Abstract
The ATR-Chk1 signaling pathway mediates cellular responses to DNA damage and replication stress and is composed of a number of core factors that are conserved throughout eukaryotic organisms. However, humans and other higher eukaryotic species possess additional factors that are implicated in the regulation of this signaling network but that have not been extensively studied. Here we show that RHINO (for Rad9, Rad1, Hus1 interacting nuclear orphan) forms complexes with both the 9-1-1 checkpoint clamp and TopBP1 in human cells even in the absence of treatments with DNA damaging agents via direct interactions with the Rad9 and Rad1 subunits of the 9-1-1 checkpoint clamp and with the ATR kinase activator TopBP1. The interaction of RHINO with 9-1-1 was of sufficient affinity to allow for the purification of a stable heterotetrameric RHINO-Rad9-Hus1-Rad1 complex in vitro. In human cells, a portion of RHINO localizes to chromatin in the absence of DNA damage, and this association is enriched following UV irradiation. Furthermore, we find that the tethering of a Lac Repressor (LacR)-RHINO fusion protein to LacO repeats in chromatin of mammalian cells induces Chk1 phosphorylation in a Rad9- and Claspin-dependent manner. Lastly, the loss of RHINO partially abrogates ATR-Chk1 signaling following UV irradiation without impacting the interaction of the 9-1-1 clamp with TopBP1 or the loading of 9-1-1 onto chromatin. We conclude that RHINO is a bona fide regulator of ATR-Chk1 signaling in mammalian cells.
Abbreviations
| 9-1-1 | = | Rad9-Hus1-Rad1 |
| UV | = | ultraviolet |
| RHINO | = | Rad9, Hus1, Rad1 interacting nuclear orphan |
| TopBP1 | = | Topoisomerase binding protein 1 |
| ATR | = | Ataxia telangiectasia-mutated and Rad3-related |
| RPA | = | Replication Protein A |
| IP | = | immunoprecipitation |
| ssDNA | = | single-stranded DNA |
Introduction
In response to DNA damage by endogenous or exogenous sources, eukaryotic cells activate DNA damage response signaling pathways that promote DNA repair, slow or arrest cell cycle progression, and maintain cellular and organismal viability.Citation1 Genetic studies from a variety of model systems ranging from budding yeast to mouse models and human cells have demonstrated a key role for a heterotrimeric complex known as the 9-1-1 (Rad9-Hus1-Rad1) clamp in the cellular response to DNA damage and in preventing tumorigenesis.Citation2-4
Structural analyses of the 9-1-1 complex demonstrated that 9-1-1 resembles PCNA,Citation5-10 a homotrimeric sliding clamp protein that facilitates the activities of a multitude of DNA metabolic enzymes on DNA,Citation11,12 including DNA synthesis by DNA polymerases. Though 9-1-1 is also capable of binding to many PNCA-interacting proteins,Citation9,Citation13-18 the best characterized function of the 9-1-1 clamp is in ATR-mediated DNA damage checkpoint signaling, where it is loaded onto primer-template junctions at sites of DNA damage and replication stress by an alternative clamp loader known as Rad17-Replication Factor C.Citation19-21
A key feature of 9-1-1 that differentiates it from PCNA is the presence of an unstructured, highly phosphorylated extension on the C-terminus of the Rad9 subunit.Citation22,23 This domain binds to a protein known as TopBP1, which serves as a direct stimulator of ATR kinase activity through DNA-independent and DNA-dependent mechanisms.Citation24-26 Once active, ATR phosphorylates a number of proteins to maintain genomic stability, including the DNA damage checkpoint effector kinase Chk1.Citation1,27 The role of the 9-1-1 clamp in activation of ATR-mediated DNA damage checkpoint signaling is therefore thought to involve the stabilization of TopBP1 at sites of damage so that it can activate ATR. Though biochemical studies using recombinant proteins of the yeast homologs of 9-1-1, TopBP1, and ATR support this general modelCitation28 and also a direct role for Rad9 in stimulating ATR kinase activity,Citation28,29 experimental validation of the model using human proteins is currently lacking.
Interestingly, a recent DNA damage response screen in human cells identified a novel factor termed RHINO (for Rad9, Hus1, Rad1 interacting nuclear orphan) that localized to sites of DNA damage, mediated cell sensitivity and/or cell cycle checkpoint response to ionizing radiation (IR) and other agents that induce double-strand breaks in DNA.Citation30 Furthermore, mass spectrometric analysis of RHINO protein complexes following exposure of cells to IR identified both the 9-1-1 checkpoint clamp and the ATR activator TopBP1.Citation30 These interactions were validated by co-immunoprecipitation approaches with ectopically expressed proteins in irradiated cells.Citation30 The observation that the RHINO gene is only present in vertebrate genomes indicates the existence of a unique regulatory factor of the ATR-Chk1 pathway in higher eukaryotes.
Here, we examined the interactions of RHINO with 9-1-1 and TopBP1 in vitro and in vivo and its role as a mediator of ATR DNA damage checkpoint signaling in mammalian cells. We find that RHINO directly binds to TopBP1 and forms a stable, heterotetrameric complex with 9-1-1. Knockdown of RHINO in human cells partially abrogated ATR-Chk1 kinase signaling following UV irradiation but did not impact the loading of 9-1-1 on chromatin or the association of 9-1-1 with TopBP1. Furthermore, we find that tethering RHINO to chromatin directly activates ATR-Chk1 signaling. Our results therefore validate RHINO as a component of the 9-1-1 checkpoint clamp complex and as a mediator of ATR kinase signaling in mammalian cells.
Results
RHINO interacts with the 9-1-1 clamp and TopBP1 in the absence of DNA damage
Though a recent report identified RHINO as a novel DNA damage checkpoint gene and 9-1-1 checkpoint clamp-interacting protein in human cells exposed to ionizing radiation,Citation30 it did not address the expression of the endogenous RHINO protein in human cells. Moreover, an earlier study was unable to detect the expression of full-length RHINO protein (also known as (C12orf32) in a panel of breast cancer cell lines and instead observed the expression of only a 16-kDa N-terminal fragment of the protein.Citation31 Using rabbit antisera that we raised against recombinant, full-length RHINO protein, we were similarly unable to detect a single, specific band of the expected molecular weight (27 kDa) of RHINO in whole cell lysates from various human cell lines (data not shown). The absence of a specific immunoreactive protein band by SDS-PAGE and immunoblotting may be due to either poor immunoreactivity or to a low level of RHINO protein expression in human cells.
To clarify the expression of RHINO in human cells and its interaction with the DNA damage checkpoint clamp, we therefore generated cell lines that expressed either FLAG-tagged Rad9 or RHINO under the control of a tetracycline-inducible promoter and then isolated co-precipitating proteins by anti-FLAG immunoprecipitation. As expected, both Rad1, which forms a stable heterotrimeric complex with Rad9 and Hus1, and TopBP1, which interacts with the C-terminal tail of Rad9 to mediate ATR kinase activation at sites of DNA damage,Citation22,23 were observed in the anti-FLAG-Rad9 immunoprecipitate (, lane 2). We also probed the Rad9 IP with our anti-RHINO antisera, and we detected a protein that was slightly larger than the expected molecular weight for RHINO and slightly smaller than a FLAG-tagged RHINO protein that was produced in a similar tetracycline-inducible cell line. Transfection with RHINO siRNAs verified that this band was indeed RHINO (see below). We conclude that endogenous, full-length RHINO protein is indeed expressed in human cells but that its immunodetection may require enrichment through isolation of the 9-1-1 checkpoint clamp.
We next used cells expressing FLAG-RHINO to verify RHINO's protein-protein interactions. As shown in (lane 3), we observed that endogenous TopBP1, Rad9, and Rad1 proteins were present in the anti-FLAG-RHINO immunoprecipitate. These results indicate that RHINO forms one or more complexes with TopBP1 and with the 9-1-1 checkpoint clamp in human cells in the absence of overt DNA damage. Though these protein-protein interactions were observed in whole cell lysates prepared from sonicated cells, the addition of Benzonase nuclease to the lysates did not abrogate the interactions (data not shown). These results indicate that RHINO's interactions with checkpoint proteins are not mediated through non-specific interactions with DNA or RNA.
Figure 1. RHINO interacts with the 9-1-1 clamp and TopBP1 in human cells in the absence of DNA damage. (A) Flp-In T-REx 293 cells expressing the indicated construct were induced with tetracycline for 2 days, and cell lysates were subjected to anti-FLAG immunoprecipitation. Vector indicates an empty vector that does not express a FLAG-tagged protein. IPs were immunoblotted with the indicated antibodies. Antibody heavy chain was detected by staining the membrane with Ponceau S prior to immunoblotting. Molecular weight markers show the approximate molecular weight of the proteins that were detected with the indicated antibodies. (B) FLAG-Rad9 was immunoprecipitated from cells 1 hr following mock irradiation or irradiation with 20 J/m2 of UV-C, and then co-precipitating proteins were examined by immunoblotting. (C) FLAG-RHINO-expressing cells were examined as in (B).
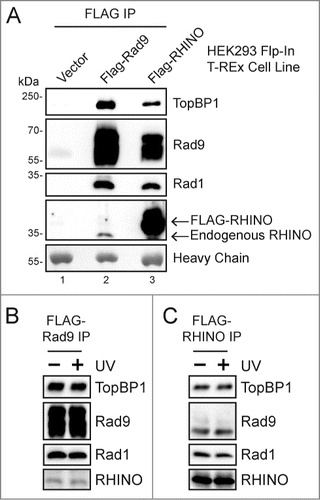
A previous report characterized RHINO's association with DNA damage checkpoint proteins in cells exposed to ionizing radiation but did not examine these interactions in the absence of genotoxins.Citation30 To determine whether the interactions of RHINO with 9-1-1 and TopBP1 are impacted by the presence of DNA damage in the cell, we exposed FLAG-Rad9 and FLAG-RHINO expressing cells to UV radiation and then isolated the FLAG-tagged protein complexes by immunoprecipitation. As shown in and , the interactions of RHINO with Rad9, Rad1, and TopBP1 were not affected by UV irradiation. These findings suggest that the interactions of RHINO with the 9-1-1 complex and with TopBP1 occur constitutively in human cells in the absence of DNA damage.
RHINO directly binds to TopBP1
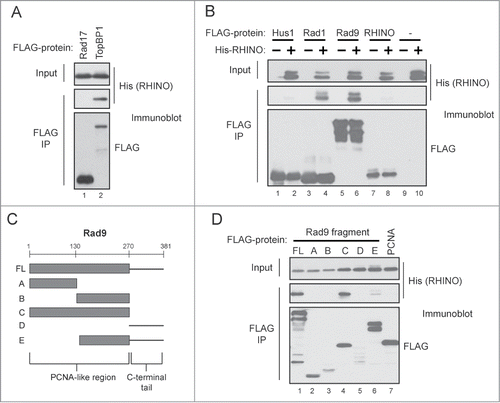
To determine whether RHINO's interactions with 9-1-1 and TopBP1 are direct or are instead mediated by additional factors in human cells, we generated baculoviruses to express RHINO in insect cells and then examined the ability of RHINO to co-immunoprecipitate with TopBP1 and the 9-1-1 clamp subunits. As shown in , we observed that His-tagged RHINO co-immunoprecipitated with FLAG-TopBP1 but not with FLAG-Rad17, which is required to load the 9-1-1 clamp on primer-template junctions.Citation19-21 We conclude that RHINO is capable of directly binding to TopBP1 in the absence of the 9-1-1 complex.
Figure 2. RHINO directly binds to Rad9, Rad1, and TopBP1. (A) Insect cells were infected with His-RHINO baculovirus in the presence of either FLAG-Rad17 or FLAG-TopBP1 baculovirus. Cell lysates were subjected to anti-FLAG immunoprecipitation, and the IPs were examined by immunoblotting with anti-FLAG or anti-His antibodies. (B) Insect cells were co-infected with baculoviruses expressing the indicated FLAG-tagged protein in the absence or presence of His-RHINO baculovirus. Cell lysates were immunoprecipitated with anti-FLAG affinity resin and examined by immunoblotting. (C) Schematic of Rad9 domains. (D) Insect cells were infected with baculoviruses expressing the indicated FLAG-tagged fragment of Rad9 along with His-RHINO baculovirus. Cell lysates were examined as described in (B).
RHINO directly binds the Rad9 and Rad1 subunits of the 9-1-1 complex
RHINO likely associates with the 9-1-1 checkpoint clamp through direct interactions with specific subunits of the 9-1-1 complex. We therefore co-infected insect cells with His-tagged RHINO and either FLAG-tagged Hus1, Rad1, Rad9, or RHINO and then isolated the various protein complexes with anti-FLAG affinity resin. As shown in , RHINO stably co-immunoprecipitated with Rad1 and Rad9. However, His-RHINO did not interact significantly with either FLAG-Hus1 or with a FLAG-tagged RHINO protein. We conclude that RHINO makes direct contacts with the Rad9 and Rad1 subunits of the 9-1-1 complex.
Rad9 contains both a PCNA-like region and an unstructured, C-terminal tail that undergoes phosphorylation, which is important in binding to the ATR-activating protein TopBP1 (). To examine the domains of Rad9 that bind to RHINO, we used a series of baculoviruses encoding various FLAG-tagged fragments of Rad9 and then co-infected insect cells with these viruses in conjunction with the His-tagged RHINO virus. As shown in , the His-tagged RHINO stably co-immunoprecipitated with both the full-length Rad9 construct and the PCNA-like segment of Rad9 but interacted only weakly with the other fragments of Rad9. Though RHINO interacted with the PCNA-like region of Rad9, it did not bind to FLAG-tagged PCNA (, lane 7).
RHINO forms a stoichiometric complex with the 9-1-1 checkpoint clamp
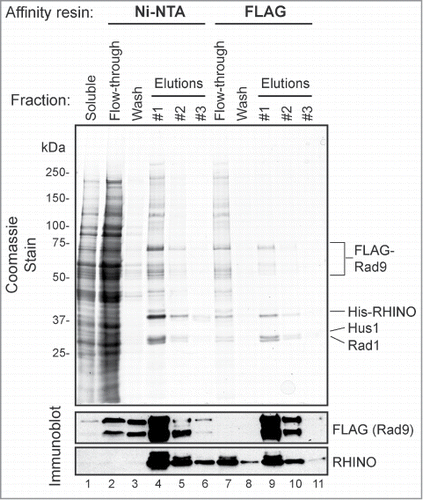
To examine the functional significance of RHINO's interactions with Rad9 and Rad1, we co-infected Sf21 insect cells with baculoviruses encoding FLAG-tagged Rad9, His-tagged RHINO, and untagged Rad1 and Hus1 and then subjected the cell lysates to sequential affinity purification with Ni-NTA agarose and anti-FLAG affinity resin. As shown in , following this 2-step purification scheme, we were able to isolate a stable, heterotetrameric RHINO-Rad9-Hus1-Rad1 (RHINO-9-1-1) complex. Quantification of 4 independent preparations of the RHINO-Rad9-Hus1-Rad1 complex showed an average stoichiometry of 1:1.6:1:0.7 (RHINO:Rad9:Hus1:Rad1; standard deviations 0:0.4:0.3:0.1). We conclude that RHINO forms a stable, near stoichiometric complex with the 9-1-1 checkpoint clamp.
Figure 3. RHINO forms a stoichiometric complex with the 9-1-1 clamp. Insect cells were infected with FLAG-Rad9, His-RHINO, and untagged Rad1 and Hus baculoviruses, and then the cell lysates were subjected to Ni-NTA and FLAG affinity purification. The soluble, flow-through, wash, and elution fractions from these steps were subjected to SDS-PAGE, coomassie staining, and immunoblotting with anti-FLAG and anti-RHINO antibodies.
Though RHINO binds to both the 9-1-1 complex and TopBP1 ( and ), attempts to purify a stable, heteropentameric RHINO-9-1-1-TopBP1 complex in baculovirus-infected insect cells were not successful. However, we have similarly been unable to purify a stable TopBP1-9-1-1 complex in this expression system, which indicates that the interaction of TopBP1 with these factors is dynamic or that post-translational modifications may be necessary to form a stable intermediate complex during ATR activation.
Having purified the RHINO-9-1-1 complex, we wished to test its interaction with various DNA substrates and its effect on ATR kinase activity in the in vitro system developed in our lab.Citation25,26,Citation32-36 However, we observed that though the complex is stable in buffers of high ionic strength, it precipitated under reaction conditions necessary to measure kinase activity. Thus we were unable to test the contribution of RHINO to ATR-Chk1 signaling in vitro. We therefore performed experiments in vivo to address this issue. Furthermore, we attempted to study the DNA binding and DNA loading properties of RHINO and the RHINO-9-1-1 clamp in vitro but were hampered by the observation that significant amounts of RHINO precipitated out of solution when the proteins were incubated in buffers containing divalent cations (data not shown). Thus, although we were able to isolate a stable, stoichiometric RHINO-9-1-1 complex in vitro, we have thus far been unable to study its biochemical properties in vitro.
RHINO localizes to chromatin in human cells
In an attempt to characterize the biochemical properties of RHINO and the RHINO-9-1-1 complex in vivo, we made use of the FLAG-RHINO-inducible cells described in . Because proteins involved in DNA damage responses, such as TopBP1 and the 9-1-1 clamp, are often found enriched in the chromatin fraction of cells, we first examined whether RHINO similarly localized to chromatin. We therefore fractionated FLAG-RHINO-expressing cells to yield a Triton-soluble fraction (containing cytosolic and soluble nuclear proteins) and a Triton-resistant, chromatin-enriched fraction, which was then treated with the nuclease Benzonase to solubilize proteins that associate with DNA or other nucleic acids within chromatin. As shown in , this fractionation method led to a clear separation of the cytosolic protein MEK2 from the chromatin-associated proteins in the Triton-resistant pellet. Moreover, treatment of the Triton-resistant chromatin fraction with Benzonase readily solubilized most of the RHINO, 9-1-1, and TopBP1 proteins from the chromatin pellet. These findings indicate that these proteins associate directly or indirectly with nucleic acid components of chromatin and not with the nuclear matrix. Quantification of these experiments showed that approximately 40–50% of the total cellular RHINO associates with the chromatin fraction in asynchronously growing cells. We therefore conclude that RHINO associates with chromatin in human cells even in the absence of exogenous DNA damage.
Figure 4. RHINO associates with chromatin. (A) FLAG-RHINO-expressing Flp-In T-REx 293 cells were fractionated to yield a Triton-soluble fraction (Sol1). The chromatin pellet was then incubated or not with Benzonase to separate soluble DNA/nucleic acid-bound proteins (Sol2) from insoluble material (Pel). Fractions from equivalent numbers of cells were examined by immunoblotting with antibodies against the indicated proteins. Core histones were visualized by staining the membrane with Ponceau S. (B) FLAG-RHINO-expressing cells were exposed to UV in the presence of HU/AraC to block gap filling DNA synthesis, and then the chromatin fraction of the cells were examined by immunoblotting. (C) The Triton-soluble and chromatin fraction of FLAG-RHINO-expressing cells were subjected to anti-FLAG immunoprecipitation and then examined by immunoblotting.
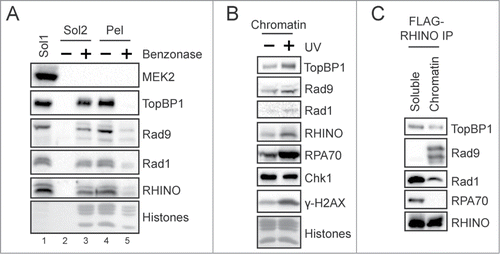
We next exposed cells to UV radiation and then repeated the fractionation procedure. As shown in , like TopBP1 and the 9-1-1 complex, the level of RHINO protein on chromatin increased following exposure to UV, which indicates that the presence of DNA damage leads to the increased association of RHINO with chromatin.
Our previous data that TopBP1 and the 9-1-1 complex co-immunoprecipitate with RHINO in whole cell lysates from human cells (see ) did not differentiate between soluble complexes or interactions that take place on chromatin. We therefore repeated the immunoprecipitation of FLAG-RHINO but from the soluble and chromatin fraction of cells shown in . As shown in , though TopBP1 and Rad1 were found in anti-FLAG-RHINO immunoprecipitates from both the soluble and chromatin fractions, Rad9 was only present in anti-FLAG-RHINO immunoprecipitates from the chromatin fraction of cells. These results indicate that functional RHINO-9-1-1 complexes likely only exist within the chromatin fraction of human cells.
Knockdown of RHINO partially abrogates ATR-Chk1 signaling
The increased association of RHINO with UV-damaged chromatin suggests that RHINO may play a role in the DNA damage checkpoint response to UV. Notably, siRNA-mediated knockdown of RHINO was recently shown to lead to a partial abrogation of IR-induced checkpoint response and a modest reduction in Chk1 phosphorylation.Citation30 To determine whether RHINO mediates Chk1 phosphorylation in response to UV irradiation, we transfected cells with RHINO siRNAs and then exposed cells to UV radiation. As shown in , cells transfected with RHINO siRNAs showed a substantial reduction in the level Chk1 phosphorylation following UV irradiation.
Figure 5. Knockdown of RHINO partially abrogates UV-induced ATR-Chk1 signaling but does not impact 9-1-1 complex association with TopBP1 or 9-1-1 loading on chromatin. A, Flp-In T-REx 293 cells were transfected with control or RHINO siRNAs and then exposed to 10 J/m2 of UV-C radiation. Cell lysates were examined by immunoblotting with anti-phospho-Chk1 (Ser345) or anti-Chk1 antibodies. The graph shows the average and standard deviation from 3 independent experiments. B, FLAG-Rad9-expressing cells transfected with control or RHINO siRNAs and were exposed to UV prior to preparation of whole cell lysates. The lysates were subjected to immunoprecipitation with anti-FLAG agarose and then examined by immnoblotting with the indicated antibodies. C The chromatin fraction of Flp-In T-REx 293 cells transfected with control or RHINO siRNAs and exposed to UV-C were examined by immunoblotting with antibodies against the indicated proteins. Core histones were visualized by staining the membrane with Ponceau S.
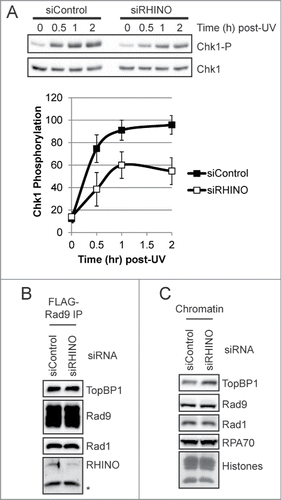
It has been suggested that because RHINO co-immunoprecipitates with both TopBP1 and the 9-1-1 complex,Citation30 the function of RHINO in IR-induced checkpoint signaling may be to bridge these proteins and help to recruit TopBP1 to the 9-1-1 complex. However, our baculovirus co-infection and protein purification approaches failed to produce evidence that RHINO stabilizes an interaction of TopBP1 with the 9-1-1 clamp (data not shown). Furthermore, when we examined the association of TopBP1 with Rad9 in human cells transfected with either control or RHINO siRNAs, we observed similar levels of TopBP1 in the Rad9 immunoprecipitates (). These results indicate that RHINO likely mediates Chk1 phosphorylation by ATR by a mechanism other than through facilitating interactions between the ATR activator TopBP1 and the 9-1-1 checkpoint clamp.
It was also formally possible that RHINO is required to load the 9-1-1 clamp at sites of DNA damage. However, we observed similar levels of Rad9 and Rad1 on chromatin following transfection with either control or RHINO siRNAs (). Though knockdown of RHINO did not impact RPA association with chromatin, we did observe a small, approximately 60% increase in TopBP1 levels on chromatin () when RHINO was depleted from cells. This increase may indicate that other factors and DNA damage response pathways that facilitate TopBP1 association with chromatin following DNA damage, including the MRN complexCitation37,38 and MDC1,Citation39 may be stimulated when RHINO is unable to carry out its function in ATR-Chk1 signaling. Nonetheless, our results demonstrate that RHINO is neither required for 9-1-1 association with chromatin () nor for the interaction of 9-1-1 with TopBP1 ().
Tethering of RHINO to chromatin activates ATR-Chk1 signaling
We and others have used an E. coli LacR-LacO system to target and tether proteins to chromatin in mammalian cells to study the mechanism of DNA damage checkpoint signaling.Citation33,40,41 We therefore made use of this methodology to further validate RHINO as a bona fide mediator of ATR-Chk1 signaling in human cells. We thus fused the cDNA encoding the Lac repressor (LacR) to the RHINO cDNA and then introduced the construct into NIH3T3 cells containing hundreds of repeats of the LacO DNA sequence.Citation40 A schematic of this approach is shown in .
Figure 6. Artificial tethering of RHINO to chromatin induces Chk1 phosphorylation in a Claspin- and Rad9-dependent manner. (A) NIH2/4 cells containing the LacO array were transfected with the indicated combinations of plasmids encoding LacR-Claspin, LacR-RHINO, or LacR-FLAG fusion proteins. Cell lysates were examined by immunoblotting with anti-phospho-Chk1 (Ser345) and anti-Chk1 antibodies. The arrow indicates the specific phospho-Chk1 signal, which migrates slightly above a non-specific band that is present in NIH2/4 cell lysates. The strength of the specific phospho-Chk1 signal relative to the non-specific band varied depending on the cell transfection efficiency. The graph shows the average and standard deviation from 3 independent experiments, in which the maximum phospho-Chk1 signal for each experiment was set to a value of 100. (B) Cells were transfected and analyzed as described in (A). (C) Cells were transfected with the indicated LacR-fusion construct in the absence or presence of LacR-Claspin. (D) Cells were transfected or not with Rad9 siRNA and were then transfected with the indicated LacR-fusion constructs.
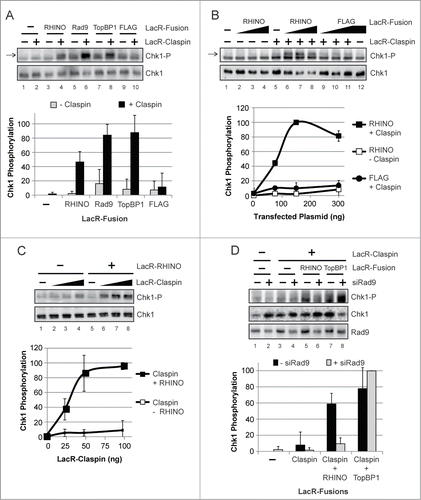
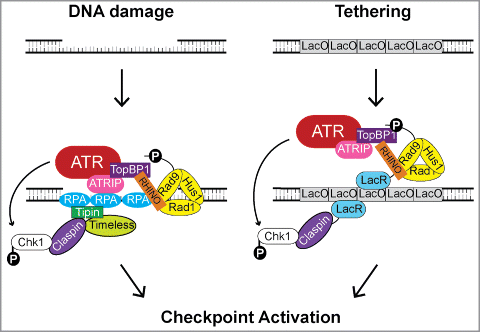
Figure 7. Model for the role of RHINO in ATR-Chk1 signaling following DNA damage or artificial tethering to chromatin. Following the induction of DNA damage, DNA repair processing events and/or replication fork stalling leads to the generation of stretches of ssDNA that become bound by RPA and to the formation of primer-template junctions onto which the 9-1-1 checkpoint clamp is loaded. The RPA-coated ssDNA is recognized by the ATR-ATRIP complex and by the Timeless-Tipin complex, which interacts with the Chk1-binding and mediator protein Claspin. TopBP1, which serves to activate ATR, binds to the C-terminal tail of Rad9. Recruitment of RHINO requires the prior recruitment of the 9-1-1 complex. RHINO directly binds to Rad9, Rad1, and TopBP1 and may alter the topology of these checkpoint proteins to facilitate efficient checkpoint signaling. In the absence of overt DNA damage, the targeting of RHINO to chromatin through fusion to LacR and recruitment to the LacO array mediates Chk1 phosphorylation by ATR when Claspin is also targeted to the LacO array.
As shown in (lane 3), the targeting of LacR-RHINO alone to the LacO array yielded only background levels of Chk1 phosphorylation, which were similar to that observed when cells were transfected with LacR-Rad9 or LacR-TopBP1 constructs (lanes 5 and 7). However, when these proteins were targeted to the LacO array in combination with Claspin, which directly binds Chk1 and serves as an adaptor to mediate the phosphorylation of Chk1 by ATR, robust phosphorylation of Chk1 was observed (, lanes 4, 6, and 8). We conclude that like Rad9 and TopBP1, the accumulation of RHINO at LacO repeats within chromatin is capable of inducing Chk1 phosphorylation in the absence of overt DNA damage.
To further characterize this response, we transfected cells with increasing amounts of the LacR-RHINO construct in the absence or presence of LacR-Claspin. As shown in , the level of Chk1 phosphorylation was highly dependent on the amount of LacR-RHINO construct that was transfected and was completely dependent upon the co-transfection with the LacR-Claspin construct.
Similarly, when we used a fixed amount of LacR-RHINO construct with increasing amounts of the LacR-Claspin construct, we observed that the level of Chk1 phosphorylation was highly correlated with the amount of LacR-Claspin that was expressed in cells containing the LacO array ().
Based on the known network of protein-protein interactions that constitute the ATR-Chk1 signaling event (), we predicted that the ability of RHINO to stimulate Chk1 phosphorylation was dependent on its interaction with Rad9. We therefore repeated the LacO targeting experiments in the absence or presence of Rad9 siRNA to reduce the expression of Rad9 in the cells. As shown in (lanes 5 and 6), knockdown of Rad9 lead to a significant abrogation of RHINO-dependent Chk1 phosphorylation with the LacR-LacO tethering system. As expected, the tethering of TopBP1, which directly activates ATR and is the sole ATR-activating protein in mammalian cells,Citation42 was not affected by Rad9 siRNA because the direct tethering is expected to bypass the function of Rad9 in ATR-Chk1 signaling ( lanes 7 and 8).
We conclude from our studies using the LacR-LacO tethering and enrichment approach that RHINO is a genuine regulator of ATR-Chk1 signaling in mammalian cells.
Discussion
The core molecular mechanism of ATR-Chk1 signaling is reasonably well-established and is based on studies using a wide range of model eukaryotic organisms and systems. However, the recent identification of RHINO as a novel ATR-Chk1 pathway geneCitation30 that is apparently only found in vertebrates indicates the existence of unique regulators of the ATR kinase in higher eukaryotes. Though RHINO was demonstrated to interact with the 9-1-1 checkpoint clamp and the ATR kinase activator TopBP1 and to mediate ATR-dependent DNA damage signaling in cells exposed to IR and other double-strand break-inducing genotoxins,Citation30 the expression of RHINO protein in undamaged cells, the relative stability of its association with checkpoint proteins, and its role in ATR-Chk1 signaling in response to other genotoxins that induce base damage remained unknown.
In this report, we have demonstrated that full-length RHINO protein is expressed in human cells and localizes to chromatin even in the absence of overt DNA damage (). In both human cells and baculovirus-infected insect cells, we found that RHINO interacted with TopBP1 and subunits of the 9-1-1 checkpoint clamp (). Our ability to purify a stable, stoichiometric RHINO-9-1-1 complex () indicates that the checkpoint clamp may function not as a heterotrimer but instead as a heterotetramer. Interestingly, we observed that the loss of RHINO neither impacted 9-1-1 loading onto chromatin nor the interaction between 9-1-1 and the ATR-activating protein TopBP1 (). In contrast, the recruitment of RHINO to sites of DNA damage required the 9-1-1 complex.Citation30
A schematic summarizing our findings and our current understanding of RHINO's role in ATR DNA damage checkpoint signaling is shown in . According to the standard model for ATR activation, the stalling of DNA polymerases by DNA lesions and the processing of DNA damage by DNA repair systems lead to the formation of regions of ssDNA and to the generation of primer-template junctions.Citation1,27,36 9-1-1, which exists in a heterotetrameric complex with RHINO, is loaded onto the primer-template junctions by Rad17-RFC.Citation19-21 This loading then facilitates the stable association of TopBP1 with the damage site.Citation22,23,28 RPA coats the ssDNA and recruits the ATR kinase through an interaction with ATRIP (ATR-interacting protein).Citation32,35,43,44 Through a network of protein-protein interactions involving the Chk1-binding protein Claspin and the Timeless-Tipin complex,Citation34,Citation45-48 RPA also recruits the ATR substrate Chk1 to the damage site.
We also showed that many of the protein-protein and protein-DNA interactions shown in the canonical ATR checkpoint model in (left panel) can be bypassed by fusing RHINO and Claspin to the LacR to target these factors to LacO sequences within chromatin (right panel). Thus, the co-tethering of RHINO (or Rad9 or TopBP1) with Claspin is sufficient to induce Chk1 phosphorylation in the absence of direct DNA damage (). However, RHINO's ability to mediate ATR activation in this system is dependent upon Rad9 (), which highlights non-redundant roles for RHINO and Rad9 in ATR activation.
It was also recently suggested that RHINO may help to stabilize checkpoint proteins at sites of DNA damage or to allosterically regulate specific factors such TopBP1 during the activation of ATR.Citation30 Such a function is consistent with existing data and the model shown in . However, additional biochemical studies using purified components and reaction conditions that are favorable for RHINO stability will therefore be necessary to fully elucidate the function of RHINO in ATR-mediated DNA damage checkpoint signaling in human cells.
Materials and Methods
Cell lines—Human Flp-In T-REx 293 (Invitrogen) and mouse NIH2/4 (NIH3T3 cells containing LacO arrays)Citation40 were cultured at 37°C in a 5% CO2 humidified incubator in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin as previously described.Citation40,41 Flp-In T-REx 293 cell lines expressing FLAG-Rad9 and FLAG-RHINO were generated using protocols provided by the manufacturer (Invitrogen). FLAG-Rad9 and FLAG-RHINO expression was induced by addition of tetracycline (1 μg/ml) to the medium for 2 d prior to experimentation. To expose cells to UV radiation, cells were placed under a GE germicidal lamp that emits primarily 254-nm UV light (UV-C) connected to a digital timer. Following irradiation, the cells were incubated for the indicated periods of time before harvesting.
Plasmid constructs—RHINO cDNA was obtained from the Human ORFeome clone set and PCR sub-cloned into pFastBac1 and pcDNA3 for expression in insect and mammalian cells. For generation of Flp-In T-REx 293 cell lines, the Rad9 and RHINO cDNAs were sub-cloned into pcDNA5/FRT/TO (Invitrogen).
Chemicals and antibodies—Hydroxyurea (HU; 10 mM) and cytosine β-D-arabinofuranoside (AraC; 100 μM) were purchased from Sigma and were added to cell culture medium, where indicated, 30 min prior to UV irradiation. Anti-FLAG affinity gel (Sigma A2220) was used to immunoprecipitate FLAG-Rad9 and FLAG-RHINO. Custom anti-RHINO antisera was generated by injecting rabbits with 6xHis-tagged RHINO produced in E. coli (Cocalico Biologicals Inc.) and was used to detect RHINO expression by immunoblotting. Additional antibodies used for immunoblotting included anti-MEK2 (catalog no. 610235) from BD Transduction Laboratories; anti-RPA70 (catalog no. A300-241A) from Bethyl Laboratories; anti-TopBP1 (catalog no. AB3245) from Millipore; anti-FLAG (catalog no. F-3165) from Sigma; anti-His antibody (catalog no. AM1010a) from Abgent; anti-Rad9 (catalog no. sc-8324), anti-Rad1 (catalog no. sc-22783), and anti-Chk1 (catalog no. sc-8408) from Santa Cruz Biotechnology; and anti-phospho-Chk1 (Ser345) (catalog no. 2348) from Cell Signaling Technology. Secondary antibodies included horseradish peroxidase-linked anti-mouse and anti-rabbit IgG (catalog nos. NA931V and NA934V) from GE Healthcare.
Human cell extracts and fractionation—Following UV irradiation or other treatments, cells were harvested by scraping the cells from the plate into cold PBS. Cells were then pelleted by gentle centrifugation (800 × g, 5 min, 4°C) and washed twice with cold PBS. Cells were then either frozen on dry ice and stored at −80×C or directly used for preparation of cell extracts. Whole cell lysates for immunoprecipitation were prepared by lysing the cells in TBS (50 mM Tris pH 7.4, 150 mM NaCl) containing 0.5% NP-40 substitute Igepal CA-630, 0.1 mM PMSF and phosphatase inhibitors (10 mM NaF, 1 mM Na2VO3, and 10 mM glycerophosphate), sonication to shear chromatin, and centrifugation at maximum speed in a microcentrifuge for 15 min at 4°C. For preparation of soluble and insoluble chromatin fractions, cells were resuspended in Buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 10% glycerol, 0.34 M sucrose, 1 mM DTT, 0.1 mM PMSF) containing 0.1% Triton X-100 and phosphatase inhibitors. Cells were extracted 3 times with Buffer A containing Triton X-100 to separate the soluble cytosolic and nuclear material from the chromatin faction. Soluble cytosolic extracts were centrifuged for 15 min at maximum speed in a microcentrifuge at 4°C to remove any contaminating nuclei. DNA/nucleic acid-associated proteins were solubilized by a 5 min incubation with Benzonase (250 U; Sigma) at 37°C in Buffer A containing 5 mM MgCl2. The remaining chromatin pellet was solubilized by sonication in 1X SDS-PAGE sample buffer (50 mM Tris pH 6.8, 100 mM DTT, 1% SDS, 5% glycerol, and 0.005% bromophenol blue).
Immunoprecipitation—Cell extracts were incubated on a rotary device at 4°C with anti-FLAG agarose (Sigma) for 2–4 hr to immunoprecipitate (IP) the indicated FLAG-tagged proteins. IPs were washed 3 times with TBS containing 0.5% Igepal CA-630. To elute immunoprecipitated proteins, the immunocomplexes were incubated for 2 hr at 4°C in TBS containing 200 μg/ml FLAG peptide (Sigma F-3290) or were boiled for 5 min in 1X SDS-PAGE sample buffer.
Baculovirus vectors and expression—Baculoviruses expressing Rad9 (full-length and fragments), Rad1, Hus1, and Rad17 were previously described.Citation5,19,49 Baculoviruses for expressing FLAG-RHINO, His-RHINO, FLAG-PCNA, and FLAG-TopBP1 were generated using the Bac-to-Bac expression system (Invitrogen).
Purification of RHINO-9-1-1 complex—Insect cells were infected with baculoviruses expressing FLAG-Rad9, His-RHINO, and untagged Rad1 and Hus1. Infected cells were lysed in Buffer B (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 10% glycerol, 0.1% NP-40, 1% Tween-20, and 2.5 mM β-mercaptoethanol) and centrifuged to pellet insoluble material. The cell lysate was then incubated and rotated with Ni-NTA agarose (Qiagen) overnight at 4°C, washed extensively with Nickel Wash Buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 10% glycerol, 10 mM imidazole, and 2.5 mM β-mercaptoethanol), and then bound proteins were eluted with Nickel Elution Buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 10% glycerol, 400 mM imidazole, and 2.5 mM β-mercaptoethanol). The elution fractions were then incubated with anti-FLAG affinity gel for 3 hr at 4°C, washed with FLAG wash buffer (50 mM Tris-HCl, 100 mM NaCl, 10% glycerol, and 5 mM β-mercaptoethanol), and eluted with the same buffer containing 250 μg/ml FLAG peptide.
Plasmid and siRNA transfection—Control (sc-37007), RHINO (sc-95847), and Rad9 (sc-36365) siRNAs were obtained from Santa Cruz Biotechnology and were transfected with Lipofectamine RNAiMax according to the manufacturer's recommendations (Invitrogen). Cell lines were transfected twice (16 and 40 hr after plating) with siRNAs at a concentration of 40 nM and were analyzed approximately 24 hr after the second transfection.
Plasmid constructs expressing LacR-fusion proteins were transfected into NIH2/4 cells with Lipofectamine 2000 as previously described.Citation41
Immunoblotting—Proteins from immunoprecipitations and cell lysates were subjected to SDS-PAGE and then transferred to Hybond ECL membranes (GE Healthcare). The blots were probed with the indicated antibodies and incubated with Clarity Western ECL Substrate (Bio-Rad) before exposure to X-ray film or visualization with a Molecular Imager Chemi-Doc XRS+ system (Bio-Rad).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by National Institutes of Health grant GM32833 (A.S.) and T32CA009156 (C.C.).
References
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 2004; 73:39-85; PMID:15189136; http://dx.doi.org/10.1146/annurev.biochem.73.011303.073723
- Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 2004; 3:1009-14; PMID:15279787; http://dx.doi.org/10.1016/j.dnarep.2004.03.032
- Helt CE, Wang W, Keng PC, Bambara RA. Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle 2005; 4:529-32; PMID:15876866; http://dx.doi.org/10.4161/cc.4.4.1598
- Broustas CG, Lieberman HB. Contributions of Rad9 to tumorigenesis. J Cell Biochem 2012; 113:742-51; PMID:22034047; http://dx.doi.org/10.1002/jcb.23424
- Griffith JD, Lindsey-Boltz LA, Sancar A. Structures of the human Rad17-replication factor C and checkpoint rad 9-1-1 complexes visualized by glycerol spraylow voltage microscopy. J Biol Chem 2002; 277:15233-6; PMID:11907025; http://dx.doi.org/10.1074/jbc.C200129200
- Shiomi Y, Shinozaki A, Nakada D, Sugimoto K, Usukura J, Obuse C, Tsurimoto T. Clamp and clamp loader structures of the human checkpoint protein complexes, Rad9-1-1 and Rad17-RFC. Genes Cells 2002; 7:861-8; PMID:12167163; http://dx.doi.org/10.1046/j.1365-2443.2002.00566.x
- Kemp M, Sancar A. DNA distress: just ring 9-1-1. Curr Biol 2009; 19:R733-4; PMID:19833574; http://dx.doi.org/10.1016/j.cub.2009.07.026
- Dore AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex–implications for clamp loading and regulation. Mol Cell 2009; 34:735-45; PMID:19446481; http://dx.doi.org/10.1016/j.molcel.2009.04.027
- Xu M, Bai L, Gong Y, Xie W, Hang H, Jiang T. Structure and functional implications of the human rad9-hus1-rad1 cell cycle checkpoint complex. J Biol Chem 2009; 284:20457-61; PMID:19535328; http://dx.doi.org/10.1074/jbc.C109.022384
- Sohn SY, Cho Y. Crystal structure of the human rad9-hus1-rad1 clamp. J Mol Biol 2009; 390:490-502; PMID:19464297; http://dx.doi.org/10.1016/j.jmb.2009.05.028
- Mailand N, Gibbs-Seymour I, Bekker-Jensen S. Regulation of PCNA-protein interactions for genome stability. Nat Rev Mol Cell Biol 2013; 14:269-82; PMID:23594953; http://dx.doi.org/10.1038/nrm3562
- Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol 2004; 78:227-60; PMID:15210332; http://dx.doi.org/10.1016/S0079-6603(04)78006-X
- Dore AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex–implications for clamp loading and regulation. Mol Cell 2009; 34:735-45; PMID:19446481; http://dx.doi.org/10.1016/j.molcel.2009.04.027
- Friedrich-Heineken E, Toueille M, Tannler B, Burki C, Ferrari E, Hottiger MO, Hubscher U. The two DNA clamps Rad9Rad1Hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J Mol Biol 2005; 353:980-9; PMID:16216273; http://dx.doi.org/10.1016/j.jmb.2005.09.018
- Querol-Audi J, Yan C, Xu X, Tsutakawa SE, Tsai MS, Tainer JA, Cooper PK, Nogales E, Ivanov I. Repair complexes of FEN1 endonuclease, DNA, and Rad9-Hus1-Rad1 are distinguished from their PCNA counterparts by functionally important stability. Proc Natl Acad Sci U S A 2012; 109:8528-33; PMID:22586102; http://dx.doi.org/10.1073/pnas.1121116109
- Song W, Pascal JM, Ellenberger T, Tomkinson AE. The DNA binding domain of human DNA ligase I interacts with both nicked DNA and the DNA sliding clamps, PCNA and hRad9-hRad1-hHus1. DNA Repair (Amst) 2009; 8:912-9; PMID:19523882; http://dx.doi.org/10.1016/j.dnarep.2009.05.002
- Wang W, Brandt P, Rossi ML, Lindsey-Boltz L, Podust V, Fanning E, Sancar A, Bambara RA. The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc Natl Acad Sci U S A 2004; 101:16762-7; PMID:15556996; http://dx.doi.org/10.1073/pnas.0407686101
- Wang W, Lindsey-Boltz LA, Sancar A, Bambara RA. Mechanism of stimulation of human DNA ligase I by the Rad9-rad1-Hus1 checkpoint complex. J Biol Chem 2006; 281:20865-72; PMID:16731526; http://dx.doi.org/10.1074/jbc.M602289200
- Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci U S A 2003; 100:1633-8; PMID:12578958; http://dx.doi.org/10.1073/pnas.0437927100
- Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol 2003; 1:E33; PMID:14624239; http://dx.doi.org/10.1371/journal.pbio.0000033
- Majka J, Binz SK, Wold MS, Burgers PM. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem 2006; 281:27855-61; PMID:16864589; http://dx.doi.org/10.1074/jbc.M605176200
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 2007; 21:1472-7; PMID:17575048; http://dx.doi.org/10.1101/gad.1547007
- Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem 2007; 282:28036-44; PMID:17636252; http://dx.doi.org/10.1074/jbc.M704635200
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell 2006; 124:943-55; PMID:16530042; http://dx.doi.org/10.1016/j.cell.2005.12.041
- Choi JH, Lindsey-Boltz LA, Sancar A. Reconstitution of a human ATR-mediated checkpoint response to damaged DNA. Proc Natl Acad Sci U S A 2007; 104:13301-6; PMID:17686975; http://dx.doi.org/10.1073/pnas.0706013104
- Choi JH, Lindsey-Boltz LA, Sancar A. Cooperative activation of the ATR checkpoint kinase by TopBP1 and damaged DNA. Nucleic Acids Res 2009; PMID: 19139065; http://dx.doi.org/10.1093/nargkn1075
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 2008; 9:616-27; PMID:18594563; http://dx.doi.org/10.1038/nrm2450
- Navadgi-Patil VM, Burgers PM. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1ATR via two distinct mechanisms. Mol Cell 2009; 36:743-53; PMID:20005839; http://dx.doi.org/10.1016/j.molcel.2009.10.014
- Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell 2006; 24:891-901; PMID:17189191; http://dx.doi.org/10.1016/j.molcel.2006.11.027
- Cotta-Ramusino C, McDonald ER 3rd, Hurov K, Sowa ME, Harper JW, Elledge SJ. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science 2011; 332:1313-7; PMID:21659603; http://dx.doi.org/10.1126/science.1203430
- Kim JW, Fukukawa C, Ueda K, Nishidate T, Katagiri T, Nakamura Y. Involvement of C12orf32 overexpression in breast carcinogenesis. Int J Oncol 2010; 37:861-7; PMID:20811708
- Choi JH, Lindsey-Boltz LA, Kemp M, Mason AC, Wold MS, Sancar A. Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proc Natl Acad Sci U S A 2010; 107:13660-5; PMID:20616048; http://dx.doi.org/10.1073/pnas.1007856107
- Hassan BH, Lindsey-Boltz LA, Kemp MG, Sancar A. Direct role for the replication protein treslin (ticrr) in the ATR kinase-mediated checkpoint response. J Biol Chem 2013; 288:18903-10; PMID:23696651; http://dx.doi.org/10.1074/jbc.M113.475517
- Lindsey-Boltz LA, Sercin O, Choi JH, Sancar A. Reconstitution of human claspin-mediated phosphorylation of Chk1 by the ATR (ataxia telangiectasia-mutated and rad3-related) checkpoint kinase. J Biol Chem 2009; 284:33107-14; PMID:19828454; http://dx.doi.org/10.1074/jbc.M109.064485
- Lindsey-Boltz LA, Reardon JT, Wold MS, Sancar A. In vitro analysis of the role of replication protein A (RPA) and RPA phosphorylation in ATR-mediated checkpoint signaling. J Biol Chem 2012; 287:36123-31; PMID:22948311; http://dx.doi.org/10.1074/jbc.M112.407825
- Lindsey-Boltz LA, Kemp MG, Reardon JT, DeRocco V, Iyer RR, Modrich P, Sancar A. Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system. J Biol Chem 2014; 289:5074-82; PMID:24403078; http://dx.doi.org/10.1074/jbc.M113.542787
- Duursma AM, Driscoll R, Elias JE, Cimprich KA. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell 2013; 50:116-22; PMID:23582259; http://dx.doi.org/10.1016/j.molcel.2013.03.006
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. The Mre11-Rad50-Nbs1 complex mediates activation of TopBP1 by ATM. Mol Biol Cell 2009; 20:2351-60; PMID:19279141; http://dx.doi.org/10.1091/mbc.E08-12-1190
- Wang J, Gong Z, Chen J. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J Cell Biol 2011; 193:267-73; PMID:21482717; http://dx.doi.org/10.1083/jcb.201010026
- Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science 2008; 320:1507-10; PMID:18483401; http://dx.doi.org/10.1126/science.1159051
- Lindsey-Boltz LA, Sancar A. Tethering DNA damage checkpoint mediator proteins topoisomerase II{beta}-binding protein 1 (TopBP1) and claspin to DNA activates ataxia-telangiectasia mutated and RAD3-related (ATR) phosphorylation of checkpoint kinase 1 (Chk1). J Biol Chem 2011; 286:19229-36; PMID:21502314; http://dx.doi.org/10.1074/jbc.M111.237958
- Zhou ZW, Liu C, Li TL, Bruhn C, Krueger A, Min W, Wang ZQ, Carr AM. An essential function for the ATR-activation-domain (AAD) of TopBP1 in mouse development and cellular senescence. PLoS Genet 2013; 9:e1003702; PMID:23950734; http://dx.doi.org/10.1371/journal.pgen.1003702
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: Partners in checkpoint signaling. Science 2001; 294:1713-6; PMID:11721054; http://dx.doi.org/10.1126/science.1065521
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300:1542-8; PMID:12791985; http://dx.doi.org/10.1126/science.1083430
- Kemp MG, Akan Z, Yilmaz S, Grillo M, Smith-Roe SL, Kang TH, Cordeiro-Stone M, Kaufmann WK, Abraham RT, Sancar A, et al. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J Biol Chem 2010; 285:16562-71; PMID:20233725; http://dx.doi.org/10.1074/jbc.M110.110304
- Sercin O, Kemp MG. Characterization of functional domains in human claspin. Cell Cycle 2011; 10:1599-606; PMID:21478680; http://dx.doi.org/10.4161/cc.10.10.15562
- Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in xenopus egg extracts. Mol Cell 2000; 6:839-49; PMID:11090622; http://dx.doi.org/10.1016/S1097-2765(05)00092-4
- Kumagai A, Dunphy WG. Repeated phosphopeptide motifs in claspin mediate the regulated binding of Chk1. Nat Cell Biol 2003; 5:161-5; PMID:12545175; http://dx.doi.org/10.1038/ncb921
- Lindsey-Boltz LA, Bermudez VP, Hurwitz J, Sancar A. Purification and characterization of human DNA damage checkpoint rad complexes. Proc Natl Acad Sci U S A 2001; 98:11236-41; PMID:11572977; http://dx.doi.org/10.1073/pnas.201373498
