Abstract
HIRA is an evolutionarily conserved histone chaperone that mediates replication-independent nucleosome assembly and is important for a variety of processes such as cell cycle progression, development, and senescence. Here we have used a chromatin sequencing approach to determine the genome-wide contribution of HIRA to nucleosome organization in Schizosaccharomyces pombe. Cells lacking HIRA experience a global reduction in nucleosome occupancy at gene sequences, consistent with the proposed role for HIRA in chromatin reassembly behind elongating RNA polymerase II. In addition, we find that at its target promoters, HIRA commonly maintains the full occupancy of the −1 nucleosome. HIRA does not affect global chromatin structure at replication origins or in rDNA repeats but is required for nucleosome occupancy in silent regions of the genome. Nucleosome organization associated with the heterochromatic (dg-dh) repeats located at the centromere is perturbed by loss of HIRA function and furthermore HIRA is required for normal nucleosome occupancy at Tf2 LTR retrotransposons. Overall, our data indicate that HIRA plays an important role in maintaining nucleosome architecture at both euchromatic and heterochromatic loci.
Introduction
Nucleosome assembly is believed to occur in a step-wise manner whereby the deposition of an (H3-H4)2 tetramer is followed by the assembly of 2 flanking H2A-H2B dimers.Citation1,2 This process is regulated by a structurally diverse group of proteins termed histone chaperones.Citation1,2 Traditionally, these proteins have been classified as either H3-H4 or H2A-H2B chaperones based upon their histone binding specificity, although some chaperones such as FACT are able to bind both H3-H4 and H2A-H2B.Citation1 During S-phase nucleosomes are removed ahead of the replication fork and then reassembled onto newly synthesized DNA. However other processes such as transcription, recombination and repair also result in the loss of nucleosomes from DNA which necessitates histone chaperones that mediate replication–independent nucleosome assembly.Citation3 Furthermore, it is now established that in addition to their traditional assembly function, histone chaperones can also mediate nucleosome disassembly and histone exchange. Indeed the central role played by histone chaperones in nucleosome dynamics is becoming increasingly recognized.Citation2
The HIRA (or HIR) complex is an evolutionarily conserved H3-H4 histone chaperone that is implicated in a range of processes including embryonic development, angiogenesis, cellular senescence and aging.Citation4 The human complex is composed of HIRA in association with UBN1 and CABIN1Citation5-7 and similarly yeast HIRA proteins (Hir1 and Hir2 in Saccharomyces cerevisiae and Hip1 and Slm9 in Schizosaccharomyces pombe), are stably associated with orthologs of CABIN1 and UBN1.Citation8-11 HIRA co-operates with another H3-H4 chaperone, Asf1 to mediate replication-independent nucleosome assembly.Citation4 Consistent with this, in higher eukaryotes HIRA is associated with the histone variant H3.3 which is deposited into chromatin independently of DNA synthesis.Citation6
The modulation of chromatin structure by HIRA has been implicated in multiple aspects of transcriptional regulation. In some contexts HIRA is necessary for transcriptional activation. For example the induction of Vegfr1 in human endothelial cells in response to angiogenic signals is HIRA-dependent.Citation12 Similarly in fission yeast, HIRA subunits are recruited to promoters of specific genes in response to environmental stress. Inactivation of HIRA compromises nucleosome eviction and transcriptional induction at these genes.Citation13 Conversely, HIRA has also been shown to be required for transcriptional repression. S. cerevisiae Hir1 and Hir2 were initially characterized as repressors of histone gene expression,Citation14 a role which is conserved in other organisms.Citation15,16 Furthermore, HIRA is required for the integrity of silent chromatin in a variety of systems. In fission yeast HIRA/Asf1 spreads across heterochromatic regions via association with the Heterochromatin Protein (HP1) ortholog, Swi6, to maintain a silent state.Citation10,15,17 In human fibroblasts HIRA/Asf1a is required for the formation of senescence associated heterochromatin,Citation18 and furthermore HIRA interacts with PRC2 and is implicated in the maintenance of the repressive H3K27me mark at developmentally regulated genes in mouse embryonic stem cells.Citation19 HIRA also suppresses the expression of retroelements. Mutation in HIRA components alleviates silencing of S. pombe Tf2 LTR retrotransposonsCitation10,20 and human HIRA was revealed as one of a group of chromatin assembly factors that suppresses HIV-1 proviral expression to maintain latency.Citation21 Other studies have also suggested a global role for HIRA in transcriptional elongation and the suppression of cryptic promoters. In S. cerevisiae hir mutations are synthetically lethal when combined with mutations in the yFACT complex which facilitates transcription elongation.Citation22 Furthermore inactivation of the HIRA complex results in increased levels of spurious transcripts from cryptic promoters in ORFs.Citation20,23,24 The genomes of cells defective in HIRA function also exhibit increased accessibility to DNA damaging agents and nucleases.Citation20,25 Taken together the data indicate that the HIRA histone chaperone plays an important role in maintaining the global integrity of chromatin.Citation20,25 Given this we have identified the impact of the fission yeast HIRA complex on genome-wide nucleosome architecture. Using a chromatin-sequencing approachCitation26 we have mapped changes to nucleosome position and occupancy in cells lacking HIRA function. We find that HIRA is required for normal nucleosome occupancy over ORFs, at some promoters, and also at heterochromatic repeats. As such, HIRA plays an important role in the maintenance of global nucleosomal architecture.
Results
We employed a chromatin sequencing technologyCitation26 to determine the impact of the HIRA histone chaperone complex on genome-wide nucleosome occupancy and positioning. With this approach, chromatin is treated with micrococcal nuclease (MNase) to generate ladders of MNase-resistant DNA which is then subjected to sequencing. The resulting datasets are then stratified based on paired read end-to-end distance into ranges representing the expected sizes of MNase resistant DNA species in eukaryotic chromatin. Thus read pairs of 150 bp (+/− 20%) derive primarily from mono-nucleosomes, whereas read pairs of 300 bp (+/− 20%) derive from di-nucleosomes. Frequency distributions of the read midpoints can then be mapped to the genome and the peaks in these distributions used to infer the presence of positioned chromatin particles in the cell population.Citation26 Therefore chromatin derived from fission yeast cells lacking HIRA function (hip1Δ) was digested with MNase to generate a DNA ladder with a highly similar molecular weight distribution to our wild type control sample (). Three biological replicate samples were pooled and sequenced which generated data sets for wild type and hip1Δ comprising of 56.3 and 49.6 million reads, respectively.
Figure 1. Paired-end mode chromatin-seq of wild type and hip1Δ mutant S. pombe. (A) Ethidium-stained gel separation of DNA pools extracted from MNase digested S. pombe chromatin used for chromatin sequencing in this study. Mono-, di- and tri-nucleosomal bands are visible. (B) Frequency distribution of paired read end-to-end size values after chromatin-seq of DNA shown in (A). (C) Nucleosomes in wild type cells (wt chromatin) were defined as the positions of 150 bp size class particle frequency peak summits (frequency value >25). This procedure marked 60, 658 putative nucleosome positions in the S. pombe genome. The 150 bp size class particle frequency distribution centered on, and surrounding (+/−1200 bp) each of these positions was then summed and normalized to the average frequency value occurring in the +/−1200 bp window. The wavelength of the peak pattern should be equal to the S. pombe nucleosome repeat length. Comparison to a previously published MNase-treated naked DNA (genomic DNA) dataset and a wt chromatin data set Citation27 is shown.
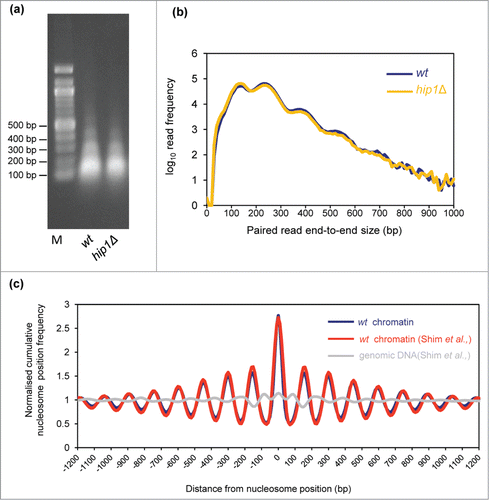
We first compared the average distribution of nucleosomes mapped in our wild-type data-set with that from a previously published study.Citation27 A cumulative frequency distribution of nucleosome position at, and surrounding nucleosome positions was plotted to assess how closely the datasets matched. This revealed that the distribution from our wild-type data set is coincident with the previously published wild-type nucleosome data-set and is distinct from a MNase-digested genomic DNA controlCitation27 (). These control comparisons, suggest that the nucleosome positions we map are accurate and that our wild-type data set agrees well with published work.
HIRA and the integrity of chromatin associated with ORFs
As HIRA has been linked to a variety of aspects of transcriptional control,Citation4 we examined the impact of deletion of hip1+ upon the chromatin surrounding the transcription start-sites (TSS) of protein coding genes. Typically chromatin in these regions is organized with a nucleosome depleted region (NDR) followed by a well ordered nucleosome array that extends from the TSS and packages the transcribed region.Citation28 In comparison, promoter regions are generally associated with lower nucleosome levels. shows a comparison of average nucleosome positions surrounding TSS in wild-type and hip1Δ cells. Loss of HIRA did not result in any changes to the NDR or the +1 nucleosome peak indicating that HIRA is not required for the maintenance of chromatin structure around the 5′ end of genes, at least at a global level. Nonetheless, the amplitudes of the nucleosome peaks from +4 onwards were reduced indicating that HIRA does contribute to the maintenance of chromatin associated with ORFs.
Figure 2. HIRA is required for normal nucleosome occupancy at Pol II transcribed genes. (A) Average nucleosome (150 bp size class particle) sequence read frequency profiles for 4013 S. pombe genes aligned at the transcription start site (TSS). (B) Whole cell extracts were subjected to western blotting with histone H3 (Abcam) and tubulin antibodies. An example of the primary data is shown along with a quantification of histone H3 levels normalized to tubulin (right). Data are the mean of 9 independent repeats and error bars represent ±SEM. *** indicates P < 0.001 t-test. (C) Strains, NT5 (wt), AW046 (hrp3Δ), SW700 (hip1Δ), CsG349 (hip1Δ hrp3Δ) were grown in YE5S medium until they reached an OD595 = 0.2–0.3. Cultures were subjected to five-fold serial dilution, spotted onto YE5S agar and incubated for 4 d at 30°C.
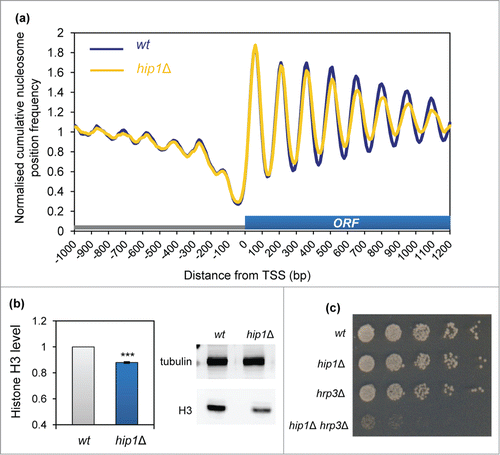
The reduction in the average peak height in the hip1Δ mutant was suggestive of a decrease in nucleosome occupancy and consistent with this view western blotting revealed a significant reduction in histone protein levels in cells lacking HIRA (). Based upon this finding we predicted that the hip1Δ allele would show a strong genetic interaction with mutations in hrp3+ which encodes a CHD ATP-dependent remodeler that controls nucleosome spacing.Citation27,29 Analysis of a hip1Δ hrp3Δ double mutant revealed that this strain was extremely slow growing and had severely elongated cell morphology (; Fig. S1) Therefore as predicted, loss of correct nucleosome spacing exacerbates the growth defects associated with HIRA inactivation.
HIRA suppresses aberrant transcription from the bodies of genesCitation20 and so the global perturbation to genic chromatin that is observed in the hip1Δ strain is consistent with this finding. To further investigate this we analyzed the nucleosome profiles of a group of genes which have been shown to produce cryptic transcripts when HIRA function is absent.Citation17,20 At the hrp1+ locus loss of HIRA function resulted in marked changes to the MNase profile which extended throughout the entire gene and into the neighboring genes (atg12+ and pap1+) (; Fig. S2). In contrast, the other genes we inspected exhibited relatively modest changes to their nucleosome profiles in the hip1Δ background (; Fig. S3). An example of this is the dbp7+ gene, where changes to nucleosome occupancy were mainly observed at the 3′-end of the gene and downstream of the transcription termination site. It therefore appears that relatively small changes to nucleosome architecture may be sufficient to result in increased levels of cryptic transcripts.
Figure 3. HIRA and nucleosome architecture in gene sequences. (A) Nucleosome (150 bp) sequence read frequency profiles of a 6.1 kb region of chromosome 1 (bp 2194720 to 2200820). The positions and orientation of the hrp1+, atg12+ and pap1+ genes are indicated below. (B) Nucleosome (150 bp) sequence read frequency profiles of the dbp7+ gene relative to the TSS. (C) The occupancy of specific nucleosomes was estimated by qPCR analysis of mononucleosomal DNA as described in the Materials and Methods. An equivalent amount of genomic DNA was analyzed as a control. The positions of the nucleosome peaks under analysis and the PCR primers are indicated in (A and B). The level of occupancy in hip1Δ relative to wild type is shown. Data is the mean of 2 technical qPCR repeats.
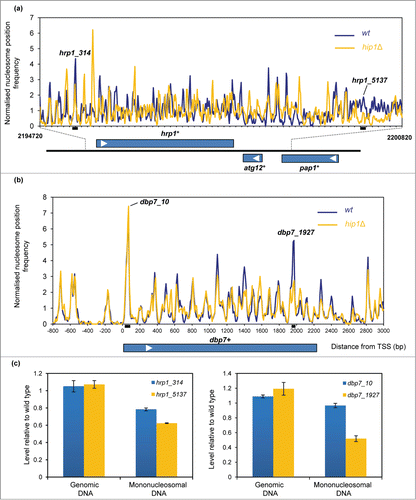
In order to confirm some of the differences in the nucleosome profiles that were observed in , mononucleosomal DNA was isolated from independent pools of MNase-digested DNA and a quantitative PCR (qPCR) approachCitation30 was used to compare the occupancy of some specific nucleosomes in the wild-type and hip1Δ samples. Similar to the results from the genome-wide mapping studies, the qPCR analysis also indicated that occupancy of nucleosomes near hrp1+ (designated hrp1_5137) and the 3′ end of the dbp7+ gene (dbp7_1927) were reduced in hip1Δ whereas the occupancy of nucleosome located at the 5′ (dbp7_10) was similar in wild type and hip1Δ.
Impact of HIRA on chromatin at promoters
In addition to the suppression of spurious transcription initiation, HIRA represses expression from numerous bona fide RNA polymerase II (Pol II) promoters. Indeed the expression of approximately 4% of fission yeast genes is increased by loss of HIRA function.Citation20 We first compared the chromatin organization of these ‘HIRA-repressed’ genes with the complete set of S. pombe coding genes. HIRA-repressed genes were found to have obvious differences in their chromatin organization as average nucleosome peaks associated with the coding sequences of these genes were poorly ordered and the height of the peaks was lower than the global average (). This suggests that the coding sequences of HIRA-repressed genes are associated with a lower than average nucleosome occupancy. Furthermore, the NDR of the HIRA-repressed gene set was both narrower and shallower when compared to the average promoter. These features are known to be characteristic of genes that have a low level of expression,Citation31 a finding which is in agreement with our previous microarray analyses which revealed that HIRA target genes overlap significantly with lowly expressed genes.Citation20 We next determined the impact of hip1+ deletion upon chromatin architecture of HIRA-repressed genes. Loss of hip1+ resulted in a reduction in the height of the −1 nucleosome peak and subtle shift in its position which suggests that HIRA promotes the proper occupancy of the −1 nucleosome, a finding which is consistent with the repressive function of HIRA at these promoters ().
Figure 4. Impact of HIRA on promoter nucleosome profiles. (A) Average nucleosome profiles for 4013 S. pombe genes aligned at the transcription start site (TSS) compared to the nucleosome sequence read frequency profile of a set of 107 HIRA-repressed protein-coding genes.Citation20 (B) Average nucleosome profiles of a set of 107 HIRA-repressed genes in wild type and hip1Δ cells. (C) Comparison of the nucleosome profile at the hht2+-hhf2+ locus in wild type and hip1Δ cells. The positions of the coding sequences are indicated by solid blue boxes while 5′ and 3′ UTRs are represented by open boxes. Positions of hht2+-hhf2+ transcription start-sites, termination sites and the AACCCT box are as described by Takayama and Takahashi.Citation30 (D) Occupancy of the hht2+-hhf2+ −1 nucleosome (−1 nuc) was determined by qPCR analysis of mononucleosomal DNA. The position of the PCR primers and peak are indicated in (C). The level of occupancy in hip1Δ relative to wild type is shown. Data is the mean of 2 technical qPCR repeats.
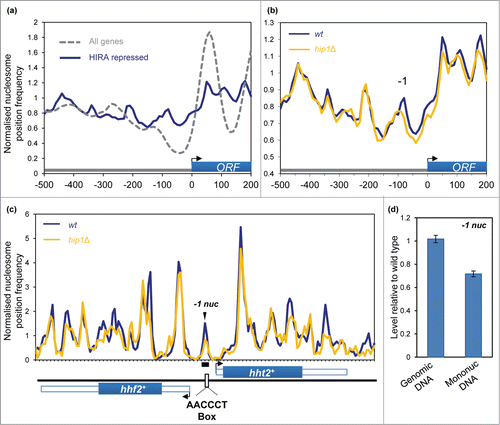
We next examined the histone H3-H4 genes hht2+-hhf2+ as their expression outside of S-phase is repressed in a HIRA-dependent manner.Citation32 hht2+-hhf2+ are divergently transcribed from a short promoter, and analysis of our MNase profiles revealed the presence of a −1 nucleosome peak in the center of this region. Since the majority of fission yeast cells in an asynchronous culture will be in G2, this chromatin configuration is likely to represent a repressed promoter state. Consistent with this hypothesis the promoter nucleosome peak occludes the AACCCT box which is a binding site for the GATA-type factor, Ams2 and the Myb domain protein, Teb1, which activate the transcription of histone genes.Citation32,33 Interestingly, both the MNase profiles and qPCR analysis of mononucleosomal DNA indicated that deletion of hip1+ resulted in a marked reduction of this peak, suggesting that HIRA promotes the occupancy of this promoter nucleosome to suppress the inappropriate expression of histone genes ().
HIRA is not required for chromatin architecture at Pol I- and Pol III-transcribed genes
As HIRA has a global impact upon the chromatin associated with Pol II-transcribed genes we examined whether it was also required for chromatin organization at Pol I and Pol III genes. We analyzed the chromatin configuration surrounding Pol III-transcribed tRNA genes. In S. cerevisiae tRNA genes are typically nucleosome free and flanked by nucleosomes positioned upstream (US) and downstream (DS).Citation34 In comparison, it has been suggested that many tRNA genes in S. pombe (like those in resting human CD4+ T cells) are associated with nucleosomes.Citation34 Plots of the average mono-nucleosome (150 bp) profile of 171 S. pombe tRNA genes aligned by TSS are consistent with this earlier report as we detected a peak centered at +20 relative to the TSS (). At this global level we were able to detect an upstream (US) nucleosome peak positioned at −160 bp but we found little evidence of a downstream nucleosome array. Loss of HIRA did not impact upon the US nucleosome although we did note some reduction in the height of the peak located at +20 bp. tRNA genes have internal promoter elements that are binding sites for TFIIIC which in turn directs the assembly of TFIIIB upstream of the transcription start-site. TFIIIB acts as the initiation factor by bringing Pol III to DNA.Citation35 In order to see if HIRA has any global impact upon Pol III transcription factor binding we examined the profile of 75 bp particles as it has been demonstrated that these particles result from the protection of DNA by transcription factors rather than by nucleosomes.Citation26 Comparison of average 75 bp profiles revealed the presence of a prominent peak immediately upstream of the TSS which given its position is likely to result from TFIIIB binding (). Loss of HIRA function did not impact upon this peak suggesting that it does not globally affect TFIIIB occupancy at tRNA genes.
Figure 5. Nucleosome architecture at Pol III and Pol (I)genes. (A) Average nucleosome (150 bp size class particle) sequence read frequency profiles for 171 S. pombe tRNA genes aligned at the transcription start site (TSS). (B) Average 75 bp size class particle sequence read frequency profiles for 171 S. pombe tRNA genes aligned at the transcription start site (TSS). (C) Nucleosome (150 bp size class particle) read profile over an rDNA repeat. The positions of the 28S, 5.8S and 18S rRNA genes are indicated.
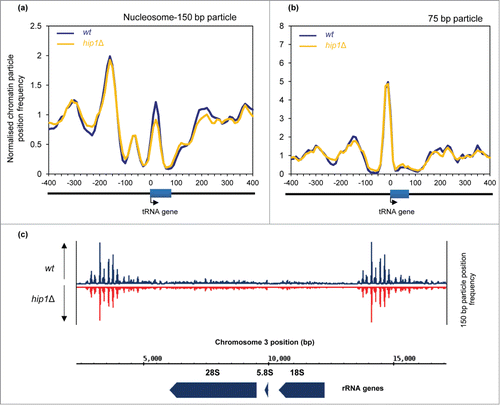
Recently, the HIR complex has been implicated in the repression of rDNA transcription in S. cerevisiae.Citation36 We therefore analyzed the MNase profiles of S. pombe rDNA repeat sequences. This suggested that Pol I-transcribed genes are nucleosome free while the intergenic regions are associated with well-positioned nucleosomes. However, loss of HIRA did not have a marked affect upon the MNase profiles of these regions () and so we find no evidence to suggest that HIRA is required for the maintenance of nucleosome architecture at Pol I genes in fission yeast. We also examined nucleosome profiles surrounding replication origins which were aligned as described previously.Citation37 In agreement with previous studies,Citation31,37 a wide nucleosome depleted region (NDR) was detectable over the origin center. This feature was also readily detectable in hip1Δ cells and indeed the average MNase profile of wild-type and hip1Δ cells surrounding origins was strikingly similar (Fig. S4) suggesting that HIRA does not contribute to the global organization of chromatin at replication origins.
Impact of HIRA upon nucleosome organization in silent chromatin
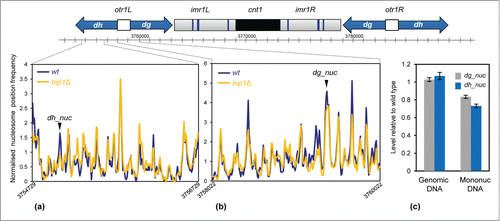
Loss of any one of the subunits of the HIRA complex alleviates heterochromatic silencing at the cryptic mating (mat) type locus and also at pericentromeric repeats.Citation8,10 These heterochromatin domains are enriched for methylation of histone H3 lysine 9 (H3K9me) which directs the assembly of chromodomain proteins such as the HP1 ortholog, Swi6.Citation38 Hip1 interacts with Swi6 and also the histone chaperone Asf1, which is required for nucleosome occupancy in heterochromatin.Citation17 We therefore examined the impact of hip1+ deletion upon the nucleosome profile at pericentromeric dg-dh repeats. This revealed that loss of HIRA resulted in changes to specific peaks rather than a uniform reduction in occupancy across the entire repeat region (). Consistent with this, qPCR analysis also indicated that occupancy of specific dg and dh nucleosomes (designated dh_nuc and dg_nuc) were reduced in the absence of HIRA (). This suggests that HIRA is required to maintain the proper occupancy of a subset of nucleosomes within heterochromatic domains and that this is required for transcriptional silencing in this region.
Figure 6. Loss of HIRA perturbs nucleosome architecture at centromeric repeats. (A and B) A schematic diagram of centromere 1 is shown along with the average nucleosome (150 bp) sequence read frequency profiles of the indicated regions of the dg and dh repeats. (C) The occupancy of specific dh and dg repeat nucleosomes was estimated by qPCR analysis of mononucleosomal DNA as described in the Materials and Methods. An equivalent amount of genomic DNA was analyzed as a control. The positions of the nucleosome peaks under analysis are indicated in (A and B). The level of occupancy in hip1Δ relative to wild type is shown. Data is the mean of 2 technical qPCR repeats.
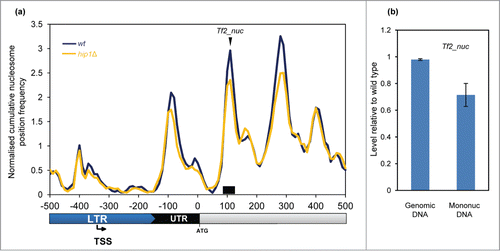
HIRA is also required for silencing the expression of all 13 intact Tf2 LTR retrotransposons.Citation20 The mechanism of silencing of these elements is distinct from heterochromatin as although it requires HIRA, it is independent of H3K9me.Citation10,39 Plots of average nucleosome profiles of these LTR retrotransposons showed that they have a nucleosome architecture which is distinct from typical RNA Pol II-transcribed genes. At Tf2 promoters (5′ LTRs), a peak overlapped the TSS and the NDR was located downstream (rather than upstream) of the TSS (). Interestingly, deletion of hip1+ had very little affect on the nucleosome peak adjacent to the TSS however we noted that hip1Δ cells had reduced +1, +2, and +3 nucleosome peaks (relative to the NDR). qPCR analysis also indicated that deletion of hip1+ resulted in a reduction in the occupancy of the +2 nucleosome (). These findings suggest that the nucleosomes downstream of the TSS may play a role in repression of Tf2 retrotransposons.
Figure 7. Nucleosome structure of Tf2 LTR retrotransposons. (A) Average nucleosome (150 bp) sequence read frequency profile for the 5′ region of Tf2 elements aligned relative to the translation start site (ATG). (B) The occupancy of a specific Tf2 nucleosome was estimated by qPCR analysis of mononucleosomal DNA as described in the Materials and Methods. An equivalent amount of genomic DNA was analyzed as a control. The position of the nucleosome peak and the PCR primers are indicated in (A). The level of occupancy in hip1Δ relative to wild type is shown. Data is the mean of 2 technical qPCR repeats.
Discussion
Here we have determined the global impact of HIRA upon nucleosome architecture and in agreement with previous evidence, find that this histone chaperone plays roles in the maintenance of both euchromatic and heterochromatic regions of the genome. We demonstrate that cells lacking HIRA (hip1Δ) experience a global reduction in nucleosome occupancy. This is consistent with previous studies which revealed that the genomes of fission yeast HIRA mutants are more accessible to DNA damaging agents.Citation20 Similarly in mammalian cells, HIRA depletion results in increased sensitivity of the genome to nucleases.Citation25
The co-ordinated replacement of nucleosomes that are displaced by elongating Pol II is necessary for maintaining the integrity of chromatin structures associated with gene sequences.Citation40 The finding that cryptic intragenic transcripts increase in the absence of HIRACitation20 is consistent with its proposed role in chromatin reassembly in the wake of Pol II. Our data adds further support to this hypothesis, as average MNase profiles revealed that hip1Δ cells have a global reduction in nucleosome occupancy which was most pronounced toward the 3′ end of gene sequences. Reduced levels of specific nucleosome peaks were also detectable at individual genes where loss of HIRA results in increased cryptic transcription. However there are also some regions of the genome which are more severely perturbed in the absence of HIRA. Indeed marked changes to both the occupancy and positioning of nucleosome peaks were evident surrounding the hrp1+ locus. Why specific regions of the genome show a greater dependency upon HIRA than others is currently not clear.
Comparison of average nucleosome plots aligned by TSS in wild-type and hip1Δ cells did not reveal a global impact of HIRA upon chromatin at promoters. However when we analyzed the profiles of a set of HIRA repressed genesCitation20 we found that hip1+ is required for the normal occupancy and positioning of the −1 nucleosome. This set of genes has a chromatin structure that is characteristic of lowly expressed genesCitation31 which is consistent with transcript profiling of HIRA mutant cells.Citation20 Taken together this suggests that HIRA commonly functions to maintain a closed/repressive chromatin structure at these genes. This was also evident at the hht2+-hhf2+ promoter where HIRA is required for full occupancy of the −1 nucleosome peak. S. pombe has 3 H3-H4 gene pairs however HIRA-mediated repression of histone gene expression is believed to operate predominantly through this gene pair.Citation32 Interestingly, the −1 nucleosome peak occludes the proposed binding site for the Ams2 and Teb1 activators suggesting that remodelling of this nucleosome may be required during transcription activation. Reduced occupancy of this nucleosome in the absence of HIRA would be expected to facilitate binding of activating transcriptions factors. While the expression of ams2+ is limited to G1/S,41 the expression of teb1+ is constitutive.Citation42 Therefore the absence of HIRA may allow increased Teb1 binding and thus expression of hht2+-hhf2+ outside of S phase. Our results suggest that HIRA is required for a ‘closed’ chromatin configuration at some promoters and similarly in S. cerevisiae Hir1 has been shown to be necessary for chromatin re-assembly at the PHO5 promoter during the switch from active transcription to repression.Citation43 While these results indicate that HIRA plays roles in promoting nucleosome occupancy, in other contexts it is also involved in mediating nucleosome eviction. HIRA subunits are recruited to specific stress-responsive promoters to facilitate nucleosome removal and gene induction.Citation13 Furthermore, the MNase profiles suggested that the occupancy of some nucleosomes is increased in the absence of HIRA. Therefore, in common with other histone chaperones, HIRA seems capable of mediating both nucleosome assembly and disassembly.Citation2
Chromodomain HP1 proteins such as Swi6 are hallmarks of heterochromatin. It has been proposed that these factors provide a platform for the assembly of other chromatin modifying proteins (including HDACs, ATP-dependent remodelers and histone chaperones) which enforce silencing of the underlying repeat sequences.Citation38 HIRA is one of these effectors as its correct localization at heterochromatic repeats is dependent upon Swi6.Citation17 Previous studies have shown that changes to nucleosome positioning perturb heterochromatin function.Citation44 Here we present data which indicates that the changes in nucleosome occupancy associated with loss of HIRA negatively impact upon heterochromatin silencing. Our MNase profiles indicate that loss of HIRA results in changes to specific nucleosomes rather than a uniform reduction across pericentromeric dg-dh repeats. Nonetheless, that cells lacking HIRA have increased levels of centromeric ncRNAs and defective trans-gene silencing,Citation10,15,17 implies that these changes are sufficient to impair heterochromatin function and allow increased access of Pol II to repeat sequences. We note that a similar situation has been reported for Hrp3 because loss of this CHD remodeler results in dysfunctional heterochromatin without producing dramatic changes upon nucleosome architecture.Citation27
HIRA has been linked to the regulation of both LTR retrotransposons and retroviruses.Citation20,45,46 That loss of HIRA function leads to a dramatic increase in expression of Tf2 LTR retrotransposons prompted comparison of MNase profiles of these elements. Tf2 5′-LTR regions are associated with a single nucleosome which overlaps the TSS. HIRA does not have an effect upon this nucleosome but is required for the full occupancy of the nucleosomes downstream of the transcription start-site. This suggests that chromatin structure in this region is important for maintaining silencing of Tf2 retrotransposons. Interestingly, analysis of HIV-1 expression has demonstrated that a nucleosome downstream of the TSS is important for mediating Pol II pausing and suppressing basal expression.Citation47 While the integrity of this region is dependent upon the FACT histone chaperone, other analyses revealed that HIRA is also required for the suppression of HIV-1 proviral expression and the maintenance of latency.Citation45 Given the parallels between Tf2 and HIV-1 it will be interesting to determine whether Pol II pausing is required for silencing of Tf2 elements.
Materials and Methods
S. pombe strains
Routine culture and genetic manipulation was performed as previously described.Citation48 The strains used in this study were 972 (h−), SW577 (h− hip1::ura4+), NT5 (h− ade6− ura4-D18 leu1–32), AW046 (h+ hrp3::kanMX ade6− leu1–32 ura4-D18), SW700 (h− hip1::ura4+ ade6− ura4-D18 leu1–32), CsG349 (h− hip1::ura4+ hrp3::kanMX ade6− ura4-D18 leu1–32).
Histone levels
Approximately ∼4 × 107 cells were harvested following addition of trichloroacetic acid (TCA) to a final concentration of 10%. Cells were resuspended in 200 μl 10% TCA and then disrupted using a beadbeater with 0.75 ml of glass beads using 2 pulses of 15 sec with 1 min on ice in between. A 500 μl aliquot of 10% TCA was added, the lysate was recovered from the beads which was then clarified by spinning at 13 000 rpm in a microcentrifuge. The resulting pellet was washed 3 times in acetone, dried and resuspended in 30 μl 100 mM Tris-HCl (pH 8.0), 1% w/v SDS, and 1 mM EDTA. Samples were analyzed on SDS-PAGE gels and subjected to protein gel blotting using anti-histone H3 (Abcam ab1791) and anti-tubulin (TAT-1) antibodies.
MNase digestion of chromatin
Cells (100 ml) were grown to OD595 = 0.75–8.0 in YE5S at 30°C, crosslinked for 20 min at 30°C using 1% formaldehyde and quenched by the addition of glycine to 125 mM. Cells were washed once in CES buffer (50 mM citric acid/50 mM Na2HPO4 [pH 5.6], 40 mM EDTA [pH 8.0], 1.2 M sorbitol and 10 mM β-mercaptoethanol) and resuspended in 500 μl of CES buffer with 0.5mg Zymolase 100-T. Cells were spheroplasted at 30°C for up to 1 h and then washed twice with ice cold 1.2 M sorbitol. Spheroplasts were then resuspended in 800 μl NP-S buffer (1.2 M sorbitol, 10 mM CaCl2, 100 mM NaCl, 1 mM EDTA pH 8.0, 14 mM β-mercaptoethanol, 50 mM Tris [pH 8.0], 0.075% NP-40, 5 mM spermidine, 0.1 mM PMSF, 1% Sigma Protease inhibitors cocktail [Sigma P8215]). Spheroplasts were then divided into 4 200 μl aliquots and each aliquot was mixed with 300 μl of NP-S buffer. Three aliquots were digested with between 75–187.5 units of MNase (USB) for 10 min at 37°C. The fourth was retained as an undigested control. MNase digestion was terminated by adding EDTA [pH 8.0] to a final concentration of 50 mM and SDS to 0.2%. Reactions were incubated at 65°C overnight with 0.2 mg/ml proteinase K and 10 μg RNAse. DNA was purified by extracting twice with phenol:chloroform followed by ethanol precipitation (0.1 volumes of 3 M sodium acetate followed by 2 volumes of ethanol). Pellets were washed in 70% ethanol and resuspended in water containing 10 μg/ml RNase and incubated at 37°C for 30 min. Triplicate digests were pooled and treated with 100 U unmodified T4 polynucleotide kinase (NEB) for 30 min at 37°C to remove 3′-phosphate groups left by MNase. DNA was extracted once more with phenol:chloroform, re-precipitated with sodium acetate and ethanol, washed with 70% ethanol, dried and re-suspended in TE (pH 7.5).
Chromatin-seq
DNA fragments were end repaired, 3′-adenylated and ligated to indexed adapters without size selection using Nextflex reagents (Newmarket Scientific, UK). Libraries were amplified with 8 cycles PCR using Kapa HiFi PCR master mix (Anachem), primers removed with GeneRead size selection protocol (QIAgen) before quantification by Bioanalyser DNA 7500 assay. Libraries were pooled, denatured, diluted to 6 nM before clustering in a single lane of a high output Illumina flowcell. Sequencing (100 nt) was undertaken on a HiSeq 2500 using TruSeq SBS v3 reagents (Illumina).
Bioinformatics
Paired reads were aligned to the ASM294v1.17 reference genome using Bowtie 0.12.7 Citation49 with command line flags: -n 0 –trim3 75 –maxins 5000 –fr -k 1 –sam. Aligned read pairs were sorted according to chromosome and then into a range of size classes based on the SAM format ISIZE value (difference between 5′ end of the mate read and the 5′ end of the first mapped read) plus or minus 20%. Mono-nucleosome-sized reads are, therefore, represented as 150 bp ± 30 bp. To define the genomic position of MNase-resistant chromatin entities we mapped the mid-point position of the read pairs in a particular size class. Frequency distributions of the mid-point positions were then calculated using 10 bp bins. Frequency distributions were lightly smoothed by taking a 3-bin moving average. All frequency distributions were output in the zero-referenced, chromosome base, 3-column .sgr format (chromosome number, feature/bin position, mid-point frequency value) for rendering with the Integrated Genome BrowserCitation50 and for further processing. Average cumulative chromatin particle position frequency distributions at, and surrounding, genomic features were calculated using the script SiteWriterCFD as described previously,Citation26,51 with values for each bin normalized to the average cumulative frequency value obtained for all bins within the feature window. To provide the comparison () of our data with the smoothed nucleosome position map of Shim and co-workersCitation27 the positions of 33874 unambiguous peak summit bins were marked in our wild type KDE mono-nucleosome data set (using script PeakMarkerEpKDE) and compared with GSM994397_WT.wig replicate and GSM994402_genomicDNA.wig data (converted to 10bp binned .sgr format). Protein-coding gene transcription start sites (TSS) positions were as described by Lantermann et al.Citation31 Replication origin positions were as described by Givens et al.Citation37
qPCR analysis of mononucleosomal DNA
MNase digests of wild type and hip1Δ cells were performed as described above. For each strain 3 biological replicate samples (independent from those used for sequencing) were pooled and analyzed on 1% TAE agarose gels. Gel slices containing mononucleosomal DNA were excised, frozen at -80°C and spun through 0.45 μM Spin-X columns (Costar). Samples were phenol extracted and ethanol precipitated and resuspended in TE (pH7.5). dsDNA concentration was measured using a Qubit fluorometer (Life Technologies). 20 ng of mononucleosomal DNA and was used in qPCR reactions using the PrimerDesign Mastermix kit. Reactions using the equivalent amount of genomic DNA were included as a control. The primers used for this analysis are listed in Table S1.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
967123_Supplementary_Materials.zip
Download Zip (1.7 MB)Funding
CG was supported by the Medical Research Council and the National Institute for Health Research (NIHR) Newcastle Biomedical Research Center based at Newcastle Upon Tyne Hospitals NHS Foundation Trust and Newcastle University. The views expressed are those of the authors and not necessarily those of the NHS, NIHR, or the Department of Health. KP and SKW acknowledge support from the Wellcome Trust Institutional Strategic Support Fund to the University of Exeter (WT097835MF) and Newcastle University (WT 097823/Z/11/Z), respectively.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol 2013; 20:14-22; PMID:23288364; http://dx.doi.org/10.1038/nsmb.2461
- Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol 2008; 18:282-9; PMID:18534842; http://dx.doi.org/10.1016/j.sbi.2008.04.003
- Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell 2010; 140:183-95; PMID:20141833; http://dx.doi.org/10.1016/j.cell.2010.01.004
- Amin AD, Vishnoi N, Prochasson P. A global requirement for the HIR complex in the assembly of chromatin. Biochimica Et Biophysica Acta 2013; 1819:264-76; PMID:24459729; http://dx.doi.org/10.1016/j.bbagrm.2011.07.008
- Banumathy G, Somaiah N, Zhang R, Tang Y, Hoffmann J, Andrake M, Ceulemans H, Schultz D, Marmorstein R, Adams PD. Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA-ASF1a chromatin-remodeling pathway in senescent cells. Mol Cell Biol 2009; 29:758-70; PMID:19029251; http://dx.doi.org/10.1128/MCB.01047-08
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 2004; 116:51-61; PMID:14718166; http://dx.doi.org/10.1016/S0092-8674(03)01064-X
- Balaji S, Iyer LM, Aravind L. HPC2 and ubinuclein define a novel family of histone chaperones conserved throughout eukaryotes. Mol Biosyst 2009; 5:269-75.
- Anderson HE, Kagansky A, Wardle J, Rappsilber J, Allshire RC, Whitehall SK. Silencing mediated by the Schizosaccharomyces pombe HIRA complex is dependent upon the Hpc2-like protein, Hip4. PLoS One 2010; 5:e13488; PMID:20976105; http://dx.doi.org/10.1371/journal.pone.0013488
- Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR 3rd, Kaufman PD. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol 2005; 15:2044-9; PMID:16303565; http://dx.doi.org/10.1016/j.cub.2005.10.053
- Greenall A, Williams ES, Martin KA, Palmer JM, Gray J, Liu C, Whitehall SK. Hip3 interacts with the HIRA proteins Hip1 and Slm9 and is required for transcriptional silencing and accurate chromosome segregation. J Biol Chem 2006; 281:8732-9; PMID:16428807; http://dx.doi.org/10.1074/jbc.M512170200
- Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL. The HIR corepressor complex binds to nucleosomes generating a distinct proteinDNA complex resistant to remodeling by SWISNF. Genes Dev 2005; 19:2534-9; PMID:16264190; http://dx.doi.org/10.1101/gad.1341105
- Dutta D, Ray S, Home P, Saha B, Wang S, Sheibani N, Tawfik O, Cheng N, Paul S. Regulation of angiogenesis by histone chaperone HIRA-mediated incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J Biol Chem 2010; 285:41567-77; PMID:21041298; http://dx.doi.org/10.1074/jbc.M110.190025
- Chujo M, Tarumoto Y, Miyatake K, Nishida E, Ishikawa F. HIRA, a conserved histone chaperone, plays an essential role in low-dose stress response via transcriptional stimulation in fission yeast. J Biol Chem 2012; 287:23440-50; PMID:22589550; http://dx.doi.org/10.1074/jbc.M112.349944
- Spector MS, Raff A, DeSilva H, Lee K, Osley MA. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol Cell Biol 1997; 17:545-52; PMID:9001207
- Blackwell C, Martin KA, Greenall A, Pidoux A, Allshire RC, Whitehall SK. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol Cell Biol 2004; 24:4309-20; PMID:15121850; http://dx.doi.org/10.1128/MCB.24.10.4309-4320.2004
- Hall IM, Noma K, Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci U S A 2003; 100:193-8; PMID:12509501; http://dx.doi.org/10.1073/pnas.232688099
- Yamane K, Mizuguchi T, Cui B, Zofall M, Noma K, Grewal SI. Asf1HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell 2011; 41:56-66; PMID:21211723; http://dx.doi.org/10.1016/j.molcel.2010.12.009
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell 2005; 8:19-30; PMID:15621527; http://dx.doi.org/10.1016/j.devcel.2004.10.019
- Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, Elsässer SJ, Chapgier A, Goldberg AD, Canaani E, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 2013; 155:107-20; PMID:24074864; http://dx.doi.org/10.1016/j.cell.2013.08.061
- Anderson HE, Wardle J, Korkut SV, Murton HE, Lopez-Maury L, Bahler J, Whitehall SK. The fission yeast HIRA histone chaperone is required for promoter silencing and the suppression of cryptic antisense transcripts. Mol Cell Biol 2009; 29:5158-67; PMID:19620282; http://dx.doi.org/10.1128/MCB.00698-09
- Gallastegui E, Millan-Zambrano G, Terme JM, Chavez S, Jordan A. Chromatin reassembly factors are involved in transcriptional interference promoting HIV latency. J Virol 2011; 85:3187-202; PMID:21270164; http://dx.doi.org/10.1128/JVI.01920-10
- Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, Rhoades AR, Kaufman PD, Stillman DJ. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the HirHpc pathway: polymerase passage may degrade chromatin structure. Genetics 2002; 162:1557-71; PMID:12524332
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 2008; 6:e277; PMID:18998772; http://dx.doi.org/10.1371/journal.pbio.0060277
- Nourani A, Robert F, Winston F. Evidence that Spt2Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol Cell Biol 2006; 26:1496-509; PMID:16449659; http://dx.doi.org/10.1128/MCB.26.4.1496-1509.2006
- Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LE, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell 2011; 44:928-41; PMID:22195966; http://dx.doi.org/10.1016/j.molcel.2011.12.006
- Kent NA, Adams S, Moorhouse A, Paszkiewicz K. Chromatin particle spectrum analysis: a method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic Acids Res 2011; 39:e26; PMID:21131275; http://dx.doi.org/10.1093/nar/gkq1183
- Shim YS, Choi Y, Kang K, Cho K, Oh S, Lee J, Grewal SI, Lee D. Hrp3 controls nucleosome positioning to suppress non-coding transcription in eu- and heterochromatin. EMBO J 2012; 31:4375-87; PMID:22990236; http://dx.doi.org/10.1038/emboj.2012.267
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 2009; 10:161-72; PMID:19204718; http://dx.doi.org/10.1038/nrg2522
- Pointner J, Persson J, Prasad P, Norman-Axelsson U, Stralfors A, Khorosjutina O, Krietenstein N, Svensson JP, Ekwall K, Korber P. CHD1 remodelers regulate nucleosome spacing in vitro and align nucleosomal arrays over gene coding regions in S. pombe. EMBO J 2012; 31:4388-403; PMID:23103765; http://dx.doi.org/10.1038/emboj.2012.289
- Infante JJ, Law GL, Young ET. Analysis of nucleosome positioning using a nucleosome-scanning assay. Methods Mol Biol 2012; 833:63-87; PMID:22183588; http://dx.doi.org/10.1007/978-1-61779-477-3_5
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol 2010; 17:251-7; PMID:20118936; http://dx.doi.org/10.1038/nsmb.1741
- Takayama Y, Takahashi K. Differential regulation of repeated histone genes during the fission yeast cell cycle. Nucleic Acids Res 2007; 35:3223-37; PMID:17452352; http://dx.doi.org/10.1093/nar/gkm213
- Valente LP, Dehe PM, Klutstein M, Aligianni S, Watt S, Bahler J, Promisel Cooper J. Myb-domain protein Teb1 controls histone levels and centromere assembly in fission yeast. EMBO J 2013; 32:450-60; PMID:23314747; http://dx.doi.org/10.1038/emboj.2012.339
- Kumar Y, Bhargava P. A unique nucleosome arrangement, maintained actively by chromatin remodelers facilitates transcription of yeast tRNA genes. BMC Genomics 2013; 14:402; PMID:23767421; http://dx.doi.org/10.1186/1471-2164-14-402
- Kassavetis GA, Geiduschek EP. Transcription factor TFIIIB and transcription by RNA polymerase III. Biochem Soc Trans 2006; 34:1082-7; PMID:17073756; http://dx.doi.org/10.1042/BST0341082
- Neumuller RA, Gross T, Samsonova AA, Vinayagam A, Buckner M, Founk K, Hu Y, Sharifpoor S, Rosebrock AP, Andrews B, et al. Conserved regulators of nucleolar size revealed by global phenotypic analyses. Sci Signaling 2013; 6:ra70; PMID:23962978; http://dx.doi.org/10.1126/scisignal.2004145
- Givens RM, Lai WK, Rizzo JM, Bard JE, Mieczkowski PA, Leatherwood J, Huberman JA, Buck MJ. Chromatin architectures at fission yeast transcriptional promoters and replication origins. Nucleic Acids Res 2012; 40:7176-89; PMID:22573177; http://dx.doi.org/10.1093/nar/gks351
- Reyes-Turcu FE, Grewal SI. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev 2012; 22:156-63; PMID:22243696; http://dx.doi.org/10.1016/j.gde.2011.12004
- Hansen KR, Burns G, Mata J, Volpe TA, Martienssen RA, Bahler J, Thon G. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol Cell Biol 2005; 25:590-601; PMID:15632061; http://dx.doi.org/10.1128/MCB.25.2.590-601.2005
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell 2007; 128:707-19; PMID:17320508; http://dx.doi.org/10.1016/j.cell.2007.01.015
- Chen ES, Saitoh S, Yanagida M, Takahashi K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol Cell 2003; 11:175-87; PMID:12535531; http://dx.doi.org/10.1016/S1097-2765(03)00011-X
- Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bähler J. Periodic gene expression program of the fission yeast cell cycle. Nat Genet 2004; 36:809-17; PMID:15195092; http://dx.doi.org/10.1038/ng1377
- Schermer UJ, Korber P, Horz W. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol Cell 2005; 19:279-85; PMID:16039596; http://dx.doi.org/10.1016/j.molcel.2005.05.028
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 2007; 128:491-504; PMID:17289569; http://dx.doi.org/10.1016/j.cell.2006.12.035
- Gallastegui E, Marshall B, Vidal D, Sanchez-Duffhues G, Collado JA, Alvarez-Fernandez C, Luque N, Terme JM, Gatell JM, Sánchez-Palomino S, et al. Combination of biological screening in a cellular model of viral latency and virtual screening identifies novel compounds that reactivate HIV-1. J Virol 2012; 86:3795-808; PMID:22258251; http://dx.doi.org/10.1128/JVI.05972-11
- Qian Z, Huang H, Hong JY, Burck CL, Johnston SD, Berman J, Carol A, Liebman SW. Yeast Ty1 retrotransposition is stimulated by a synergistic interaction between mutations in chromatin assembly factor I and histone regulatory proteins. Mol Cell Biol 1998; 18:4783-92; PMID:9671488
- Vanti M, Gallastegui E, Respaldiza I, Rodriguez-Gil A, Gomez-Herreros F, Jimeno-Gonzalez S, Jordan A, Chávez S. Yeast genetic analysis reveals the involvement of chromatin reassembly factors in repressing HIV-1 basal transcription. PLoS Genet 2009; 5:e1000339; PMID:19148280; http://dx.doi.org/10.1371/journal.pgen.1000339
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 1991; 194:795-823; PMID:2005825; http://dx.doi.org/10.1016/0076-6879(91)94059-L
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10:R25; PMID:19261174; http://dx.doi.org/10.1186/gb-2009-10-3-r25
- Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 2009; 25:2730-1; PMID:19654113; http://dx.doi.org/10.1093/bioinformatics/btp472
- Maruyama H, Harwood JC, Moore KM, Paszkiewicz K, Durley SC, Fukushima H, Atomi H, Takeyasu K, Kent NA. An alternative beads-on-a-string chromatin architecture in Thermococcus kodakarensis. EMBO Rep 2013; 14:711-7; PMID:23835508; http://dx.doi.org/10.1038/embor.2013.94
