglia D, Nerini IF, Mangioni C, Cattoretti G, Clivio L, Beltrame L, et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncol 2011; 12:273-85; PMID:21345725; http://dx.doi.org/10.1016/S1470-2045(11)70012-2
- Vaksman O, Stavnes HT, Kaern J, Trope CG, Davidson B, Reich R. miRNA profiling along tumour progression in ovarian carcinoma. J Cell Mol Med 2011; 15:1593-602; PMID:20716115; http://dx.doi.org/10.1111/j.1582-4934.2010.01148.x
- Kobayashi H, Kajiwara H, Kanayama S, Yamada Y, Furukawa N, Noguchi T, Haruta S, Yoshida S, Sakata M, Sado T, et al. Molecular pathogenesis of endometriosis-associated clear cell carcinoma of the ovary (review). Oncol Rep 2009; 22:233-40; PMID:19578761; http://dx.doi.org/10.3892/or_00000417
- Lv L, Zhang T, Yi Q, Huang Y, Wang Z, Hou H, Zhang H, Zheng W, Hao Q, Guo Z, et al. Tetraploid cells from cytokinesis failure induce aneuploidy and spontaneous transformation of mouse ovarian surface epithelial cells. Cell cycle 2012; 11:2864-75; PMID:22801546; http://dx.doi.org/10.4161/cc.21196
- Tanner MM, Grenman S, Koul A, Johannsson O, Meltzer P, Pejovic T, Borg A, Isola JJ. Frequent amplification of chromosomal region 20q12-q13 in ovarian cancer. Clin Cancer Res : an official J Am Assoc Cancer Res 2000; 6:1833-9; PMID:10815905
- Roschke AV, Stover K, Tonon G, Schaffer AA, Kirsch IR. Stable karyotypes in epithelial cancer cell lines despite high rates of ongoing structural and numerical chromosomal instability. Neoplasia 2002; 4:19-31; PMID:11922387; http://dx.doi.org/10.1038/sj.neo.7900197
- Hall J. The Ataxia-telangiectasia mutated gene and breast cancer: gene expression profiles and sequence variants. Cancer Lett 2005; 227:105-14; PMID:16112413; http://dx.doi.org/10.1016/j.canlet.2004.12.001
- Khanna KK. Cancer risk and the ATM gene: a continuing debate. J Natl Cancer Inst 2000; 92:795-802; PMID:10814674; http://dx.doi.org/10.1093/jnci/92.10.795
- Foulkes WD, Shuen AY. In brief: BRCA1 and BRCA2. J Pathol 2013; 230:347-9; PMID:23620175; http://dx.doi.org/10.1002/path.4205
- Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol 2001; 11:105-9; PMID:11231126; http://dx.doi.org/10.1016/S0960-9822(01)00019-7
- Michor F, Iwasa Y, Vogelstein B, Lengauer C, Nowak MA. Can chromosomal instability initiate tumorigenesis? Semin Cancer Biol 2005; 15:43-9; PMID:15613287; http://dx.doi.org/10.1016/j.semcancer.2004.09.007
- Nowak MA, Komarova NL, Sengupta A, Jallepalli PV, Shih Ie M, Vogelstein B, Lengauer C. The role of chromosomal instability in tumor initiation. Proc Nat Acad Sci USA 2002; 99:16226-31; PMID:12446840; http://dx.doi.org/10.1073/pnas.202617399
- Xu CX, Xu M, Tan L, Yang H, Permuth-Wey J, Kruk PA, Wenham RM, Nicosia SV, Lancaster JM, Sellers TA, et al. MicroRNA miR-214 regulates ovarian cancer cell stemness by targeting p53/Nanog. J Biol Chem 2012; 287:34970-8; PMID:22927443; http://dx.doi.org/10.1074/jbc.M112.374611
- Deng M, Ye Q, Qin Z, Zheng Y, He W, Tang H, Zhou Y, Xiong W, Zhou M, Li X, et al. miR-214 promotes tumorigenesis by targeting lactotransferrin in nasopharyngeal carcinoma. Tumour Biol : the J Int Soc Oncodevelop Biol Med 2013; 34:1793-800; PMID:23479198; http://dx.doi.org/10.1007/s13277-013-0718-y
- Wang Z, Cai H, Lin L, Tang M, Cai H. Upregulated expression of microRNA-214 is linked to tumor progression and adverse prognosis in pediatric osteosarcoma. Pediatric Blood & Cancer 2014; 61:206-10; PMID:24038809; http://dx.doi.org/10.1002/pbc.24763
- Yamane K, Jinnin M, Etoh T, Kobayashi Y, Shimozono N, Fukushima S, Masuguchi S, Maruo K, Inoue Y, Ishihara T, et al. Down-regulation of miR-124/-214 in cutaneous squamous cell carcinoma mediates abnormal cell proliferation via the induction of ERK. J Mol Med 2013; 91:69-81; PMID:22828925; http://dx.doi.org/10.1007/s00109-012-0935-7
- Wang X, Chen J, Li F, Lin Y, Zhang X, Lv Z, Jiang J. MiR-214 inhibits cell growth in hepatocellular carcinoma through suppression of beta-catenin. Biochem Biophys Res Commun 2012; 428:525-31; PMID:23068095; http://dx.doi.org/10.1016/j.bbrc.2012.10.039
- Derfoul A, Juan AH, Difilippantonio MJ, Palanisamy N, Ried T, Sartorelli V. Decreased microRNA-214 levels in breast cancer cells coincides with increased cell proliferation, invasion and accumulation of the Polycomb Ezh2 methyltransferase. Carcinogenesis 2011; 32:1607-14; PMID:21828058; http://dx.doi.org/10.1093/carcin/bgr184
- Huang SD, Yuan Y, Zhuang CW, Li BL, Gong DJ, Wang SG, Zeng ZY, Cheng HZ. MicroRNA-98 and microRNA-214 post-transcriptionally regulate enhancer of zeste homolog 2 and inhibit migration and invasion in human esophageal squamous cell carcinoma. Mol Cancer 2012; 11:51; PMID:22867052; http://dx.doi.org/10.1186/1476-4598-11-51
- Li B, Han Q, Zhu Y, Yu Y, Wang J, Jiang X. Down-regulation of miR-214 contributes to intrahepatic cholangiocarcinoma metastasis by targeting Twist. FEBS J 2012; 279:2393-8; PMID:22540680; http://dx.doi.org/10.1111/j.1742-4658.2012.08618.x
- Salim H, Akbar NS, Zong D, Vaculova AH, Lewensohn R, Moshfegh A, Viktorsson K, Zhivotovsky B. miRNA-214 modulates radiotherapy response of non-small cell lung cancer cells through regulation of p38MAPK, apoptosis and senescence. British J Cancer 2012; 107:1361-73; PMID:22929890; http://dx.doi.org/10.1038/bjc.2012.382
- Zhang ZC, Li YY, Wang HY, Fu S, Wang XP, Zeng MS, Zeng YX, Shao JY. Knockdown of miR-214 promotes apoptosis and inhibits cell proliferation in nasopharyngeal carcinoma. PloS One 2014; 9:e86149; http://dx.doi.org/10.1371/journal.pone.0086149
- Yang TS, Yang XH, Wang XD, Wang YL, Zhou B, Song ZS. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int 2013; 13:68; PMID:23834902; http://dx.doi.org/10.1186/1475-2867-13-68
- Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. British J Cancer 2013; 108:479-85; PMID:23299535; http://dx.doi.org/10.1038/bjc.2012.581
- Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 2012; 19:201-6; PMID:22266820; http://dx.doi.org/10.1038/nsmb.2211
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res 1988; 16:7351-67; PMID:3045756; http://dx.doi.org/10.1093/nar/16.15.7351
- Zhang H, Jiang X, Zhang Y, Xu B, Hua J, Ma T, Zheng W, Sun R, Shen W, Cooke HJ, et al. microRNA 376a Regulates Follicle Assembly by Targeting Pcna in Fetal and Neonatal Mouse Ovaries. Reproduction 2014.
- Zhang Y, Yang Y, Zhang H, Jiang X, Xu B, Xue Y, Cao Y, Zhai Q, Zhai Y, Xu M, et al. Prediction of novel pre-microRNAs with high accuracy through boosting and SVM. Bioinformatics 2011; 27:1436-7; PMID:21436129; http://dx.doi.org/10.1093/bioinformatics/btr148
- Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nature Protoc 2006; 1:2315-9; PMID:17406473; http://dx.doi.org/10.1038/nprot.2006.339
- Wang Z, Yin H, Lv L, Feng Y, Chen S, Liang J, Huang Y, Jiang X, Jiang H, Bukhari I, et al. Unrepaired DNA damage facilitates elimination of uniparental chromosomes in interspecific hybrid cells. Cell Cycle 2014; 13.
