Abstract
We recently reported that the p12 subunit of human DNA polymerase δ (Pol δ4) is degraded by CRL4Cdt2 which regulates the licensing factor Cdt1 and p21WAF1 during the G1 to S transition. Presently, we performed multiparameter laser scanning cytometric analyses of changes in levels of p12, Cdt1 and p21WAF1, detected immunocytochemically in individual cells, vis-à-vis the initiation and completion of DNA replication. The latter was assessed by pulse-labeling A549 cells with the DNA precursor ethynyl-2′-deoxyribose (EdU). The loss of p12 preceded the initiation of DNA replication and essentially all cells incorporating EdU were p12 negative. Completion of DNA replication and transition to G2 phase coincided with the re-appearance and rapid rise of p12 levels. Similar to p12 a decline of p21WAF1 and Cdt1 was seen at the end of G1 phase and all DNA replicating cells were p21WAF1 and Cdt1 negative. The loss of p21WAF1 preceded that of Cdt1 and p12 and the disappearance of the latter coincided with the onset of DNA replication. Loss of p12 leads to conversion of Pol δ4 to its trimeric form, Pol δ3, so that the results provide strong support to the notion that Pol δ3 is engaged in DNA replication during unperturbed progression through the S phase of cell cycle. Also assessed was a correlation between EdU incorporation, likely reflecting the rate of DNA replication in individual cells, and the level of expression of positive biomarkers of replication cyclin A, PCNA and Ki-67 in these cells. Of interest was the observation of stronger correlation between EdU incorporation and expression of PCNA (r = 0.73) than expression of cyclin A (r = 0.47) or Ki-67 (r = 0.47).
Abbreviations
| Cdt1 | = | Cdc10-dependent transcript 1 |
| Cdt2 | = | Cdc10-dependent transcript 2 |
| Cdk | = | cyclin-dependent kinase |
| CRL | = | cullin-ring ligase |
| Pol δ | = | DNA polymerase δ |
| PCNA | = | proliferating cell nuclear antigen |
| CDK inhibitor p21WAF1 | = | imaging cytometry |
Introduction
DNA polymerase δ (Pol δ), together with Pol ϵ, are the primary DNA polymerases responsible for the synthesis of genomic DNA in eukaryotes.Citation1,2 In yeast, it has been established that Pol δ is largely responsible for synthesis of the lagging strand, while Pol ϵ is involved in synthesis of the leading strand.Citation3 Human Pol δ consists of 4 subunits, the p125 catalytic subunit, p68, p50 and p12.Citation4-6 p12, the smallest subunit, is absent in S. cerevisiae.Citation2 The targeted degradation of p12 in response to DNA damage is an important regulatory mechanism that leads to the conversion of Pol δ4 to Pol δ3, the trimer lacking p12.Citation7-10 Reconstitution of human Pol δ and its subassembliesCitation11,12 have allowed detailed biochemical comparisons of the properties of Pol δ4 and Pol δ3. These studies have revealed that the removal of p12 leads to fundamental alterations in the kinetic properties of Pol δ such that Pol δ3 appears to be adapted for a role in DNA repair processes, and in fact is endowed with greater fidelity.Citation9,13,14 Biochemical analysis of Pol δ3 in a reconstituted assay for Okazaki fragment processing showed that its properties are also well suited for a role in lagging strand synthesis, and support the hypothesis that Pol δ3 is involved in DNA replication.Citation10,15
Recently, we identified two E3 ligases, RNF8 and CRL4Cdt2, which participate in the targeting of p12 for degradation in response to DNA damage.Citation16,17 CRL4Cdt2 plays a central role in the control of the licensing of origins during the G1/S transition, providing one of the crucial mechanisms for the prevention of re-replication.Citation18,19 Thus, CRL4Cdt2 targets Cdt1, p21 (p21WAF1) and Set8 for degradation. In the case of CRL4Cdt2, however, we have demonstrated that it also targets p12 for degradation during the normal progression of the cell cycle during the G1/S transition, and observed that both p12 and p21 levels decline on entry into S phase.Citation10,17 Using synchronized cells, we showed that p12 levels fall during the G1/S transition, so that Pol δ3 is formed during the S phase.Citation17 The fall in the level of p12 during the S-phase was also observed in individual exponentially growing cells by DNA content analysis by cytometry.Citation8,17 The evidence that Pol δ3 is formed during the S phase, together with studies of Pol δ3 in a reconstituted assay for Okazaki fragment processing, lends support to the hypothesis that Pol δ3 is involved in DNA replication.Citation10,15
The degradation of p12 therefore emerges as an important mechanism for regulating the interconversion between Pol δ4 and Pol δ3 during the entry into S phase, perhaps regulating the balance of these two forms of Pol δ which, by virtue of their biochemical differences, may serve different or complementary roles in cellular replication. Moreover, the G1/S transition is tightly regulated during cell cycle progression and the placement of p12 degradation under CRL4Cdt2, concurrently with p21, Cdt1 and Set8, suggests an integration of controls essential for initiation of DNA synthesis that involves the conversion of Pol δ4 to Pol δ3 during the G1/S transition. However, the exact sequence of the CRL4Cdt2 mediated protein degradation events during G1/S is still not well defined, and there remain questions as to whether this is purely a stochastic process dictated by kinetic aspects of their affinities for PCNA or whether there is a requirement for their ordered degradation that serves a regulatory function.
In this study, we have performed a more precise cytometric analysis of the presence and degradation of p12 during the cell cycle, specifically in the individual cells that initiate and complete DNA replication during S phase. In parallel, cytometric analysis of the level of expression of p21 and Cdt1, the 2 key substrates also regulated by CRL4Cdt2, was performed as well. All three proteins possess a PIP degron.Citation17-19 p21 is transcriptionally regulated during the DNA damage response, controlled by the tumor suppressor protein p53 through which, in reaction to a variety of stress stimuli, it mediates the p53-dependent cell cycle G1 phase arrest.Citation20,21 p21 binds and inhibits the activity of cyclin-CDK2, -CDK1, and -CDK4/6 complexes and functions as a regulator of cell cycle progression at G1 and S phase.Citation20-22 It is now also well established that p21 is also degraded during the cell cycle and by exposure to low UV flux by its targeting for proteosomal degradation by E3 ligases. Because p21 binds to PCNA with high affinity, it is capable of inhibiting a number of PCNA requiring processes and thereby constrains multiple cellular activities where to activate them its removal is required.Citation23-25 These actions include the potential for the inhibition of Pol δ with which it exhibits a direct interaction,Citation9,26 and for the inhibition of Pol η.Citation23 Thus, the concept is that p21 degradation may be required to allow access of these polymerases to PCNA. Cdt1 binds to the origin recognition complex and with Cdc6, loads the MCM2–7 (minichromosome maintenance subunits 2–7) and licenses the complex for replication; its destruction during the G1/S transition is required to prevent re-replication.Citation25 The PIP-degrons as a group have higher affinities for PCNA than PIP-boxes,Citation19 and thus Cdt1, in addition to its functions as a licensing factor, may also have inhibitory effects on PCNA binding processes, as reported recently for the translesion polymerases Pol η and Pol κ.Citation27
The degradation of p12 co-incident with the S phase suggests that p12 may serve as a biomarker whose absence is highly specific for S phase cells. Identification and quantification of cells replicating DNA, the key identifier of cell proliferation, is of significant importance in biology and medicine. Also of importance is the accurate identification of cells that are just initiating and/or completing DNA replication. The most direct means of identification of such cells is their ability to incorporate DNA precursors, which historically has been achieved by using precursors tagged with radioactive (3H, 14C) or halogen (BrdU, IdU) markers that were subsequently detected by autoradiography, cytochemical or immunocytochemical means, respectively (reviewCitation28). The incorporation of 5-ethynyl-2′-deoxyuridine (EdU), detected with fluorochrome-tagged azides by a copper (I) catalyzed [3 + 2] cycloaddition reaction, defined as "click chemistry," has been recently introduced as a novel DNA precursor.Citation28,29 By offering several advantages over the prior methods EdU has now become the preferred DNA precursor applicable in flow and imaging cytometry.Citation30-33 The major advantage of EdU is its compatibility with the concurrent immunocytochemical detection of other cell attributes. However its application in experiments that require DNA labeling followed by long-term incubation is limited because of the EdU-induced perturbation of the cell cycle progression and cytotoxicity.Citation33-35 In the present studies we pulse-labeled human pulmonary adenocarcinoma epithelial A549 cells with EdU, detected p12, p21 and Cdt1 immunocytochemically and used laser scanning cytometry followed by gating analysis to correlate the incorporation of EdU with a loss of these proteins in individual cells during the G1 to S- progression and with their reappearance at the cells traversed from S to G2.
In parallel to correlating expression of p12, Cdt1 and p21 with DNA replication we also correlated and compared the expression of other markers associated with DNA replication and cell cycle progression. These were Cyclin A, PCNA, and Ki-67. Cyclin A regulates progression through the cell cycle at 2 distinct phases. Its association with CDK2 is required for passage through S phase whereas association with CDK1 drives the cell into mitosis.Citation36-38 During duration of S phase cyclin A resides in the nucleus where it is involved in the initiation and completion of DNA replication.Citation39,40 The level of expression of cyclin A in relation of DNA replication is thus reverse to that of p12 or p21.Citation41,42
Proliferating Cell Nuclear Antigen (PCNA)Citation43 is the DNA sliding clamp that serves as the processivity factor for Pol δ and also as a docking platform where other proteins dock to carry out different DNA processes related to replication and repair.Citation44-47 PCNA, detected immunocytochemically, is a widely recognized cell proliferation marker serving also as a potential prognostic indicator for a variety of tumors.Citation48 Similar to cyclin A, the induction of its expression is expected to be correlated with the loss of the p12 subunit of Pol δ. It should also be noted that the recognition of p12 by CRL4Cdt2 requires the former to be bound to PCNA that is loaded onto DNA.Citation17,19
The protein Ki-67 is perhaps the most widely used indicator of cycling cells.Citation49-53 Although this marker has been known for over three decades,Citation49,50 its molecular structure and role in cell cycle progression has only recently begun to be elucidated. The reported data indicate that the Ki-67 protein is the key factor in RNA polymerase I dependent nucleolar rRNA synthesis.Citation52,53 Its expression thus is associated with the production and accumulation of rRNA. This finding is consistent with results of our prior studies showing that the content of cellular RNA, most of which is rRNA, is strongly correlated with cell proliferation and its abundance can be used as a marker to distinguish the cycling from non-cycling cells.Citation54,55 Many subsequent studies confirmed value of cellular RNA content as a determiner of cells progressing through the cycleCitation56-58 and also correlated with the rate of traverse through the cycle.Citation59,60
Results
Cytometric analysis of p12, p21 and Cdt1 levels vis-a-vis EdU pulse-labeling reveals a strong inverse correlation with DNA synthesis
The relationship between incorporation of EdU and expression of p12 subunit of Pol δ was examined by labeling A549 cells with EdU for 30 or 60 min, following which the cells were fixed and differentially stained for p12, EdU incorporation and DNA content, as described in Materials and Methods. Fluorescence of individual cells was analyzed by laser scanning cytometry.Citation61-63 The results of a representative experiment are shown in . The bivariate DNA content versus EdU incorporation distributions (scattergrams) are presented in panels A and E. The characteristic “horse-shoe” shaped distribution allows one to discriminate the subpopulation of cells not incorporating EdU (marked black) and as outlined by the rhombohedral red frame (gate) and by the paint-a-gate software electronically colored red, three subpopulations of cells that did incorporate the precursor (E). The cells that were initiating DNA replication (entering S; eS) during duration of the pulse-exposure to EdU show variable amount of the incorporated precursor and DNA content close to that of the G1-phase cells. The subpopulation characterized by the maximal amount of incorporated EdU represents cells that were constantly exposed to the precursor during the pulse duration (mid-S). The cells that were completing DNA synthesis and entering G2 (eG2) during of exposure to EdU are located on the right side of the “horse-shoe” distribution.
Figure 1. Relationship between expression of p12, incorporation of EdU and cellular DNA content. A549 cells were exposed to EdU for 30 (top rows) or 60 min (bottom rows), the EdU incorporation was detected by the Click-ItTM protocol, cellular expression of p12 was detected immunocytochemically, DNA was counterstained with DAPI and cellular fluorescence was measured by laser scanning cytometry.Citation60-62 During the “paint-a-gate” analysis the cells incorporating EdU were colored red (A and E) and on the bivariate distribution histograms the EdU incorporation is correlated with expression of p12 presented either as the mean integrated value of fluorescence intensity over cell nucleus (B and F) or the mean intensity of maximal pixel (C and G). The dashed skewed lines in (B, C, F, and G) show the upper level of fluorescence of the cells stained with the secondary Ab only; the cells below these lines are thus considered p12 negative. Also presented is a direct relationship between p12 and EdU incorporation (D and H). The inset in (D) shows the DNA content frequency histogram from that culture. The dashed lines in (E) outline the cells that during duration of the 60 min pulse-exposure to EdU were entering S (eS), were constantly present during pulse duration (mid-S), or were entering G2 (eG2). It is quite evident that nearly all cells with DNA content equivalent of S are incorporating EdU and are p12 negative. Presented is the percentage of cells at the G1 to S transition (with DNA content equivalent of G1) that are p12-negative and did not incorporate EdU (42% and 33%; pointed at by open arrows). As described in the text, such distribution of EdU-negative and p12-negative cells at the G1 to S transition is indicative that initiation of DNA replication starts rather rapidly after loss of p12; the amount of DNA synthesized at that time however is so small that based on DNA content these cells are still recognized as in G1. Likewise among the p12 negative cells at the S/G2 transition (with a G2M DNA content) also less frequent are the cells that did not incorporate EdU (34% and 29%). Thus termination of DNA replication appears to be shortly followed by re-expression of p12.
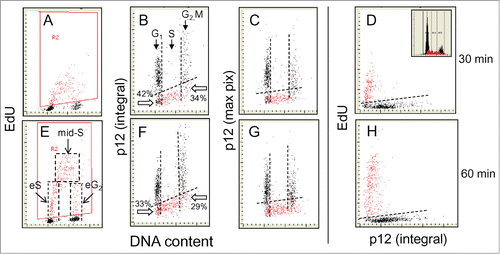
The integrated p12 fluorescence and the max pixel with DNA content are shown in and , respectively, where those cells actively synthesising DNA (incorporating EdU during the pulse) are marked by electronically coloring them red. The dashed skewed lines show the level of maximal fluorescence in controls stained with secondary antibody only, i.e., those cells below the line can be considered to have p12 levels below the threshold of discrimination by antibody staining and thus p12-negative. The p12 vs. DNA content bivariate distribution histograms () exhibit an angular “U” shaped distribution, that is inversely correlated to EdU incorporation, Most remarkably, nearly all the cells that are active in DNA replication are at the bottom of the curve, indicating that p12 is almost completely depleted during S phase, within the limits of the parameters of the antibody detection in this experiment. The data thus distinctly indicate that incorporation of EdU is associated with loss of p12 and that during S phase Pol δ3 is the predominant nuclear species of Pol δ.
Analysis of p12 expression among the cells incorporating EdU but identified as in G1 or in G2/M based on their DNA content reveals kinetics of the loss- and re-appearance of p12 during transition from G1 to S and from S to G2, respectively, throughout duration of exposure to the precursor. Specifically, these “G1” cells although appearing to have DNA content equivalent of G1 did already incorporate adequate quantity of the precursor to be detectable as entering S phase during the time-window of the EdU pulse (; eS). The amount of DNA synthesized during the pulse however was relatively small vis-à-vis the total cellular DNA content and therefore based on staining with DAPI these cells were still identified as G1. Thus, as we have previously noted,Citation61 EdU labeling allows identification of cells early in their transition to S-phase defined by active DNA synthesis. Likewise, the EdU-labeled cells with DNA content equivalent of G2/M represent the cells that during exposure to the precursor traversed from S to G2 (eG2). Examination of only those p12 negative cells below the dashed line in G1 phase (with DNA content that of G1) showed that there were 42% cells that did not incorporate EdU during the 30 min, and 33% during 60 min, of exposure to the precursor. Thus, after degradation of p12 during G1 to the level of that of negative control, the majority of cells were already incorporating EdU.
Among the cells having DNA content equivalent of G2/M (S to G2 transition; eG2) only 34% and 29% of the p12 negative cells did not incorporate EdU during 30 or 60 min exposure to the precursor, respectively. Thus, among the cells that were undergoing S to G2 transition, similar as during G1 to S, majority of the p12-negative cells were already incorporating EdU. The relative paucity of p12-negative cells that did not incorporate EdU indicates that EdU incorporation starts shortly after degradation of p12 and that the presence of p12 is restored rather rapidly after completion of EdU incorporation.
The similarity of the patterns of p12 expression measured by the integral () as compared with the maximal pixel of p12 fluorescence intensity (), the latter emphasizing the local high-density fluorophore localization,Citation62,63 indicates that distribution of this protein within the nuclear chromatin is relatively uniform and diffuse, without the presence of high intensity local p12 regions. The bivariate p12 versus EdU incorporation scatterplots () indicate a distinct separation between these 2 variables, i.e., there is a strong association between the depletion of p12 and DNA replication and additionally confirm that the cells replicating DNA are p12 negative. Overall, the data distinctly show that incorporation of EdU, viz., active DNA synthesis, is associated with loss of p12. It should be noted that in the prior studies using synchronized cells we have shown that the levels of the other three Pol δ subunits do not exhibit major fluctuations during the cell cycle,Citation17 whereas p12 is degraded in response to DNA damageCitation7 and the loss of p12 reports the formation of Pol δ3. In this context then, the changes in p12 can be viewed as the formation of Pol δ3Citation10 coinciding with active DNA synthesis during unperturbed progression through the cell cycle.
illustrates the relationship between EdU incorporation and the presence of p21. As expected from previous studies using synchronized cells the general pattern for p21 is very similar to that of p12.Citation17 Similar as in the case of p12, all EdU-labeled cells are p21 negative (B, C, F, G). However, unlike the case of p12 vs. EdU scatterplots () proportionally more p21 negative cells having a G1 DNA content did not incorporate EdU during 30 (76%) or 60 min (80%) exposure to the precursor (B, F). As mentioned, the incidence of EdU-labeled cells with a G1 DNA content reports kinetics of G1 to S transition, in this case with respect to expression of p21. The data thus indicate that following the loss of p21 the cells remain for longer periods of time in G1 prior to initiation of DNA replication than after the loss of p12. This is also consistent with previous analyses of p12 and p21 degradation in synchronized cell populations, where the fall of p21 was seen to occur prior to that of p12.Citation10,17 At the S to G2 transition also proportionally more are p21-negative cells (with a G2M DNA content) that did not incorporate EdU, during the 30 (63%) or 60 min (57%) exposure to the precursor, compared with p12 (34%, 29%). Thus, following termination of DNA replication the length of time in G2 until the cells start to re-express p21 appears to be longer than to re-express p12 and thereby after termination of DNA replication the re-expression of p12 precedes that of p21.
Figure 2. Relationship between expression of p21WAF1 and EdU incorporation. Similar as in the cells were exposed to EdU for 30 or 60 min, the EdU labeled cells were gated (colored red; A and E) and the EdU incorporation is correlated with expression of p21 and cellular DNA content (B, C, F, and G). Also presented is a direct relationship between p21 and EdU incorporation (D and E). The inset in (D) shows DNA content histogram of cells from this culture. The details on gating strategy and “paint-a-gate” data analysis are described in the legend to ; the dashed skewed lines represent the top level of fluorescence of the cells stained with the secondary Ab only and thus discriminate between the p21 negative and positive cells. As is evident, essentially all cells incorporating EdU are p21 negative. Among the p21-negative cells at the G1 to S transition during the 30 and 60 min EdU pulse there are 76% and 80% cells that did not incorporate EdU. Likewise at the S to G2 transition majority of cells (63% and 57%) were not incorporating EdU.
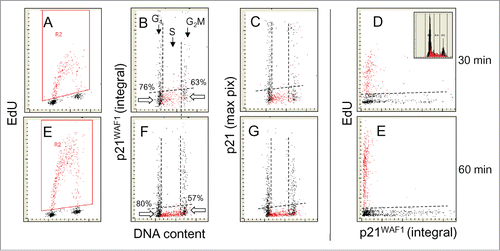
The similarity of the pattern of expression of p21 represented by the integral versus maximal pixel of intensity of the measured fluorescence (), as in the case of p12 (), indicates a rather uniform and diffuse distribution of this protein across the nucleus.Citation62,63 The bivariate plots of EdU incorporation against p21, in particular the data for the 60 min EdU pulse, show a strong separation of cells incorporating EdU and expressing p21 (D,H). These experiments reporting comparison of the expression of p12 vs. p21 were repeated three times on A549 cells, and were also carried on WI-38 cells, yielding essentially similar patterns of the bivariate DNA content versus p12 or p21 distributions (not shown).
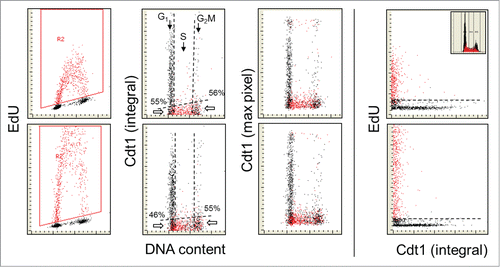
Similar results were obtained with the analysis of the changes in Cdt1 and its correlation with EdU incorporation (). Here, the analysis of the cells in the G1/S transition region showed that among the Cdt1-negative cells 55% and 46% were not incorporating EdU during the 30 and 60 min EdU pulses, while among Cdt1 negative at the G2/M transition the values 56% and 55%, respectively, were obtained (). These values are intermediate between those for p12 and p21, which would suggest that Cdt1 is degraded just after p21 but prior to p12.
Figure 3. Relationship between expression of Cdt1 and EdU incorporation. Similar as in , the cells were exposed to EdU for 30 or 60 min, the EdU labeled cells were gated (colored red; A and E) and the EdU incorporation is correlated with expression of Cdt1 and cellular DNA content (B, C, F, and G). Also presented is a direct relationship between Cdt1 and EdU incorporation (D and E). The inset in (D) shows DNA content histogram of cells from this culture. The details on gating strategy and “paint-a-gate” data analysis are described in the legend to ; the dashed skewed lines represent the top level of fluorescence of the cells stained with the secondary Ab only and thus discriminate between the Cdt1 negative and positive cells. As is evident, essentially all cells incorporating EdU are Cdt1 negative. Among the Cdt1-negative cells at the G1 to S transition the cells that did not incorporate EdU are 55% and 46% for the 30 and 60 min EdU pulses and 56% and 55% at the S to G2 transition.
Cytometric analysis of cyclin A levels in cells active in DNA synthesis by EdU pulse-labeling
The relationship between expression of cyclin A and incorporation of EdU is presented in . In contrast to expression of p12, p21 or Cdt1, the presence of cyclin A rather than its absence is associated with DNA replication. As shown by us while studying incorporation of BrdUCitation41,42 and confirmed presently with EdU () the expression of this protein is confined to cells in late G1, S and G2 and thus all DNA replicating cells are cyclin A positive. Its cellular content progressively increases with cell advancement through the S phase, peaks late in S and in G2, and is followed by degradation at the entrance to mitosis, concurrently with phosphorylation of Ser10 of histone H3.Citation64 Analysis of the cells at the G1/S interphase shows that initiation of EdU incorporation is very closely associated with the appearance and increase in expression of cyclin A. As in the case of p12, p21 and Cdt1, similarity of integral and maximal pixel scatterplots indicates a uniform nuclear distribution of cyclin A.
Figure 4. Relationship between expression of cyclin A and EdU incorporation. The cells were exposed in culture to EdU for 60 min. As in the EdU incorporating cells are gated/colored red (A). On the bivariate distributions of cyclin A versus DNA content (B and C) it is evident that nearly all cells incorporating EdU are cyclin A positive and that cell advancement through the S phase is associated with the progressively rising expression of this protein. In accordance with our prior findingsCitation41,42,64 cells in mitosis (M) are cyclin A negative (they were identified by the image analysis on the iCys cytometer)Citation62,63 while G2 cells have maximal level of this protein (B). As described in the text the correlation between the extent of EdU incorporation and expression of cyclin A among the mid-S phase cells, selected/gated as shown by the dashed vertical lines (A), is weak (r = 0.26; E) but stronger when all EdU-labeled cells are analyzed (r = 0.47). The DNA content histogram of these cells from this culture is shown as the inset in D.
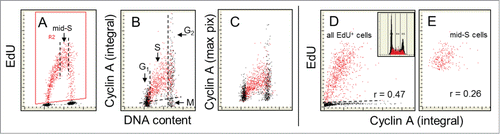
There is a distinct intercellular variability in the amount of incorporated EdU of cells in the mid-S phase, i.e., among the cells that were exposed to the precursor for the full duration of 60 min (). It is possible that this variability is due to different rates of DNA replication in individual cells. Since cyclin A is associated with the machinery of the cell cycle progression.Citation36-40,64 it may be expected that its abundance during S phase is associated with DNA replication rate. To explore such a possibility we selected the mid-S phase cells by the secondary gating () and analyzed the correlation between incorporation of EdU and the level of expression of cyclin A (). The regression analysis of this cell subpopulation revealed rather weak positive correlation between these two variables (r = 0.26). However, the correlation between EdU and cyclin A level among all EdU-positive cells which includes the cells undergoing G1 to S and S to G2 transition during duration of the pulse is distinctly stronger (; r = 0.47).
Cytometric analysis of PCNA levels in cells active in DNA synthesis by EdU pulse-labeling
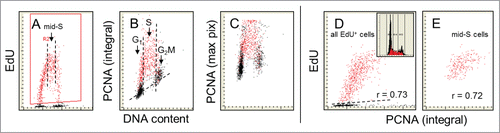
The relationship between expression of PCNA and DNA replication is presented in . Note a striking similarity between the patterns of EdU incorporation () and PCNA expression () vs. DNA content. As it is evident all cells that incorporate EdU do express PCNA and the cells initiating DNA replication have minimal content of this protein. The level of expression of PCNA as well as incorporation of EdU both peak in mid-S phase. The increased level of PCNA expression represented by the mean intensity of the maximal pixel () compared with the mean integral fluorescence intensity () indicates a more localized (punctate) distribution of this protein within nuclear chromatin compared to more uniform (diffuse) distribution of p12 (), p21 (), Cdt1 () or cyclin A (). In analogy to expression of cyclin A () the mid-S phase cells incorporating EdU were selected (A; vertical dashed lines) and their EdU incorporation was plotted against expression of PCNA (). In comparison with cyclin A (r = 0.26) the correlation between these variables is significantly stronger (; r = 0.72). Essentially the same degree of correlation between EdU incorporation and expression of PCNA is seen in the case of all cells replicating DNA i.e., including cells entering- and exiting- S phase during duration of the pulse (; r = 0.73)
Figure 5. Relationship between expression of PCNA and EdU incorporation. The cells were exposed to EdU for 60 min and similar as in the prior figures () the EdU-labeled cells were gated, colored red (A) and analyzed with respect to their expression of PCNA as measured by the integral (B) or maximal pixel (C) intensity of nuclear fluorescence. The PCNA-negative cells are below the dashed skewed line. Note striking similarity between the patterns of EdU incorporation (A) and PCNA expression (B) in relation to DNA content. All cells that incorporate EdU are expressing PCNA. The cells just initiating DNA replication have minimal expression of PCNA. Among the mid-S phase cells that all were exposed to EdU for full 60 min (E) the correlation between expression of PCNA and the amount of incorporated EdU is relatively strong (r = 0.72) and similar (r = 0.73) when assessed for all EdU-incorporating cells. The DNA frequency histogram of these cells is shown in (D).
Cytometric analysis of Ki-67 levels in cells active in DNA synthesis by EdU pulse-labeling
The relationship between DNA replication and expression of Ki-67 protein is shown in . Among the cells with a G1 DNA content there are 2 distinct cell subpopulations differing in expression of Ki-67 that do not incorporate EdU. One is Ki-67 negative (a) the other strongly expresses the protein (b). The cells initiating DNA replication at the G1/S transition have intermediate levels of Ki-67 expression (c). Interestingly, there is a striking difference in the pattern of Ki-67 expression depending on its measurement either as integral () or the mean maximal pixel of fluorescence intensity (C). In the latter case the intercellular variability in the level of Ki-67 expression is much more pronounced and the distinction between the cells expressing this protein at high and low level is more apparent. The punctate local distribution of the Ki-67 protein primarily within nucleoli is responsible for the high intensity and wide distribution of the maximal pixel of Ki-67 immunofluorescence in these bivariate scatterplots (). The cells with high intensity of Ki-67 maximal pixel were gated and their representative images are shown. The G2 cells on average had two or three Ki-67-positive nucleoli while G1 cells predominantly a single one.
Figure 6. Relationship between expression of Ki-67 protein and EdU incorporation. The cells were exposed to EdU for 60 min, and as in the prior figures the cells incorporating the precursor are gated/colored red (A). Note marked a difference when Ki-67 fluorescence intensity is plotted as integral (B) compared to that when shown as maximal pixel (C). The high intercellular variability and intensity of fluorescence of the Ki-67 maximal pixel plot (C) is due to the punctuate localization of its strong fluorescence in nucleoli. The cells in G1 and G2 with high intensity of maximal pixel of Ki-67 were gated (marked by oval dashed lines) and their representative images are shown on both sides of (C). Note the presence of 2 distinct G1 cell subpopulations, Ki-67 negative (a) and positive (b+c). The cells initiating DNA replication have the intermediate level of Ki-67 expression (c). As in the mid-S phase cells incorporating EdU cells were gated out (vertical dashed lines in A) and their EdU incorporation was plotted vs. Ki-67 expression to assess the degree of correlation (r = 0.19) between these variables (E). This correlation was stronger when all DNA replicating cells were analyzed (r = 0.47). The DNA frequency histogram of the cells is shown in (D).
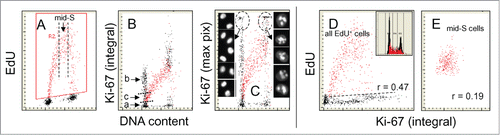
There is an evident relationship between the degree of expression of Ki-67 and advancement of cells through S phase, with much more than doubling in content of this protein in the cells at the S/G2 interphase compared to cells at G1/S transition (). Similar as in case of cyclin A () and PCNA () we have tested whether the intercellular variability in EdU incorporation of the cells in mid-S phase is correlated with Ki-67 expression. The regression analysis of this cell subpopulation shows very weak positive correlation between these variables (; r = 0.19). However, as in the case of cyclin A (), the correlation is significantly stronger while assessed for all cells incorporating EdU i.e., when including the cells entering- and exiting- S phase during the EdU pulse (r = 0.47).
Discussion
Our studies here provide the first utilization of EdU labeling and laser scanning cytometry to the parallel analysis of expression of p12, p21 and Cdt1. The results show that the ability to correlate their levels in relation to the initiation and completion of DNA replication provides a significant tool that allows examination of the early events occurring at the G1/S and S/G2 phase transitions on the cell by cell basis. This analysis extends information obtained by the use of synchronized cells in which whole populations of cells can be examined. It should be noted however that cell synchronization induces undesirable effects such as DNA damage responseCitation65 as well as growth imbalance leading to altered level of cyclin A and cyclin E66,67 and thus can cause a bias in estimates of content of various other proteins including those associated with DNA replication.
It can be argued that in addition to DNA replication the rate of EdU incorporation in individual cells may also be influenced by its rate of penetration through plasma membrane as well by equilibration and competition with the endogenous pool of dT and thus may not reflect the replication rate. We have recently observed, however, that EdU penetration through plasma membrane and incorporation into DNA is very rapid as even as short as 30 sec duration of A549 cells exposure to the precursor results in the detectable level of its incorporation.Citation68 It is very likely therefore that the presently assessed level of EdU incorporation primarily reflects DNA replication rate and is not affected by the obstructed accessibility of the precursor to replication machinery.
The cytometric analysis indicate that p12 levels fall to levels where near-complete degradation occurs, as indicated by the boundaries set by immunofluorescence detection. The cytometric analyses performed in these studies expand our previous investigations using synchronized cells, where levels of p21 and 12 were shown to be degraded during the S phase.Citation17 In the latter studies only partial degradation of p12 to levels of approximately 35% of the G1 levels were observed by western blotting. This could be due to the fact that complete cell synchronization was not achievable, and also differences in the techniques, as noted above.
Nevertheless, our data indicate that the bulk of p12 is degraded, and is consistent with similar findings for p21 and Cdt1, noting that all three proteins are degraded by the same mechanisms. These findings provide compelling evidence that Pol δ3 plays a role in cellular DNA replication, and taken together with our analyses of the properties of Pol δ3 in a reconstituted Okazaki fragment processing system, provides strong evidence for assigning a role for Pol δ3 in lagging strand DNA synthesis as we have proposed.Citation10,15 This implies an evolutionary conservation of the lagging strand polymerase enzyme, since S. cerevisiae Pol δ also consists of the 3 cognate subunitsCitation2,69 present in Pol δ3. This does not exclude the possibility that Pol δ4 plays complementary roles in either Okazaki fragment processing at the lagging strand,Citation10,15 or in synthesis at the leading strand which requires a highly processive polymerase. We have recently found that PDIP46, a Pol δ and PCNA binding proteinCitation70,71 significantly affects the processivity of Pol δ4 and could strengthen the case for its having a function in leading strand synthesis (Wang and Lee, in preparation). Here, it may be noted that our studies only measure the nuclear pool of p12 (and of Pol δ3), and do not provide direct information on the species of Pol δ at the replication fork. It has been estimated by quantitative protein gel blotting of the catalytic subunits of Pol δ and Pol ϵ in HeLa cells that there are ca. 40–100 molcules of each polymerase per origin, so that there may indeed be sufficient Pol δ4 that allow it to be recruited for specific functions in replication. Studies of the depletion of p12 have shown that it leads to genomic instability,Citation71,72 indicating that Pol δ4 must also have a significant role in DNA replication. The present studies provide significant supporting evidence that remarkably, Pol δ3 is the primary polymerase involved in lagging strand synthesis, and further underscores the importance of the regulation of p12 in the context of DNA replication and cell cycle progression.
The cell cycle regulated control of p12 levels suggest that loss of such control may lead to genomic instability, and may be of clinical significance in cancer. As noted above, studies of the expression of the POLD4 gene that encodes p12 in lung cancer tissues showed that there was a reduced mRNA expression in small cell lung cancer, as well as in non-small cell lung cancers associated with poor prognosis.Citation72 The loss of p12 implies increased levels of Pol δ3. Cellular studies showed that siRNA depletion of p12 expression resulted in cell cycle defects, checkpoint activation and DNA damage, as well as increased genomic instability.Citation71,72 More recently, it has also been shown that siRNA depletion of p12 leads to cell cycle blockage at the G1/S transition, and induction of the Cdk inhibitors p21 and p27KIP1 through suppression of the Akt-Skp2 pathway that regulate their degradation.Citation73 These considerations argue for future research into the involvement of p12 in cancer tissues as a marker, and as a potential nexus for generation of genomic instability. While no clear mechanistic evidence is available at present, it can be argued that the appropriate balance between Pol δ4 and Pol δ3 via the regulation of p12 expression is crucial for genomic stability, and it may be speculated that imbalances may lead to improper genomic replication as well as alteration of cellular mechanisms for checkpoint and/or cell cycle regulatory systems.
The present data demonstrate that the absence of p12 subunit of DNA polymerase δ is a distinct marker of DNA replication (). These findings confirm our prior observations that this subunit is degraded not only after induction of DNA damageCitation7-10 but also during unperturbed progression through the cell cycle, in G1 phase prior to entrance to S phase.Citation17 Close analysis of the cells that initiate DNA replication during the 30 or 60 min exposure to EdU reveals that nearly all of them are p12 negative (). This observation is consistent with the notion that degradation of this protein at the end of G1 phase precedes and appears to be a prerequisite, or perhaps the trigger, for DNA replication.
We observed similar results for p21 and Cdt1 as for p12. This is in support of the molecular mechanisms involving degradation of Cdt1 and p21Citation18,19 as well as p12Citation17 by CRL4Cdt2prior to S phase of the cell cycle. CRL4Cdt2 recognizes a specific PIP-degron and involves recognition of the PCNA-bound PIP-degron bound to DNA.Citation19 Our studies provide some additional insights, in that we also were able to extract a temporal sequence where p21 is the first to be completely degraded, to be followed later by Cdt1 and p12. While all three proteins possess an extended PIP-box, the PIP-degron, and are considered to be high affinity ligands for PCNA, this sequence is consistent with p21 likely having the greatest affinity. In this case, temporal aspects of their degradation could be a stochastic process governed by their affinities for PCNA. The affinity (KA = 1.21 × 107) of the p21 PCNA binding peptide has been shown by isothermal titration colorimetry to be approximately 200 fold greater than that of the PIP-box of the p68 subunit of Pol δ.74 That of p12 has not been directly measured, and is more complex in the sense that p12 is a component of the Pol δ4 enzyme, where the p125,Citation75 and p68Citation5 subunits also bind PCNA. The KD of Pol δ4 for DNA is 34 nM which is still 30-fold lower in comparison with the affinity of p21 for PCNA.Citation13
The impressive affinity of p21 for PCNA also underscores its potential for interfering with other PCNA binding proteins. For example, for a protein with a 100 fold lower affinity for PCNA than p21, it would mean that even if p21 levels were reduced to 1% of its starting levels, it would still compete equally well for PCNA, all other things being equal. The insertion of p12 into the stable of CRL4Cdt2 regulated substrates also provides a closer connection between control of p21 and Cdt1 and the initiation of DNA synthesis, particularly since our current data confirm that p12 disappearance and formation of Pol δ3 is associated with active DNA synthesis. Previously, we have notedCitation17 that the requirement of CRL4Cdt2 for PCNA bound to DNACitation19,75,76 imposes a likelihood that it is a primed DNA that is involved, based on the reasoning that RFC loading of PCNA is optimally biased toward loading at the primer terminus.Citation46,77 This would place CRL4Cdt2 action closer to the point of formation of the DNA primer by Pol α, consistent with the correlation of their disappearance and the initiation of EdU incorporation that we observed.
In addition to p12, Cdt1 and p21, the three proteins whose absence provides the “negative” marker of the S phase, we compared the positive markers cyclin A, PCNA and Ki-67, all of them with the initiation of DNA replication. Consistent with prior findingsCitation41,42 a strong relationship was observed between expression of cyclin A and DNA replication when the appearance of cyclin A and its rise at the early section of S phase was very closely associated with initiation of DNA replication (). Also a close association was observed between expression of PCNA or Ki-67 and beginning of DNA synthesis ().
The pattern of Ki-67 expression vis-à-vis DNA replication is quite unique as it reveals striking heterogeneity of the G1 subpopulation and the presence of three subgroups of the G1 cells along the Ki-67 coordinate, one Ki-67 negative, the second strongly positive but not initiating DNA replication, and the third one, intermediate in terms of expression of Ki-67 from which the cells initiate to incorporate EdU, marked respectively (a), (b) and (c) () This subdivision of G1 cells based on different expression of Ki-67 resembles their classification onto G1Q, G1A and G1B compartments based on content of cellular RNA (rRNA) and defined as quiescent (G1Q), temporarily non-cycling (in the growth phase, prior to the “restriction point”) (G1A), and cells that passed the restriction point and are undergoing transition to S (G1B).Citation55,56 In analogy to the RNA content as a marker of these subpopulations, it is likely that (i) the Ki-67 negative cells represent the cells temporarily withdrawn from the cycle (G1Q-like cells), (ii) the Ki-67 positive/EdU negative cells are in the growth phase (equivalent of G1A cells while (iii) the cells entering S phase are analogous to the G1B cells, which after accumulation of the threshold amount of rRNA or growth to the threshold size initiate DNA replication.Citation56-59 Interestingly, there is a similarity in the subdivision of G1 to the above compartments with the subdivision of G1 based on binding of Mcm6 and PCNA, which also is delineating the presence of the restriction point.Citation78
We have also assessed a correlation between the level of expression of cyclin A, PCNA and Ki-67 and the rate of EdU incorporation, considering that the latter may reflect a rate of DNA replication in individual cells. In the case of cyclin A, when the analysis was limited to mid-S phase cells, the correlation was rather weak (r = 0.26) but when all DNA replicating cells were analyzed this correlation was stronger (r = 0.47). In analogy to cyclin A weak correlation was seen between EdU incorporation and Ki-67 expression for the mid-S phase cells (r = 0.19). These variables correlated much stronger when estimated for all cells, including the ones that were initiating DNA replication (r = 0.47). This indicates that correlation between DNA replication and cyclin A as well Ki-67 content is greater for the cells initiating replication than for the cells that are already in the mid-S phase of the cycle. In contrast to cyclin A and Ki-67, however the correlation between incorporation of EdU and expression of PCNA was much stronger for the mid-S phase cells as well as for all EdU incorporating cells (r = 0.72; r = 0.73; respectively). This observation is not unexpected in light of evidence of the direct involvement of PCNA in conjunction with Pol δ in DNA replication.Citation79 It should be mentioned that our estimates of PCNA by the laser scanning cytometry, while based on quantification of immunofluorescence intensity via contouring the cell nucleus, do not distinguish between the chromatin-bound and unbound nuclear PCNA.Citation78,80-83 It is quite likely that the relationship between DNA replication and the content of chromatin-bound PCNA fraction is even stronger compared to its total nuclear content as presently measured.
The methodology presented above utilizing imaging cytometry made it possible to compare the kinetics and assess relative sequence of a loss and re-appearance of p21 versus cdt1 vs. p12, vis-a-vis initiation and completion of DNA replication at the G1/S and S/G2 transitions, respectively. It also was possible to assess direct association between the expression of positive DNA replication markers cyclin A, PCNA or Ki-67 and actual DNA replication, within the same cells. Experiments are underway in our laboratories to use this approach toward expanding the studies to compare kinetics and respective sequence of a loss/ reappearance of other critical proteins controlling the G1 restriction/checkpoint.Citation83-85 Among the studied proteins are CDK inhibitors p15, p16, p27KIP1 and p57 as well other factors such as the DNA replication licensing factor Mcm6 protein, known to be also in a control of the G1/S transition.Citation86
Materials and Methods
Cells, cell treatment
A549 cells, obtained from American Type Culture Collection (ATCC; Manassas, VA), were grown in Ham's F-12K Nutrient Mixture (Mediatech, Inc.) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine (GIBCO/BRL) in 25 ml FALCON flasks (Becton Dickinson Co.) at 37.5°C in an atmosphere of 95% air and 5% CO2. The cells were maintained in an exponential and asynchronous phase of growth by repeated trypsinization and reseeding prior to reaching sub-confluency. The cells were then trypsinized and seeded at about 5×104 cells per chamber in 2-chambered Falcon CultureSlides (Beckton Dickinson). Twenty 4 hours after seeding the cultures were treated with 20 μM EdU (Invitrogen/Molecular Probes) for the indicated times. Subsequently the cells were fixed by transferring slides into Coplin jars containing 1% methanol-free formaldehyde (Polysciences) in PBS for 15 min on ice, then rinsed with PBS and stored in 70% ethanol at −20°C for up to several days.
Fluorochrome cell labeling
The fixed cells were then washed twice in PBS and treated on slides with 0.1% Triton X-100 (Sigma) in PBS for 15 min, and then in a 1% (w/v) solution of bovine serum albumin (BSA; Sigma) in PBS for 30 min to suppress nonspecific antibody (Ab) binding. The cells were then incubated in 100 μl volume of 1% BSA containing 1:100 dilution of p21 rabbit polyclonal Ab (Santa Cruz Biotechnology), or cyclin A mouse monoclonal antibody (mAb) (Abcam), or Ki-67 rabbit polyclonal Ab (Abcam), or 1:100 dilution of p12 rabbit polyclonal Ab (described inCitation7) or 1:1000 dilution of PCNA mAb 74B1,Citation87 or rabbit mAb Cdt1 (Cell Signaling Technology). After overnight incubation at 4°C, the slides were washed twice with PBS and then incubated with the fluorochrome-tagged secondary Abs: 100 μl of 1:100 dilution of AlexaFluor488 (AF488) goat anti-mouse (cyclin A2 and PCNA) or AF488 goat anti-rabbit (p12, p21WAF1 Cdt1, and Ki-67), both from Invitrogen/Molecular Probes for 45 min at room temperature in the dark. The Click-iTTM EdU AF633 imaging kit (Life Technologies) was used to detect EdU incorporation. The cells were then counterstained with 2.8 μg/ml 4,6-diamidino-2-phenylindole (DAPI; Sigma) in PBS for 15 min to be analyzed by laser scanning cytometry. The cells incubated with secondary Ab only served as negative control; to estimate the percent of positive cells expressing particular antigen the upper threshold of the control was set at the level of +3SD of the mean of respective negative control and the cells above this level were considered positive.
Analysis of cellular fluorescence
A549 cells: Cellular immunofluorescence representing the binding of the respective Abs, EdU incorporation, as well as the blue emission of DAPI stained DNA was measured by Laser Scanning Cytometry (LSC) (iCys, formerly CompuCyte; Thorlabs Imaging Systems, Sterling, VA) utilizing standard filter settings; fluorescence was excited with helium neon (633 nm, for detection of EdU), 488-nm argon (for Abs), and violet (405 nm, for DAPI) lasers. Intensities of maximal pixel and integrated fluorescence were measured and recorded for each cell. At least 3,000 cells were measured per sample, the experiments were 3 times repeated and the representative results are shown in . Gating analysis and other details are described in Figure legends and in our recent reports.Citation8,10,17,88
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Supported by NIH NCI RO128 704, the Robert A. Welke Foundation for Cancer Research and “This Close” Cancer Research Foundation (ZD), and NIH GM31973 and ES14737 to MYWTL.
References
- Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem 2009; 284:4041-5; PMID:18835809; http://dx.doi.org/10.1074/jbc.R800062200
- Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol 2005; 40:115-28; PMID:15814431; http://dx.doi.org/10.1080/10409230590935433
- Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol 2008; 18:521-7; PMID:18824354; http://dx.doi.org/10.1016/j.tcb.2008.08.005
- Lee MY, Jiang YQ, Zhang SJ, Toomey NL. Characterization of human DNA polymerase delta and its immunochemical relationships with DNA polymerase alpha and epsilon. J Biol Chem 1991; 266:2423-9; PMID:1703528
- Mo J, Liu L, Leon A, Mazloum N, Lee MY. Evidence that DNA polymerase delta isolated by immunoaffinity chromatography exhibits high-molecular weight characteristics and is associated with the KIAA0039 protein and RPA. Biochemistry 2000; 39:7245-54; PMID: 10852724; http://dx.doi.org/10.1021/bi0000871
- Liu L, Mo J, Rodriguez-Belmonte EM, Lee MY. Identification of a fourth subunit of mammalian DNA polymerase delta. J Biol Chem 2000; 275:18739-44; PMID: 10751307; http://dx.doi.org/10.1074/jbc.M001217200
- Zhang S, Zhou Y, Trusa S, Meng X, Lee EY, Lee MY. A novel DNA damage response: rapid degradation of the p12 subunit of DNA polymerase delta. J Biol Chem 2007; 282:15330-40; PMID:17317665; http://dx.doi.org/10.1074/jbc.M610356200
- Chea J, Zhang S, Zhao H, Zhang Z, Lee EY, Darzynkiewicz Z, Lee MY. Spatiotemporal recruitment of human DNA polymerase delta to sites of UV damage. Cell Cycle 2012; 11:2885-95; PMID:22801543; http://dx.doi.org/10.4161/cc.21280
- Lee MY, Zhang S, Lin SH, Chea J, Wang X, LeRoy C, Wong A, Zhang Z, Lee EY. Regulation of human DNA polymerase delta in the cellular responses to DNA damage. Environ Mol Mutagen 2012; 53:683-98; PMID:23047826; http://dx.doi.org/10.1002/em.21743
- Lee MY, Zhang S, Lin SH, Wang X, Darzynkiewicz Z, Zhang Z, Lee EY. The tail that wags the dog: p12, the smallest subunit of DNA polymerase delta, is degraded by ubiquitin ligases in response to DNA damage and during cell cycle progression. Cell Cycle 2013; 13:23-31; PMID:24300032; http://dx.doi.org/10.4161/cc.27407
- Xie B, Mazloum N, Liu L, Rahmeh A, Li H, Lee MY. Reconstitution and characterization of the human DNA polymerase delta four-subunit holoenzyme. Biochemistry 2002; 41:13133-42; PMID:12403614; http://dx.doi.org/10.1021/bi0262707
- Zhou Y, Meng X, Zhang S, Lee EY, Lee MY. Characterization of human DNA polymerase delta and its subassemblies reconstituted by expression in the multibac system. PLoS One 2012; 7:e39156; PMID:22723953; http://dx.doi.org/10.1371/journal.pone.0039156
- Meng X, Zhou Y, Lee EY, Lee MY, Frick DN. The p12 subunit of human polymerase delta modulates the rate and fidelity of DNA synthesis. Biochemistry 2010; 49:3545-54; PMID:20334433; http://dx.doi.org/10.1021/bi100042b
- Meng X, Zhou Y, Zhang S, Lee EY, Frick DN, Lee MY. DNA damage alters DNA polymerase delta to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res 2009; 37:647-57; PMID:19074196; http://dx.doi.org/10.1093/nar/gkn1000
- Lin SH, Wang X, Zhang S, Zhang Z, Lee EY, Lee MY. Dynamics of enzymatic interactions during short flap human okazaki fragment processing by two forms of human DNA polymerase δ. DNA Repair (Amst) 2013; 12:922-35; PMID:24035200; http://dx.doi.org/10.1016/j.dnarep.2013.08.008
- Zhang S, Zhou Y, Sarkeshik A, Yates JR, Thomson T, Zhang Z, Lee EY, Lee MY. Identification of RNF8 as a ubiquitin ligase involved in targeting the p12 subunit of DNA polymerase δ for degradation in response to DNA damage. J Biol Chem 2013; 288:2941-50; PMID: 23233665; http://dx.doi.org/10.1074/jbc.M112.423392
- Zhang S, Zhao H, Darzynkiewicz Z, Zhou P, Zhang Z, Lee EY, Lee MY. A novel function of CRL4Cdt2: regulation of the subunit structure of DNA polymerase delta in response to DNA damage and during the S phase. J Biol Chem 2013; 288:29950-61; PMID: 23913683; http://dx.doi.org/10.1074/jbc.M113.490466
- Abbas T, Dutta A. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle 2011; 10:241-9; PMID:21212733; http://dx.doi.org/10.4161/cc.10.2.14530
- Havens CG, Walter JC. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev 2011; 25:1568-82; PMID:21828267; http://dx.doi.org/10.1101/gad.2068611
- Rodriguez R, Meuth M. Chk1 and p21 cooperate to prevent apoptosis during DNA replication fork stress Mol Biol Cell 2006; 17:402-12; PMID:16280359; http://dx.doi.org/10.1091/mbc.E05-07-0594
- Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res 2005; 65:3980-5; PMID:15899785; http://dx.doi.org/10.1158/0008-5472.CAN-04-3995
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell 1993; 75:817-25; PMID: 8242752; http://dx.doi.org/10.1016/0092-8674(93)90500-P
- Soria G, Gottifredi V. PCNA-coupled p21 degradation after DNA damage: The exception that confirms the rule? DNA Repair (Amst) 2010; 9:358-64; PMID: 20060369; http://dx.doi.org/10.1016/j.dnarep.2009.12.003
- Mansilla SF, Soria G, Vallerga MB, Habif M, Martinez-Lopez W, Prives C, Gottifredi V. UV-triggered p21 degradation facilitates damaged-DNA replication and preserves genomic stability. Nucleic Acids Res 2013; 41:6942-51; PMID:23723248; http://dx.doi.org/10.1093/nar/gkt475
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009; 9:400-14; PMID:19440234; http://dx.doi.org/10.1038/nrc2657
- Li H, Xie B, Rahmeh A, Zhou Y, Lee MY. Direct interaction of p21 with p50, the small subunit of human DNA polymerase delta. Cell Cycle 2006; 5:428-36; PMID:16479163; http://dx.doi.org/10.4161/cc.5.4.2425
- Tsanov N, Kermi C, Coulombe P, Van der Laan S, Hodroj D, Maiorano D. PIP degron proteins, substrates of CRL4Cdt2, and not PIP boxes, interfere with DNA polymerase eta and kappa focus formation on UV damage. Nucleic Acids Res 2014; 42:3692-706; PMID:24423875; http://dx.doi.org/10.1093/nar/gkt1400
- Darzynkiewicz Z, Traganos F, Zhao H, Halicka HD, Li J. Cytometry of DNA replication and RNA synthesis: historical perspective and recent advances based on “click chemistry.” Cytometry A 2011; 79:328-37; PMID:21425239; http://dx.doi.org/10.1002/cyto.a.21048
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A 2008; 105:2415-20; PMID:18272492; http://dx.doi.org/10.1073/pnas.0712168105
- Hamelik RM, Krishan A. Click-iT assay with improved DNA distribution histograms. Cytometry A 2009; 75:862-5; PMID:19658154; http://dx.doi.org/10.1002/cyto.a.20780
- Sun Y, Sun Y, Lin G, Zhang R, Zhang K, Xie J, Wang L, Li J. Multicolor flow cytometry analysis of the proliferations of T-lymphocyte subsets in vitro by EdU incorporation. Cytometry A 2012; 81:901-9; PMID:22930591; http://dx.doi.org/10.1002/cyto.a.22113
- Buck SB, Bradford J, Gee KR, Agnew BJ, Clarke ST, Salic A. Detection of S-phase cell cycle progression using 5-ethynyl-2′-deoxyuridine incorporation with click chemistry, an alternative to using 5-bromo-2′-deoxyuridine antibodies. BioTechniques 2008; 44:927-9; PMID:18533904; http://dx.doi.org/10.2144/000112812
- Diermeier-Daucher S, Clarke ST, Hill D, Vollmann-Zwerenz A, Bradford JA, Brockhoff G. Cell type specific applicability of 5-ethynyl-2′-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytometry A 2009; 75:535-46; PMID:19235202; http://dx.doi.org/10.1002/cyto.a.20712
- Kohlmeier F, Maya-Mendoza A, Jackson DA. EdU induces DNA damage response and cell death in mESC in culture. Chromosome Res 2013; 21:87-100; PMID:23463495; http://dx.doi.org/10.1007/s10577-013-9340-5
- Zhao H, Halicka HD, Li J, Biela E, Berniak K, Dobrucki J, Darzynkiewicz Z. DNA damage signaling, impairment of cell cycle progression, and apoptosis triggered by 5-ethynyl-2′-deoxyuridine incorporated into DNA. Cytometry A 2013; 83:979-88; PMID: 24115313; http://dx.doi.org/10.1002/cyto.a.22396
- Yam CH, Fung TK, Poon RY. Cyclin A in cell cycle control and cancer. Cell Mol Life Sci 2002; 59:1317-26; PMID:12363035; http://dx.doi.org/10.1007/s00018-002-8510-y
- Pagano M, Pepperkok R, Verde F Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J 1992; 11:961-71; PMID:1312467
- De Biasio A, Blanco FJ. Proliferating cell nuclear antigen structure and interactions: too many partners for one dancer? Adv Protein Chem Struct Biol 2013; 91:1-36; PMID:23790209; http://dx.doi.org/10.1016/B978-0-12-411637-5.00001-9
- Bendris N, Lemmers B, Blanchard JM, Arsic N. Cyclin A2 mutagenesis analysis: a new insight into CDK activation and cellular localization requirements. PLoS One 2011; 6:e22879; PMID:21829545; http://dx.doi.org/10.1371/journal.pone.0022879
- Woo, RA, Poon, RY. Cyclin-dependent kinases and S phase control in mammalian cells. Cell Cycle 2003; 2:316-24; PMID:12851482; http://dx.doi.org/10.4161/cc.2.4.468
- Juan G, Li X, Darzynkiewicz Z. Correlation between DNA replication and expression of cyclins A and B1 in individual MOLT-4 cells. Cancer Res 1997; 57:803-7; PMID:9041174
- Darzynkiewicz Z, Juan G, Traganos F. Cytometry of cell cycle regulatory proteins. Prog Cell Cycle Res 2003; 5:533-42; PMID:14593748
- Celis JE, Madsen P, Nielsen S, Celis A. Nuclear patterns of cyclin (PCNA) antigen distribution subdivide S-phase in cultured cells-some applications of PCNA antibodies. Leuk Res 1986; 10:237-49; PMID: 2419706; http://dx.doi.org/10.1016/0145-2126(86)90021-4
- Kelman Z. PCNA: structure, functions and interactions. Oncogene 1997; 14:629-40; PMID:9038370; http://dx.doi.org/10.1038/sj.onc.1200886
- Kuriyan J, O'Donnell M. Sliding clamps of DNA polymerases. J Mol Biol 1993; 234:915-25; PMID: 7903401; http://dx.doi.org/10.1006/jmbi.1993.1644
- Bowman GD, Goedken ER, Kazmirski SL, O'Donnell M, Kuriyan J. DNA polymerase clamp loaders and DNA recognition. FEBS Lett 2005; 579:863-7; PMID: 15680964; http://dx.doi.org/10.1016/j.febslet.2004.11.038
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell 2007; 129:665-79; PMID:17512402; http://dx.doi.org/10.1016/j.cell.2007.05.003
- Matsushita K, Cha EK, Matsumoto K, Baba S, Chromecki TF, Fajkovic H, Sun M, Karakiewicz PI, Scherr DS, Shariat SF. Immunohistochemical biomarkers for bladder cancer prognosis. Int J Urol 2011; 18:616-29; PMID:21771101
- Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer 1983; 31:13-20; PMID:6339421; http://dx.doi.org/10.1002/ijc.2910310104
- Bruno S, Darzynkiewicz Z. Cell cycle dependent expression and stability of the nuclear protein detected by Ki-67 antibody in HL-60 cells. Cell Prolif 1992; 25:31-40; PMID:1540682; http://dx.doi.org/10.1111/j.1365-2184.1992.tb01435.x
- Endl E, Gerdes J. The Ki-67 protein: fascinating forms and an unknown function. Exp Cell Res 2000; 257:231-7; PMID:10837136; http://dx.doi.org/10.1006/excr.2000.4888
- Rahmanzadeh R, Huttmann G, Gerdes J, Scholzen T. Chromophore-assisted light inactivation of pKi-67 leads to inhibition of ribosomal RNA synthesis. Cell Prolif 2007; 40:422-30; PMID:17531085; http://dx.doi.org/10.1111/j.1365-2184.2007.00433.x
- Bullwinkel J, Baron-Luhr B, Ludemann A, Wohlenberg C, Gerdes J, Scholzen T. Ki-67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J Cell Physiol 2006; 206:624-35; PMID:16206250; http://dx.doi.org/10.1002/jcp.20494
- Darzynkiewicz Z, Traganos F, Sharpless T, Melamed MR. Lymphocyte stimulation: a rapid multiparameter analysis. Proc Natl Acad Sci U S A 1976; 73:2881-4; PMID:822422; http://dx.doi.org/10.1073/pnas.73.8.2881
- Darzynkiewicz Z, Sharpless T, Staiano-Coico L, Melamed MR. Subcompartments of the G1 phase of cell cycle detected by flow cytometry. Proc Natl Acad Sci U S A 1980; 77:6696-9; PMID:6161370; http://dx.doi.org/10.1073/pnas.77.11.6696
- Darzynkiewicz Z, Traganos F, Melamed MR. New cell cycle compartments identified by multiparameter flow cytometry. Cytometry 1980; 1:98-108; PMID: 6170495; http://dx.doi.org/10.1002/cyto.990010203
- Darzynkiewicz Z, Crissman H, Traganos F, Steinkamp J. Cell heterogeneity during the cell cycle. J Cell Physiol 1982; 113:465-74; PMID:6184378; http://dx.doi.org/10.1002/jcp.1041130316
- Andreeff M, Darzynkiewicz Z, Sharpless TK, Clarkson BD, Melamed MR. Discrimination of human leukemia subtypes by flow cytometric analysis of cellular DNA and RNA. Blood 1980; 55:282-93; PMID:6928106
- Darzynkiewicz Z, Evenson DP, Staiano-Coico L, Sharpless TK, Melamed ML. Correlation between cell cycle duration and RNA content. J Cell Physiol 1979; 100:425-38; PMID:489667; http://dx.doi.org/10.1002/jcp.1041000306
- Darzynkiewicz Z. Cellular RNA content, a feature correlated with cell kinetics and tumor prognosis. Leukemia 1988; 2:777-87; PMID:2462138
- Zhao H, Traganos F, Darzynkiewicz Z. Kinetics of the UV-induced DNA damage response in relation to cell cycle phase. Correlation with DNA replication. Cytometry A 2010; 77:285-93; PMID:20014310; http://dx.doi.org/10.1002/cyto.a.20839
- Bedner E, Burfeind P, Gorczyca W, Melamed MR, Darzynkiewicz Z. Laser scanning cytometry distinguishes lymphocytes, monocytes, and granulocytes by differences in their chromatin structure. Cytometry 1997; 29:191-6; PMID:9389435; http://dx.doi.org/10.1002/(SICI)1097-0320(19971101)29: 3<191::AID-CYTO1>3.0.CO;2-F
- Pozarowski P, Holden E, Darzynkiewicz Z. Laser scanning cytometry: principles and applications. Methods Mol Biol 2006; 319:165-92; PMID:16719355; http://dx.doi.org/10.1007/978-1-59259-993-6_8
- Juan G, Traganos F, James WM, Ray JM, Roberge M, Sauve DM, Anderson H, Darzynkiewicz Z. Histone H3 phosphorylation and expression of cyclins A and B1 measured in individual cells during their progression through G2 and mitosis. Cytometry 1998; 32:71-7; PMID:9627219; http://dx.doi.org/10.1002/(SICI)1097-0320(19980601)32: 2<71::AID-CYTO1>3.0.CO;2-H
- Kurose A, Tanaka T, Huang X, Traganos F, Darzynkiewicz Z. Synchronization in the cell cycle by inhibitors of DNA replication induces histone H2AX phosphorylation, an indication of DNA damage. Cell Prolif 2006; 39:231-40; PMID:16672000; http://dx.doi.org/10.1111/j.1365-2184.2006.00380.x
- Gong JP, Traganos F, Darzynkiewicz Z. Growth imbalance and altered expression of cyclins B1, A. E and D3 in MOLT-4 cells synchronized in the cell cycle by inhibitors of DNA replication. Cell Growth & Different 1995; 6:1485-93; PMID:8562487
- Gong JP, Ardelt B, Traganos F, Darzynkiewicz Z. Unscheduled expression of cyclin B1 and cyclin E in several leukemic and solid tumor cell lines. Cancer Res 1994; 54:4285-8; PMID:8044772
- Li B, Zhao H, Rybak P, Dobrucki JW, Darzynkiewicz Z, Kimmel M. Different rates of DNA replication at early versus late S-phase sections: Multiscale modeling of stochastic events related to DNA content/EdU (5-ethynyl-2′deoxyuridine) incorporation distributions. Cytometry A 2014; 85:785-97; PMID:24894899; http://dx.doi.org/10.1002/cyto.a.22484; Epub 2014, 3 Jun
- Johansson E, Majka J, Burgers PM. Structure of DNA polymerase delta from Saccharomyces cerevisiae. J Biol Chem 2001; 276:43824-8; PMID:11568188; http://dx.doi.org/10.1074/jbc.M108842200
- Liu L, Rodriguez-Belmonte EM, Mazloum N, Xie B, Lee MY. Identification of a novel protein, PDIP38, that interacts with the p50 subunit of DNA polymerase delta and proliferating cell nuclear antigen. J Biol Chem 2003; 278:10041-7; PMID:12522211; http://dx.doi.org/10.1074/jbc.M208694200
- Huang QM, Akashi T, Masuda Y, Kamiya K, Takahashi T, Suzuki M. Roles of POLD4, smallest subunit of DNA polymerase delta, in nuclear structures and genomic stability of human cells. Biochem Biophys Res Commun 2010; 391:542-6; PMID:19931513; http://dx.doi.org/10.1016/j.bbrc.2009.11.094
- Huang QM, Tomida S, Masuda Y, Arima C, Cao K, Kasahara TA, Osada H, Yatabe Y, Akashi T, Kamiya K, et al. Regulation of DNA polymerase POLD4 influences genomic instability in lung cancer. Cancer Res 2010; 70:8407-16; PMID:20861182; http://dx.doi.org/10.1158/0008-5472.CAN-10-0784
- Huang Q, Suzuki M, Zeng Y, Zhang H, Yang D, Lin H. Downregulation of POLD4 in Calu6 cells results in G1-S blockage through suppression of the Akt-Skp2-p27 pathway. Bioorg Med Chem Lett 2014; 24:1780-3; PMID:24618301; http://dx.doi.org/10.1016/j.bmcl.2014.02.033
- Bruning JB, Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure 2004; 12:2209-19; PMID:15576034; http://dx.doi.org/10.1016/j.str.2004.09.018
- Zhang P, Mo JY, Perez A, Leon A, Liu L, Mazloum N, Xu H, Lee MY. Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase delta. J Biol Chem 1999; 274:26647-53; PMID:10480866; http://dx.doi.org/10.1074/jbc.274.38.26647
- Havens CG, Shobnam N, Guarino E, Centore RC, Zou L, Kearsey SE, Walter JC. Direct Role for proliferating cell nuclear antigen (PCNA) in substrate recognition by the E3 Ubiquitin ligase CRL4-Cdt2. J Biol Chem 2012; 287:11410-21; PMID:22303007; http://dx.doi.org/10.1074/jbc.M111.337683
- Havens CG, Walter JC. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol Cell 2009; 35:93-104; PMID:19595719; http://dx.doi.org/10.1016/j.molcel.2009.05.012
- Frisa PS, Jacobberger JW. Cytometry of chromatin bound Mcm6 and PCNA identifies two states in G1 that are separated functionally by the G1 restriction point. BMC Cell Biol 2010; 11:26; PMID:20398392; http://dx.doi.org/10.1186/1471-2121-11-26
- Hingorani MM, Coman MM. On the specificity of interaction between the Saccharomyces cerevisiae clamp loader replication factor C and primed DNA templates during DNA replication. J Biol Chem 2002; 277:47213-24; PMID:12370190; http://dx.doi.org/10.1074/jbc.M206764200
- Gonzalez-Melendi P, Testillano PS, Ahmadian P, Reyes J, Risueno MC. Immunoelectron microscopy of PCNA as an efficient marker for studying replication times and sites during pollen development. Chromosoma 2000; 109:397-409; PMID:11072795; http://dx.doi.org/10.1007/s004120000091
- Miura M. Detection of chromatin-bound PCNA in mammalian cells and its use to study DNA excision repair. Radiat Res 1999; 40:1-12; PMID:10408173; http://dx.doi.org/10.1269/jrr.40.1
- Scovassi AI, Prosperi E. Analysis of proliferating cell nuclear antigen (PCNA) associated with DNA. Methods Mol Biol 2006; 314:457-75; PMID:16673899; http://dx.doi.org/10.1385/1-59259-973-7: 457
- Wang QM, Jones JB, Studzinski GP. Cyclin-dependent kinase inhibitor p27 as a mediator of the G1-S phase block induced by 1,25-Dihydroxyvitamin D3 in HL6O cells. Cancer Res 1996; 56:264-7; PMID: 8542578
- Blagosklonny MV, Pardee AB. The restriction point of the cell cycle. Cell Cycle 2002;1:103-10; PMID: 12429916
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 1999; 13:1501-12; PMID:10385618; http://dx.doi.org/10.1101/gad.13.12.1501
- Wei Z, Liu C, Wu X, Xu N, Zhou B, Liang C, Zhu G. Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6. J Biol Chem 2010; 285:12469-73; PMID:20202939; http://dx.doi.org/10.1074/jbc.C109.094599
- Roos G, Jiang Y, Landberg G, Nielsen NH, Zhang P, Lee MY. Determination of the epitope of an inhibitory antibody to proliferating cell nuclear antigen. Exp Cell Res 1996; 226:208-13; PMID:8660957; http://dx.doi.org/10.1006/excr.1996.0220
- Zhao H, Halicka HD, Li J, Darzynkiewicz Z. Berberine suppresses gero-conversion from cell cycle arrest to senescence, Aging (Albany) 2013; 6:623-36; PMID: 23974852
