Abstract
We have previously reported the identification of a novel WD-domain protein, STRAP that plays a role in maintenance of mesenchymal morphology by regulating E-cadherin and that enhances tumorigenicity partly by downregulating CDK inhibitor p21Cip1. However, the functional mechanism of regulation of E-cadherin and p21Cip1 by STRAP is unknown. Here, we have employed STRAP knock out and knockdown cell models (mouse embryonic fibroblast, human cancer cell lines) to show how STRAP downregulates E-cadherin and p21Cip1 by abrogating the binding of Sp1 to its consensus binding sites. Moreover, ChIP assays suggest that STRAP recruits HDAC1 to Sp1 binding sites in p21Cip1 promoter. Interestingly, loss of STRAP can stabilize Sp1 by repressing its ubiquitination in G1 phase, resulting in an enhanced expression of p21Cip1 by >4.5-fold and cell cycle arrest. Using Bioinformatics and Microarray analyses, we have observed that 87% mouse genes downregulated by STRAP have conserved Sp1 binding sites. In NSCLC, the expression levels of STRAP inversely correlated with that of Sp1 (60%). These results suggest a novel mechanism of regulation of E-cadherin and p21Cip1 by STRAP by modulating Sp1-dependent transcription, and higher expression of STRAP in lung cancer may contribute to downregulation of E-cadherin and p21Cip1 and to tumor progression.
Abbreviations
| STRAP | = | serine threonine kinase receptor-associated protein |
| Sp1 | = | specificity protein 1 |
| Sp/KLF | = | specificity protein/Krüppel-like factor |
| SWI/SNF | = | SWItch/Sucrose nonfermentable |
| p300/CBP | = | p300/ CREB-binding protein |
| TSS | = | transcription start site |
| MEF | = | mouse embryonic fibroblast |
| HNF4 | = | hepatocyte nuclear factor 4 |
| TSA | = | trichostatin A |
| HDAC1 | = | histone deacetylase 1 |
| HDAC2 | = | histone deacetylase 2 |
| HDAC3 | = | histone deacetylase 3 |
| NF-YA | = | nuclear transcription factor Y subunit alpha |
| TβR I | = | II, TGF-β receptor I, II |
| RNase | = | A ribonuclease A |
| CDK2 | = | cyclin-dependent kinase 2 |
| CDK4 | = | cyclin-dependent kinase 4 |
| PARP | = | poly (ADP-ribose) polymerase |
| RhoA | = | Ras homolog gene family, member A |
Introduction
The ubiquitously expressed transcription factor Sp1 (specificity protein 1) is the first identified member of the Sp/KLF family of mammalian transcription factors.Citation1 Within KLF family the nine Sp members are distinguished by the presence of Buttonhead (BTD) domain on the N-terminal side of the DNA binding domain. Sp proteins play a important role in embryonic and early postnatal development. Sp1, Sp2, Sp3 and Sp4, which have similar modular structure, are a subgroup of the Sp members. Sp1, Sp3 and Sp4 are highly expressed in tumors and cancer cell lines. Sp1 recognizes and binds GC-rich sites of target gene promoters via three Cys2-His2 zinc finger motifs localized at its carboxyl terminus.Citation2 Sp1 binds individual Sp1 binding sites also as a multimer and is capable of synergistic activation of promoters containing multiple binding sites.Citation3 Sp1 interact directly or indirectly with transcription factors, transcriptional regulators and chromatin remodeling factors (e.g. estrogen receptor (ER) a, HDAC1, p300/CBP, SWI/SNF) to activate or repress gene expression,Citation4 thus it regulates the transcriptional activity of many genes involved in a wide range of biological processes including metabolism, cell growth, differentiation, angiogenesis, apoptosis, and immune response.Citation5-7
We have previously reported the identification of a novel WD40 domain-containing protein, STRAP (serine threonine kinase receptor-associated protein), which interacts with both TβRI and TβRII and negatively regulates TGF-β-induced gene expression. STRAP associates with Smad7, recruits it from the cytosol to the activated TβRI, stabilizes the heteromeric complex, and thus assists Smad7 in preventing Smad2 and Smad3 activation by the receptor complex.Citation8 WD40 domain-containing proteins, in general, seem to serve regulatory functions in various cellular processes, such as signal transduction, transcriptional regulation, RNA processing, vesicular trafficking, and cell cycle progression.Citation9-11 There is growing evidence to suggest that STRAP exerts its tumorigenic influence on cells, largely through TGF-ß-independent signaling. STRAP has been shown to be strong predictive marker of 5-fluorouracil-based adjuvant chemotherapy benefit in colorectal cancer and is up-regulated mostly in transformed epithelium in human colorectal and lung carcinomas.Citation12 STRAP activates mitogen activated protein (MAP) kinase (MAPK)/ extracellular signal-regulated kinase (ERK) pathway.Citation12 STRAP inhibits the transactivation function of EWS (Ewing Sarcoma Protein) by displacing p300 from the functional transcriptional complex.Citation13
We have previously reported that STRAP is involved in maintaining mesenchymal morphology by regulating E-cadherin and that it enhances tumorigenicity partly by downregulating CDK inhibitor p21Cip1.Citation9,10,12 but the functional mechanism of regulation of E-cadherin and p21Cip1 by STRAP is unknown. Homozygous deletion of STRAP gene in mice resulted in embryonic lethality between embryonic day (E) 10.5 and 12.5 due to the defects in angiogenesis, cardiogenesis, somitogenesis, neural tube closure and embryonic turning.Citation14 This wide variety of functions of STRAP suggests a broader role for it in tumorigenesis and development. As Sp1 is involved in so many biological functions during development and tumorigenesis by interacting with a large variety of proteins, it is tempting to hypothesize that STRAP and Sp1 could be somehow interconnected. However, nothing is known about how STRAP might regulate the function of Sp1 and vice versa. In this study, we set out to investigate the role of STRAP with oncogenic properties in the regulation of E-cadherin and p21Cip1 by modulating Sp1-dependent transcription. We find that STRAP inhibits the transactivation function of Sp1 either by directly blocking its DNA binding domain or destabilizing Sp1 protein through ubiquitin-proteasome pathway in cell cycle G1 phase. Notably, our observations in the cell culture studies have been supported by Microarray data and immunohistochemical analyses of non-small cell lung cancer specimens.
Results
STRAP inhibits Sp1-dependent activation of E-cadherin promoter
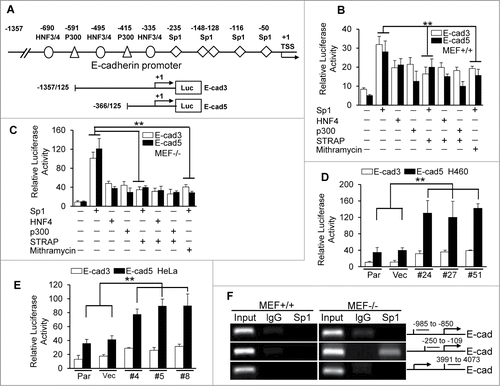
It has been shown that E-cadherin can be regulated at multiple levels including synthesis, processing and stability of mRNA; synthesis and stability of protein; localization and posttranslational modification.Citation15-16 We have previously reported STRAP plays a role in maintenance of mesenchymal morphology by downregulating E-cadherin in mRNA levels,Citation9 which raises a possibility that STRAP might interrupt some transcription factor(s) binding to the promoter resulting in reduced transcription. So we first decided to analyze the mechanism of regulation of E-cadherin by STRAP. The proximal 1.3 kb of the human E-cadherin promoter contains consensus binding sites of different transcription factors, including HNF family members, p300, and Sp1 ().Citation17 To test whether these factors could activate the E-cadherin promoter, we performed transient transfection analyses with two E-cadherin promoter constructs (E-cad3 and E-cad5) and the expression vectors as indicated. We observed a significant increase in reporter activity in the presence of Sp1 than that in the presence of p300 and HNF4 () in MEF+/+ cells when compared with the corresponding luciferase vector control. Co-expression of STRAP decreased Sp1-induced E-cadherin promoter activity by around 1.5-fold (). In contrast, STRAP had no effect on HNF4- and p300-induced promoter activity. In similar experiments with STRAP−/− MEFs, Sp1 strongly induced E-cadherin promoter activities (> 2.5-fold) compared to other transcription factors, and STRAP inhibited the effect of Sp1 by >3-fold (). These results suggest that the activity of the E-cadherin promoter highly depends on Sp1 binding to its promoter and STRAP can directly inhibit this response, which is similar to the effect of Mithramycin, a Sp1-DNA binding inhibitor. For verification of this effect of STRAP in epithelial tumor-derived cells, we established STRAP knock-down clones using H460 and HeLa cell lines (Fig. S1B and C). We observed that Sp1 induced both reporter activities, which is further enhanced in STRAP knock-down clones (), suggesting an inhibitory role of endogenous STRAP in Sp1-dependent transcription. To test whether STRAP has effect on Sp1 binding to the endogenous E-cadherin locus, we performed ChIP experiments using anti-Sp1 antibody and analyzed Sp1 recruitment to the E-cadherin promoter. Our data revealed an enrichment of Sp1 to the E-cadherin proximal promoter in STRAP−/− MEF cells () contrary to STRAP+/+ cells. To confirm the role of STRAP on Sp1/DNA binding, we performed DNA Affinity Precipitation Assay (DAPA) by immunoprecipitating the complex with biotinylated oligos containing Sp1 binding site in TGF-ß type II receptor from MEFs and then immunoblotting with anti-Sp1 antibody. We observed more Sp1 enrichment on its consensus DNA-binding sequence of TβRII promoter in the absence of STRAP (Fig. S1D). These results suggest that STRAP plays a crucial role in deregulation of Sp1-dependent activation of E-cadherin expression.
Figure 1. STRAP inhibits transcriptional activation of E-cadherin through Sp1. (A) The human E-cadherin promoter with the positions of potential transcription factor binding sites and transcription start site (TSS) has been shown. The luciferase reporters used are shown, together with base pair numbers relative to the TSS. (B) Wild type MEFs were transfected with two E-cadherin reporters and the expression plasmids for Sp1, HNF4 and p300 together with STRAP vector as indicated. Sp1 inhibitor, Mithramycin, was used to treat cells for 24 hours before harvesting. Luciferase activity was normalized to β-Gal activity and presented as mean ± sd from triplicate luciferase values. (C) Similar experiment was repeated with STRAP null MEFs. (D and E) STRAP stable knock-down clones in H460 and HeLa cells were co-transfected with the indicated E-cadherin promoter reporters and Sp1 expression plasmid. Luciferase activity was normalized to β-Gal activity and presented as mean ± sd from triplicate luciferase values. (F) Anti-Sp1 antibody was used for ChIP assays. PCR amplification was done with upstream and downstream sequences in E-cadherin promoter as indicated. Every experiment was repeated at least three times. Significance levels were determined by Student's t test. ** P < 0.01, when compared with the corresponding control.
STRAP interacts with Sp1 in the nucleus through its C-terminus
Since STRAP appeared to inhibit Sp1-induced transcriptional response of E-cadherin, we wondered whether STRAP might interact with Sp1. To test this, 293T cells were transfected with expression constructs encoding HA-tagged Sp1 and Myc-tagged STRAP. Cell lysates were used for immunoprecipitation (IP) with anti-HA antibody and STRAP was detected in the immune complex of Sp1. In a reciprocal experiment, we observed that Sp1 was co-immunoprecipitated with STRAP (), indicating the association of these two proteins. However, STRAP binds with neither c-Myc nor E2F1 under similar conditions (data not shown), suggesting the specific interaction between STRAP and Sp1. To identify the specific region of Sp1 protein that is necessary for binding with STRAP, we co-transfected a series of deletion mutants of HA-Sp1 with Myc-STRAP in 293T cells as indicated followed by co-IP assay with anti-HA antibody and immunobloted with anti-Myc antibody (). Besides the full-length Sp1 (1–788), Sp1 (1– 688) and Sp1 (622-788) showed an interaction with STRAP, whereas, the interaction was completely abolished with Sp1 (1–293), Sp1 (1– 621), and Sp1 (1– 648) although the expression levels were comparable. Taken together, amino acids 648–688 in Sp1 are sufficient for binding with STRAP, which contains the zinc finger domain. We further assessed endogenous binding in a few cell lines, such as A549, ACC-LC176, Beas2B as well as 293T. Anti-Sp1 antibody was used to immunoprecipitate Sp1 from whole cell lysates and STRAP was detected in the immune complex, suggesting in vivo binding of these two proteins ().
Figure 2. STRAP interacts with Sp1 through its C-terminal DNA binding domain. (A) 293T cells were transfected with either HA-tagged Sp1 or Myc-tagged STRAP or both together for 48 hours. Cells were lysed and protein complexes were immunoprecipitated by anti-HA or anti-Myc antibodies. Specific co-precipitating protein bands are indicated with arrows and expression of proteins in the lysates is shown below. (B) 293T cells were co-transfected with HA epitope-tagged deletion constructs of Sp1 and Myc epitope-tagged STRAP as indicated. Lysates were subjected to anti-HA immunoprecipitation and analyzed by western blotting with anti-Myc antibody. Protein expression was tested by immunoblotting. (C) Sp1 was immunoprecipitated with anti-Sp1 antibody from lysates of indicated cell lines. Immune complexes were then analyzed by immunoblotting with anti-STRAP antibody. (D) The cytoplasmic and nuclear extracts from A549 cells were used for immunoprecipitation with anti-Sp1 antibody. Co-precipitated STRAP was detected by immunoblotting with anti-STRAP antibody. Subcellular extraction was monitored by western blot analyses. (E) MEF+/+ and MEF−/− cells were treated with HDAC inhibitors MS-275 or TSA for 24 hours, harvested for the immunoprecipitation assay using anti-Sp1 antibody and then analyzed by immunoblotting with anti-pan-acetyl antibody. Specific immunoprecipitated bands are indicated with arrows and uniform levels of Sp1 in the lysates are shown below. These blots are representative of 3 independent experiments.
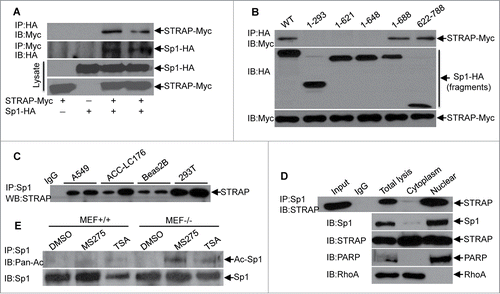
Previous report suggests that Sp1 protein localizes mostly in the nucleus.Citation18 Our previous data showed STRAP is expressed in both nuclear and cytoplasmic compartments.Citation13 To determine the subcellular binding between STRAP and Sp1, we used cytoplasmic and nuclear fractions from A549 cells to immunoprecipitate STRAP. The immune complexes were used for immunoblotting with anti-Sp1 antibody. As shown in , Sp1 was detected mostly in the immune complexes of nuclear fractions. In reverse experiments, STRAP was detected in the immune complexes of Sp1 in nuclear fractions. Complete separation of cytoplasmic and nuclear proteins was verified by immunoblotting analyses for RhoA and PARP, respectively. We repeated these experiments in wild type MEF and HeLa cell lines and obtained similar results (Fig. S2A and B). Co-localization of STRAP and Sp1 was observed in the nucleus by immunofluorescence analyses (Fig. S2C). Taken together, these data indicate that STRAP and Sp1 co-localize and interact in the nucleus.
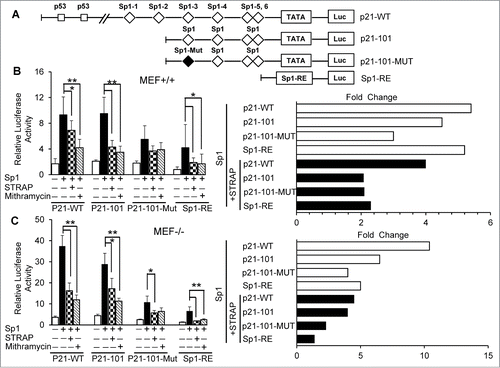
Figure 3. STRAP inhibits Sp1-dependent activation of p21Cip1. (A) Schematic representation of p21Cip1 promoter luciferase reporter plasmids: p21-WT containing the full length promoter with two p53 binding sites and six (1-6) Sp1 binding sites; p21-101 driven by four (3-6) consensus Sp1 sites in proximal promoter; p21-101-Mut containing the promoter segment with one mutated Sp1 site (third) and three consensus Sp1 sites (4-6) and Sp1-RE (artificial) construct containing three tandem repeats of consensus Sp1 binding sites. The squares represent the positions of p53 and the diamonds represent the locations of Sp1 sites, and the black diamond indicates mutation within the Sp1 site. (B) MEF+/+ cells were co-transfected with Sp1 and STRAP expression plasmids with the indicated p21Cip1 luciferase reporters. Mithramycin was treated as control. Fold changes from left panel is shown in the right panel. (C) Similar experiments were carried out as mentioned in (B) using MEF−/− cells. Data are from 3 independent experiments. Significance levels were determined by Student's t test. *, P < 0.05 **, P < 0.01, when compared with the corresponding control.
The only known acetylated residue of Sp1 was identified at lysine-703 (K703), which resides in its third zinc finger domain by Alanine scanning mutagenesis.Citation19 Since we have already demonstrated the physical interaction between STRAP and Sp1 through the latter's c-terminal amino acids, we asked whether this binding could affect the acetylation of Sp1 by blocking this domain. To address this, we treated MEF+/+ and MEF−/− cells with two HDAC inhibitors, MS-275 and TSA. The lysates were used for immunoprecipitation with anti-Sp1 antibody and the immune complexes were subjected to immunoblotting analyses with anti-acetyl lysine antibody (). These results suggest the enrichment of acetylated-Sp1 in STRAP null MEFs.
STRAP inhibits p21Cip1 promoter activity by regulating Sp1-dependent transcription
The human p21Cip1 promoter contains two p53-binding sites, and six Sp1 motifs in the proximal part before the TATA box ().Citation20,21 To examine the mechanism regulating the expression of p21Cip1, we analyzed the effect of STRAP and Sp1 on p21Cip1 promoter activity using luciferase assays. Luciferase reporter constructs containing different portions of the human p21 promoter () were transfected into MEF+/+ cells together with STRAP and/or Sp1 expression vectors. We observed that Sp1 significantly activated the p21Cip1 promoter independent of p53 (∼5 folds), which is suppressed by co-expression of STRAP (, p21-WT). Deletion of the p53-binding sites could not impair the responsiveness to p21-101, suggesting p53 had hardly any effect on the transactivation by Sp1. In addition, removal of Sp1-binding sites 1 and 2 had little effect on the promoter activation by Sp1, and STRAP inhibited this response by >2-fold (, right panel, p21-101). Mutation of the Sp1-3 site diminished activation of the p21Cip1 promoter by Sp1 (p21-101-MUT). Importantly, STRAP had less effect on the induction of p21-101-MUT in comparison to P21-101. We repeated the same assay using STRAP−/− MEFs. The p21Cip1 wt promoter is highly responsive to Sp1 (>10-fold) (, p21-WT) and co-expression of STRAP with Sp1 led to a significant reduction in response (>2.5-fold), suggesting an inhibitory role of endogenous STRAP on Sp1-dependent transcription. Together, our data demonstrate an inhibitory effect of STRAP in Sp1-dependent activation of p21Cip1 gene.
HDAC1 is recruited by STRAP to the Sp1-binding core region in p21Cip1 promoter
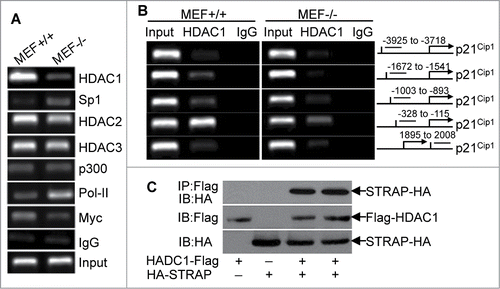
As descried above, Sp1 is important for maintaining an active p21Cip1 gene. Given this, inactivation of Sp1 transcriptional function could reduce the p21Cip1 expression in mRNA levels. To test this, we treated MEF−/− cells with Sp1 inhibitor Mithramythin, but there is little difference in p21Cip1 mRNA expression with or without treatment (data not shown). Therefore, we hypothesized that Sp1, although necessary for basal level, is not sufficient to induce transcription of p21Cip1. Previous research reported that histone deacetylation is involved in the regulation of p21Cip1 gene transcription.Citation22 Therefore, it became important to determine whether STRAP could induce alterations in the components of p21Cip1 promoter associated proteins potentially involved in its regulation. To explore this possibility, we used antibodies to HDAC1, HDAC2, HDAC3, p300, PoI-II and Myc to perform ChIP assay followed by qPCR with primers specifically targeting the upstream of TATA region (between -258 to -51) of p21Cip1promoter including Sp1-binding core region. As revealed in , there is higher occupancy of HDAC1 (>6-fold) and less enrichment of Sp1 (<5.5-fold) on the p21Cip1 promoter in MEF+/+ cells compared with that in MEF−/− cells, suggesting that STRAP can recruit epigenetic regulator HDAC1 to Sp1 binding regions in p21Cip1 promoter. In contrast, there is little difference in DNA fragments precipitated by HDAC2 and HDAC3 in STRAP+/+ and −/− MEFs. As expected, we observed higher levels of DNA fragments precipitated by c-Myc in MEF+/+ cells. However, loss of STRAP induced a marked increase in PoI-II associated with the promoter, suggesting an induction in transcription. Moreover, in STRAP-expressing cells we found enrichment of HDAC1 in the vicinity of the p21Cip1 proximal promoter upstream of the TATA box and this association of HDAC1 with the proximal promoter is dramatically decreased in MEF−/− cells (). Thus, it was necessary to determine whether STRAP binds with HDAC1 and can recruit it to the p21Cip1 promoter. Co-immunoprecipation assays after transfection of STRAP and Sp1 expression plasmids revealed that there is an interaction between STRAP and HDAC1 (). Taken together, these studies demonstrate that STRAP can recruit HDAC1 to the p21Cip1 promoter in the Sp1 binding region that is important for inhibiting Sp1-mediated expression of p21Cip1.
Figure 4. Loss of STRAP leads to alterations in the components of p21Cip1 promoter-associated complexes. (A) Anti-HDAC1, anti-HDAC2, anti-HDAC3, anti-Sp1, anti-p300, anti-PoI-II, and anti-Myc antibodies were used for ChIP assays. PCR amplification was done with proximal region of p21Cip1 promoter. Extracted DNA was used as input control. (B) PCR amplification was done with distal and proximal regions of p21Cip1 promoter. Anti-HDAC1 antibody was used for ChIP assay (left panel). A schematic graph (right panel) indicates the location of the primer pairs used for this assay. (C) 293T cells were co-transfected with Flag-tagged HDAC1 and HA-tagged STRAP as indicated. Lysates were subjected to Flag immunoprecipitation and then immunoblotted using anti-HA antibody. Protein expression was tested by immunoblotting.
STRAP mediates Sp1 stability through ubiquitin-proteasome pathway in a cell cycle-dependent manner
Since STRAP binds to Sp1 at its C-terminal domain containing one putative ubiquitin site,Citation23 we investigated whether the ubiquitin-proteosome system was involved in regulating Sp1 levels in STRAP null cells. However, we did not observe any alteration in Sp1 protein levels in the presence or absence of STRAP (data not shown). It has been reported that Sp1 protein levels fluctuate during the cell cycle especially in G1 phase and functions primarily during this phase.Citation24,25 Therefore, we sought to investigate whether STRAP has any effect on the expression of Sp1 in G1 phase. To test this, HeLa cells were first synchronized in G0/G1 phase by serum starvation for 72 h and then released into the medium with 10% FBS. Enrichment of G0/G1 phase cells (>70%, , bottom panel) was verified by flow cytometry analyses with STRAP knock-down clones and control cells. The whole cell lysate was collected at the indicated intervals, and both STRAP and Sp1 levels were assessed by immunoblot analyses. The results showed that expression of Sp1 was reduced to a basal level at 0 h after synchronization and gradually increased from 3 to 6 h until reached a peak after 6 h and stayed thereafter in control group. In contrast, loss of STRAP resulted in increasing Sp1 levels through 0 h to 9 h (, upper panel), which corresponded to an early G1 phase arrest at 3 h and subsequent delay into late G1 phase as evidenced by flow cytometric analyses (, highlighted box in bottom panel). Treatment of the control cells with the proteasome inhibitor MG132 could restore the expression of Sp1 (data not shown). Taken together, the data above suggests that the stability of Sp1 is controlled by cell cycle progression and STRAP is involved in the degradation of Sp1 protein at the early G1 phase.
Figure 5. STRAP mediates Sp1 stability during cell cycle progression. (A) STRAP knock-down stable clones from HeLa cells with control cells were synchronized by serum-starvation for 72h and released by adding 10% FBS for the indicated time-points. Total cell lystes were analysed for Sp1, STRAP and β-actin expression by immunoblotting using respective antibody (upper panel). Cell cycle phase were monitored by flow cytometry (bottom panel). (B) HeLa cells were synchronized by serum-starvation for 24h and then co-transfected with HA-ubiquitin and Sp1-Flag or STRAP-Myc expression plasmids as indicated. After total 72 hours cells were released into fresh media, cell lysates were collected at different time-points, subjected to anti-Flag immunoprecipitation and then analyzed by immunoblotting with anti-Ubiquitin antibody. Blots are representative of three independent experiments.
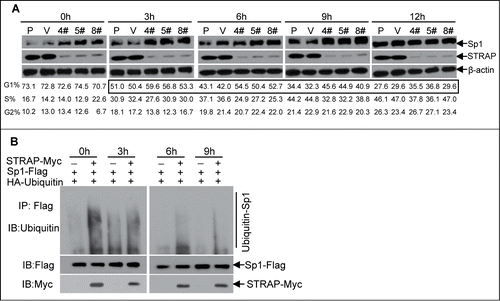
Next, we investigated whether the ubiquitin-proteasome system was involved in regulating Sp1 levels mediated by STRAP. This was accomplished by co-transfecting HeLa cells with HA-ubiquitin and Sp1-Flag expression plasmids in the presence or absence of STRAP-Myc plasmid after cells were synchronized and released at the indicated time-points, followed by incubation with proteasome inhibitor MG132. Sp1-ubiquitin complexes were isolated by anti-Flag antibody and subjected to immunoblot analysis for assessing Sp1 ubiquitination by anti-ubiquitin antibody. The results showed smear band indicating ubiquitinated Sp1 in the presence of STRAP at 0 h and gradually decreased with the time course, which is consistent with the level of Sp1 in control group (). As expected, the absence of STRAP could protect Sp1 from being ubiquitinated and degraded during 0-9 h. Taken together, these results suggest that Sp1 expression is tightly regulated by STRAP through the ubiquitin-proteasome pathway in G1 phase of the cell cycle and loss of expression of STRAP could stabilize Sp1 in this process.
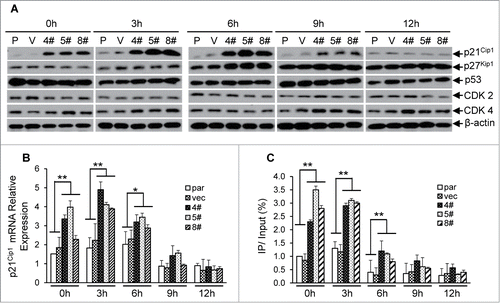
As shown in our initial experiments, loss of STRAP could induce HeLa cells to arrest at the early G1 phase of the cell cycle. To understand the mechanism, we analyzed the protein expressions of either cyclin-dependent kinase 2 and 4 (Cdk2 and Cdk4) or cyclin-dependent kinase inhibitors (p21Cip1 and p27Kip1). Compared with control cells, the expression of p21Cip1 was significantly induced (>4.5-fold) during 0 to 6 h and decreased from 9 h in STRAP knocking-down cells (). Other proteins mentioned above have no detectable alterations that correlate with STRAP expression (). qRT-PCR assays coupled with cell cycle analyses revealed a similar alteration trend in the mRNA levels of p21Cip1 as described above (), suggesting that the regulation of p21Cip1 expression is directly through transcriptional level. Since our model system demonstrates that p21Cip1 promoter is activated by Sp1, we hypothesized that knocking STRAP down causes p21Cip1 induction by Sp1 in a cell cycle dependent manner. To this end, we performed ChIP assays with samples synchronized and released at the indicated time-points to assess the Sp1 binding on the proximal region of p21Cip1 promoter during cell cycle progression. As shown in , more Sp1 associates to the promoter through 0 h to 3 h in STRAP knock-down clones when compared with control cells and results in an enhanced mRNA levels of the p21Cip1 in the same time course (). Our data also showed that Sp1 dissociates from p21Cip1 promoter after 6h in both of control and STRAP knocking-down groups (), suggesting that cell-cycle related negative regulator(s) might be recruited to replace Sp1 that is independent of STRAP control. Collectively, these data indicate that Sp1-induced transcription of p21Cip1 contributes to early G1 phase arrest in the absence of STRAP.
Figure 6. Knocking STRAP down results in cell cycle-dependent enhanced expression of p21Cip1. (A) Total cell lysates from serum-starved HeLa cell clones (as described in 5A above) were analysed for p21Cip1, p27Kip1, p53, CDK2, CDK4 and β-actin expressions by immunoblotting using respective antibody. (B) Total RNA from the cells above was prepared at indicated time points and levels of p21Cip1 mRNA were measured by qRT-PCR. (C) ChIP assay was performed from cells mentioned above using control IgG or anti-Sp1 antibody. PCR amplification was done with the proximal region of p21Cip1 promoter. The results are expressed as percentages of immunoprecipitated DNA compared to total input DNA. Significance levels were determined by Student's t test. *, P < 0.05 **, P < 0.01, when compared with the corresponding control.
STRAP expression inversely correlates with that of Sp1 in non-small cell lung cancer
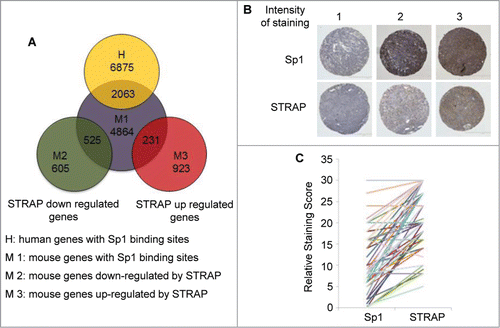
We next determined the number of genes enriched for the presence of Sp1 binding motif in their promoters (consensus Sp1 binding motif GGGGCGGGG and its variant GG[G/T]G[C/T]GGG.Citation26 The presence of Sp1 binding sites was searched using a computational approach in the promoter regions consisting of 1000 bp upstream from the transcription start site (http://www.sitesearch.mshri.on.ca/Genome/index.html). This search yielded 6875 genes from human database and 4864 genes from mouse database, respectively, out of which 2063 genes overlap in these two species (). We extended this analysis to our microarray data obtained from MEF+/+ and MEF−/− cell lines. Surprisingly, Sp1 site containing genes were highly enriched in the genes whose expression was decreased by STRAP with at least 1.5-fold change of expression (87%, 525 out of 605). A subset of genes (25%, 231 out of 923) up-regulated by STRAP also contains Sp1 sites, suggesting Sp1 may also be important for upregulation of STRAP target genes by other mechanisms. Sp1 is downregulated in lung tumor cells with high invasiveness and in patients with late stage lung adenocarcinoma.Citation27 We stained serial sections of a lung tissue microarray (TMA) from 42 lung cancer patients with anti-Sp1 and anti-STRAP antibodies. shows sample staining patterns for both STRAP and Sp1 in a pair-wise manner. Each specimen on the TMA was scored for the percentage of tumor cells showing staining (N) and for the intensity of staining (I), scored as 0 (no expression) to 3 (highest expression). These two numbers were then multiplied (N × I) to get the staining score for each spot. The score for the duplicate spots was averaged and then compared pair wise between STRAP and Sp1 staining. Using the Pearson's pair-wise comparison ratio we obtained an overall inverse correlation of 60% (P < 0.001) for STRAP and Sp1 in the lung cancer TMA (). This indicates a highly significant inverse correlation between STRAP and Sp1 levels in NSCLC and raises a possibility that STRAP might be a critical negative regulator of Sp1 in lung cancer.
Figure 7. STRAP inversely correlates with Sp1 expressions in NSCLC. (A) Venn diagrams show the overlap of 2063 genes with conserved Sp1 binding sites presented in both human and mouse. In MEFs, 525 genes (out of 605) with Sp1 sites were downregulated and 231 genes (out of 923) were upregulated by STRAP. (B) Immunohistochemical analysis of Sp1 and STRAP expressions in NSCLC TMA. Upper panel shows Sp1 expression and bottom panel shows STRAP expression in serial sections of three patients. (C) To evaluate the correlation between Sp1 and STRAP expressions in the same patient, staining score obtained from the TMA (n = 42) is shown for individual protein.
Discussion
One of the reasons of diverse protein functions of STRAP is its WD40 domains-based rigid scaffold platform that allows its interaction with a number of proteins. We have shown that STRAP is upregulated in several cancers and it functions as a putative oncogene.Citation12,13 However, little is known about the functional mechanism by which STRAP regulates the expression of tumor suppressor genes, E-cadherin and p21Cip1. As Sp1 is involved in large number of biological functions during development and tumorigenesis by interacting with a large variety of proteins and as its expression is connected with prognosis and survival of cancer patients, we hypothesize that STRAP and Sp1 might be functionally connected in regulating the expression and function of these two genes. In this study, we demonstrate for the first time that STRAP is a crucial negative regulator for the DNA-binding activity and cell cycle-dependent stabilization of Sp1. We have shown that STRAP downregulates E-cadherin by abrogating the binding of Sp1 to its consensus binding sites. STRAP binds with Sp1 in vivo in the nucleus through its C-terminal DNA binding domain and recruits epigenetic regulator HDAC1 to Sp1 binding regions in p21Cip1 promoter. Moreover, loss of STRAP can stabilize Sp1 by suppressing its ubiquitination in early G1 phase of the cell cycle, resulting in an enhanced expression of p21Cip1 and cell cycle arrest. Interestingly, microarray analyses have identified 525 genes out of 605 genes downregulated by STRAP with conserved Sp1 binding sites and the expression levels of STRAP inversely correlates with that of Sp1 in non-small cell lung cancer.
One of our novel findings is that STRAP interacts with Sp1 in the nucleus through the C-terminal DNA binding domain of Sp1 and is capable of abrogating the transcriptional activation of E-cadherin and p21Cip1 through Sp1. It is possible that STRAP is directly inhibiting the DNA binding ability of Sp1 by the mechanism of steric hindrance. Consistent with this idea, binding of the oncogene MDM2 (mouse double minute 2) to Sp1 prevents Sp1 from interacting with DNA. In turn, interaction of the tumor suppressor RB (retinoblastoma protein) with the same domain of MDM2 releases Sp1 from MDM2 and thus restores the DNA-binding and transactivation by Sp1.Citation28 Data from our microarray analyses suggest that 87% of STRAP downregulated genes has consensus Sp1 binding site/s. Therefore, it prompts to speculate that STRAP-induced steric hindrance is a potential mechanism for downregulating its target genes through Sp1 binding site/s. Future studies with a range of different STRAP target genes will generalize this mechanism.
Growing evidences indicate that phosphorylation, acetylation, sumoylation, ubiquitynation and glycosylation are among the posttranslational modifications that can influence the stability of Sp1 and its transcriptional activity.Citation29 Our data indicate that STRAP regulates the ubiquitynation modification of Sp1 in a cell cycle-dependent manner. Loss of STRAP results in stabilization of Sp1 in early G1 phase and subsequently enhances the expression of p21Cip1. However, the direct evidence that the ubiquitinated-Sp1 regulated by STRAP affects its transactivation activity remains to be defined. We have shown that STRAP interacts with Sp1 in various cell lines and promotes Sp1 degradation through ubiquitin-protesome pathway in G0/G1phase. Now the question is how Sp1 can be a target of the E3 ubiquitin ligase in the presence of STRAP. It is generally recognized that multiple WD-40 repeat proteins contain conserved tandem repeat known as the DWD box, providing specificity of ubiquitynation by interacting with the specific protein targets.Citation30,31 So far, 100 WD-40 repeat proteins containing the DWD box have been identified, indicating that 100 different E3 ubiquitin ligase complexes exist. We have shown before that one of them is the Cul4-DDB1-STRAP complex.Citation32 It is possible that in this scenario, the E3 ubiquitin ligase complex containing STRAP provide specificity of ubiquitination of Sp1 in a cell cycle dependent manner.
A computational approach comparing the promoters of a set of cell cycle regulated genes reveal that the genes expressed in G1/S phase of the cell cycle have more Sp1 binding sites in their promoters, suggesting a probability of higher Sp1 expression in the G1 phase.Citation25 Sp1 expression has also been shown to predominate in the G1 phase.Citation24 Our flow cytometry data indicated a G1 arrest in STRAP downregulated cells and immunoblotting analyses demonstrated a significant increase in the expression of Sp1 and p21Cip1 in early G1 phase. The p21Cip1 protein binds to and inhibits the activity of cyclin-CDK2 and -CDK4/6 complexes, and thus functions as a negative regulator of cell cycle progression at G1 phase.Citation33 While p21Cip1 is activated by p53-dependent mechanisms in response to DNA damage to ensure cell cycle arrest and repair, Sp1 has been implicated in the activation of the p21Cip1 gene. We found that loss of STRAP expression led to more cells arrested in the early G1 phase and a delay into the late G1 phase, suggesting STRAP is a potent positive regulator of G1 to S phase transition. Thus, we suggest a model of cell cycle regulation axis formed by STRAP, Sp1, and p21Cip1, which regulate the progression of cells through G1 phase.
Abnormal histone deacetylase (HDAC) activity and histone acetyl transferase (HAT) activity in cancers play a major role in deregulating tumor suppressor and promoter genes. HDACs modify histones through deacetylation leading to chromatin condensation and subsequent transcriptional repression. The expression of p21Cip1 has been shown to be regulated by several HDAC inhibitors.Citation34 However, little is known about the mechanism of how p21Cip1 might be regulated by HDACs through Sp1. A few mechanisms have been reported about the regulation of Sp1 transactivation activity by HDACs. E2F-1 binds to the same domain of Sp1 as HDAC1 so that they compete with each other for binding to Sp1 and consequently displaces HDAC1 from Sp1 and de-repress Sp1 target genes.Citation35 Another mechanism is that the oncogene c-Jun binds to Sp1 and strengthens the binding of HDAC1 to Sp1.Citation19 Therefore, it is not surprising that p21Cip1 activation is also tightly controlled by STRAP through recruiting HDAC1 to Sp1 core complex on the proximal p21Cip1 promoter to suppress Sp1-induced transcriptional activity. These results suggest a new mechanism of downregulation of STRAP target genes.
The transcription factor Sp1 is ubiquitously expressed in many tissues and cell lines and possesses three C2H2-type zinc fingers as a DNA-binding domain, which binds to GC-boxes with the consensus sequence. It has been suggested that there are over 12,000 Sp1-binding sites in the full human genome.Citation36 To determine the proportion of genes that are potentially transcribed via binding of Sp1 to their promoter, we have searched for Sp1-binding sites in the promoter of regulated genes in both human and mouse species. We observe that only downregulated genes by STRAP show a significant enrichment of Sp1 binding sites in their promoters. It is unlikely that this inhibition is due to the squelching of global factors required for transcription, as there is no change in expression of these genes following loss of STRAP. Analysis of the categories of genes with Sp1 site/s downregulated by STRAP includes cell cycle inhibitor, tumor suppressor WT1, cadherin family, as well as brain-specific angiogenesis inhibitor (Bai) genes (Table S1).
In summary, we have shown for the first time, the mechanisms of downregulation of the tumor suppressor genes E-cadherin and p21Cip1 by STRAP through abrogating Sp1-dependent transactivation. This study provides an important clue about how the loss of STRAP can stabilize Sp1 by repressing its ubiquitination in the G1 phase of the cell cycle, resulting in an enhanced expression of p21Cip1 and cell cycle arrest. The inverse correlation of the expressions of STRAP and Sp1 and the upregulation of STRAP in non-small cell lung cancers may explain, at least in part, the loss of expression of E-cadherin and p21Cip1.
Materials and methods
Cell culture and plasmids
Wt and STRAP-null mouse embryonic fibroblasts (MEFs), HEK-293T, and HeLa cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS). A549, ACC-LC176, Beas2B and H460 cells were cultured in RPMI1640 supplemented with 10% FBS. HA-tagged Sp1 and deleted constructs were a gift from Dr. Hans Rotheneder (University of Vienna, Austria). p53 and the human wild-type p21Cip1 expression plasmids, and deleted as well as mutant promoter luciferase reporter plasmids, were gift from Dr. Mottet Denis (University of Liege, Belgium). STRAP constructs have been described previously.Citation13 The E-cadherin luciferase constructs were a gift from Dr. Amparo Cano (Universidad Autónoma de Madrid, Spain). HA-HNF4 was kindly provided by Dr. Akiyoshi Fukamizu (Tsukuba Univeristy, Japan). p300 expressing vector and HA-Ubiquitin plasmid were obtained from Addgene.
Lentiviral transduction
1×105 cells were seeded in 6-well tissue culture plates and infected the following day with STRAP or vector control lentivirus. The cells were then selected for 7 days with puromycin. To generate stable STRAP-knockdown clones, cells were plated at high dilutions in 10 cm dishes and colonies obtained from single cells were screened for STRAP expression by immunoblot analysis.
Cell synchronization and cell cycle analysis using flow cytometry
Cells were synchronized at G0/G1 phase using serum starvation method. Briefly, 5×105 cells were cultured under serum-free medium for 72 h and then were continued to culture in 10% FBS containing medium. The synchronized cells were collected at the indicated time points after release from G0/G1 block and fixed with chilled 70% alcohol for 24 h. The cell sediment was collected and incubated with 20 μl of RNase A (20 mg/ml) (Life Technologies) for 30 min and stained with 25 μg/ml propidium iodide (Sigma-Aldrich) for 30 min. The cell cycle distribution was then evaluated using flow cytometry.
RNA isolation and quantitative PCR analysis
Total cellular RNA was extracted with TRIzol reagent (Life Technologies). One microgram of total RNA was used for reverse transcription by Moloney murine leukemia virus reverse transcriptase (Promega). The RT products were used as the template for quantitative PCR. PCR products were determined by SYBR Green reagent (Fermentas). Primers specific for detecting the human p21Cip1 mRNA levels were: forward: 5′ctgctgcaggcgccatgtca3′ and reverse: 5′ cacgctcccaggcgaagtca 3′.
Immunoblotting assay
For immunoblotting, whole-cell lysates were prepared in RIPA buffer and sonicated as described before.Citation12 The proteins were separated by 10% SDS/PAGE and probed with primary antibodies. Anti-Sp1, p21Cip1, p27Kip1, p53, CDK2, CDK4, PARP, RhoA, pan-Acetyl, HA, and Myc antibodies were purchased from Santa Cruz Biotechnology. Anti-STRAP antibody was obtained from BD Transduction Laboratories. Anti-Ubiquitin antibody was from Cell Signaling Technology and Anti-FLAG antibody was from Sigma-Aldrich. Primary antibodies were incubated for 2-3 hr at room temperature, followed by incubation with species-specific secondary antibodies for 1 hr at room temperature. The signal was visualized by enhanced chemiluminescence assay.
Co-immunoprecipitation
HEK-293T cells were plated in 60 mm dish and transfected with appropriate combination of plasmids using Lipofectamine reagent (Life Technologies) in 1:3 ratio. Cells were solubilized in 1 ml of lysis buffer as described.Citation13 Briefly, protein lysates were incubated with appropriate antibodies as indicated followed by incubation with 20 μl of protein G-Sepharose beads (Sigma-Aldrich). The immune complexes were washed with the lysis buffer and the beads were finally boiled in 25 μl of 2 × SDS sample buffer. Bound proteins were analyzed by immunoblot analysis using appropriate antibodies.
Nuclear protein extraction
Untreated and HDAC inhibitors treated cells were washed with cold PBS and harvested by scraping. The nuclei were prepared as described.Citation13 Briefly, the pellet was resuspended in 5 pellet volumes of cytoplasmic extract (CE) buffer and centrifuged at 4°C for 10 min. Wash the nuclei with 100 μl of CE buffer without detergent. Add 1 pellet volume of nuclear extraction buffer (NE) to nuclear pellet and adjust the salt concentration to 400 mM. The pellet was incubated on ice for 30 min with gentle shaking. Nuclear proteins were centrifuged at 4°C for 30 min and collected.
ChIP assay
Purification of sonicated nuclear lysates and immunoprecipitation were performed using EZ-ChIP assay kit (Upstate Biotechnology). Briefly, Cells were treated with formaldehyde to cross-link nuclear proteins with genomic DNA. Quenched Cells were lysed in SDS buffer on ice followed by sonicating to shear genomic DNA into 500- to 1000-bp fragments. Appropriate volume of supernatant was diluted (1:10) in dilution buffer and blocked with sheared salmon sperm DNA/Protein A/G-agarose. The supernatant obtained by brief centrifugation after blocking was immunoprecipitated with 2–5 ug of specific antibody at 4°C overnight. IgG was used as a negative control for IP. After incubating salmon sperm DNA/Protein A/G-agarose with IP samples at 4°C for another 2 h, beads were sequentially washed by low salt buffer, high salt buffer, LiCl buffer and TE buffer. The DNA/protein complex was eluted by elution buffer and reversely cross-linked. Purified DNA was used as the template for the qPCR analysis. Primer sequences are available upon request.
Luciferase reporter assay
Luciferase construct along with respective expression plasmids were transfected into the cells using Lipofectamine and Plus reagent (Life Technologies). In each experiment equal amounts of total DNA were transfected. Where needed, cells were treated with Sp1 inhibitor Mithramycin (150 nM) (Sigma-Aldrich) as indicated. After approximately 48 hours, cells were lysed and luciferase assays were performed using a luminometer. β-galactosidase plasmid was transfected to serve as an internal control.
Immunohistochemical analysis
Tissue Microarray (TMA) slides containing 42 duplicate samples of different lung carcinomas were obtained from the Lung SPORE project at Vanderbilt University. The slides were placed in the sodium citrate solution and heated in a pre-warmed steamer. After antigen retrieval, the specimens were treated with 3% H2O2 and further incubated with anti-STRAP antibody (BD Transduction Laboratories) and anti-Sp1 antibody (Santa Cruz Biotechnology). The specimens were then incubated with biotin-labeled goat anti-mouse immunoglobulin. Slides were lightly counterstained with Mayer's hematoxylin for nuclear staining.
Statistical analysis
Data are presented as mean ± sd, unless otherwise indicated. Statistical comparisons were made with Student's t test and ANOVA when appropriate. Values of P < 0.05 were considered to be significant. The correlation between STRAP and Sp1 levels in the lung cancer TMA was determined using the Pearson's pair-wise comparison ratio.
Disclosure of Potential of Conflicts of Interest
No potential conflicts of interest were disclosed.
973310_Supplementary_Materials.zip
Download Zip (422.1 KB)Acknowledgements
We thank Lung SPORE project at Vanderbilt University for providing lung TMA slide.
Funding
This study was supported by R01 CA095195, and Veterans Affairs Merit Review Award (to PK Datta).
Supplemental Materials
Supplemental data for this article is available online at the publisher's website.
References
- Dynan WS, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 1983; 35: 79-87; PMID:6313230; http://dx.doi.org/10.1016/0092-8674(83)90210-6
- Suske G. The Sp-family of transcription factors. Gene 1999; 238: 291-300; PMID:10570957; http://dx.doi.org/10.1016/S0378-1119(99)00357-1
- Mastrangelo IA, Courey AJ, Wall JS, Jackson SP, Hough PV. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci U S A 1991; 88: 5670-4; PMID:2062845; http://dx.doi.org/10.1073/pnas.88.13.5670
- Li L, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochem Cell Biol 2004; 82: 460-71 PMID:15284899; http://dx.doi.org/10.1139/o04-045
- Kaczynski J, Cook T, Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biol 2003; 4: 206 PMID:12620113; http://dx.doi.org/10.1186/gb-2003-4-2-206
- Opitz OG, Rustgi AK. Interaction between Sp1 and cell cycle regulatory proteins is important in transactivation of a differentiation-related gene. Cancer Res 2000; 60: 2825-30 PMID:10850422 http://dx.doi.org/10.1038/labinvest.2012.172
- Jones KA, Kadonaga JT, Luciw PA, Tjian R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science 1986; 232: 755-59 PMID:3008338; http://dx.doi.org/10.1126/science.3008338
- Datta PK, Moses HL. STRAP and Smad7 synergize in the inhibition of transforming growth factor beta signaling. Mol Cell Biol 2000; 20: 3157-67 PMID:10757800; http://dx.doi.org/10.1128/MCB.20.9.3157-3167.2000
- Kashikar ND, Reiner J, Datta A, Datta PK. Serine threonine receptor-associated protein (STRAP) plays a role in the maintenance of mesenchymal morphology. Cell Signal 2010; 22: 138-49 PMID:19781628; http://dx.doi.org/10.1016/j.cellsig.2009.09.024
- Buess M, Terracciano L, Reuter J, Ballabeni P, Boulay JL, Laffer U, Metzger U, Herrmann R, Rochlitz C. STRAP is a strong predictive marker of adjuvant chemotherapy benefit in colorectal cancer. Neoplasia 2004; 6: 813-20 PMID:15720808; http://dx.doi.org/10.1593/neo.04307
- Kashikar ND, Zhang W, Massion PP, Gonzalez AL, Datta PK. Role of STRAP in regulating GSK3β function and Notch3 stabilization. Cell Cycle. 2011 May 15; 10(10):1639-54. PMID:24626182; http://dx.doi.org/10.4161/cc.28417
- Halder SK, Anumanthan G, Maddula R, Mann J, Chytil A, Gonzalez AL, Washington MK, Moses HL, Beauchamp RD, Datta PK. Oncogenic function of a novel WD-domain protein, STRAP, in human carcinogenesis. Cancer Res 2006; 66: 6156-66 PMID:16778189; http://dx.doi.org/10.1158/0008-5472.CAN-05-3261
- Anumanthan G, Halder SK, Friedman DB, Datta PK. Oncogenic serine-threonine kinase receptor-associated protein modulates the function of Ewing sarcoma protein through a novel mechanism. Cancer Res 2006; 66: 10824-32 PMID:17108118; http://dx.doi.org/10.1158/0008-5472.CAN-06-1599
- Chen WV, Delrow J, Corrin PD, Frazier JP, Soriano P. Identification and validation of PDGF transcriptional targets by microarray-coupled gene-trap mutagenesis. Nat Genet 2004; 36: 304-12 PMID:14981515; http://dx.doi.org/10.1038/ng1306
- Bae GY, Choi SJ, Lee JS, Jo J, Lee J, Kim J, Cha HJ. Loss of E-cadherin activates EGFR-MEK/ERK signaling, which promotes invasion via the ZEB1/MMP2 axis in non-small cell lung cancer. Oncotarget. 2013 Dec; 4(12):2512-22. PMID:24318272
- Bourboulia D, Han H, Jensen-Taubman S, Gavil N, Isaac B, Wei B, Neckers L, Stetler-Stevenson WG. TIMP-2 modulates cancer cell transcriptional profile and enhances E-cadherin/beta-catenin complex expression in A549 lung cancer cells. Oncotarget. 2013 Jan; 4(1):166-76.PMID:23371049
- Liu YN, Lee WW, Wang CY, Chao TH, Chen Y, Chen JH. Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene 2005; 24: 8277-90 PMID:16116478; http://dx.doi.org/10.1038/sj.onc.1208991
- Ito T, Azumano M, Uwatoko C, Itoh K, Kuwahara J. Role of zinc finger structure in nuclear localization of transcription factor Sp1. Biochem Biophys Res Commun 2009; 380: 28-32 PMID:19138671; http://dx.doi.org/10.1016/j.bbrc.2008.12.165
- Hung JJ, Wang YT, Chang WC. Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol Cell Biol 2006; 26: 1770-85 PMID:16478997; http://dx.doi.org/10.1128/MCB.26.5.1770-1785.2006
- Mottet D, Pirotte S, Lamour V, Hagedorn M, Javerzat S, Bikfalvi A, Bellahcène A, Verdin E, Castronovo V. HDAC4 represses p21(WAF1/Cip1) expression in human cancer cells through a Sp1-dependent, p53-independent mechanism. Oncogene 2009; 28, 243-56 PMID:18850004; http://dx.doi.org/10.1038/onc.2008.371
- Li Q, Li J, Wen T, Zeng W, Peng C, Yan S, Tan J, Yang K, Liu S, Guo A, Zhang C, Su J, Jiang M, Liu Z, Zhou H, Chen X. Overexpression of HMGB1 in melanoma predicts patient survival and suppression of HMGB1 induces cell cycle arrest and senescence in association with p21 (Waf1/Cip1) up-regulation via a p53-independent, Sp1-dependent pathway. Oncotarget. 2014 Aug 30; 5(15):6387-403. PMID:25051367
- Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci U S A 2003; 101:1241-4646 PMID:14734806; http://dx.doi.org/10.1073/pnas.0307708100
- Wang YT, Yang WB, Chang WC, Hung JJ. Interplay of Posttranslational Modifications in Sp1 Mediates Sp1 Stability during Cell Cycle Progression. J Mol Biol 2011; 414: 1-14 PMID:21983342; http://dx.doi.org/10.1016/j.jmb.2011.09.027
- Grinstein E, Jundt F, Weinert I, Wernet P, Royer HD. Sp1 as G1 cell cycle phase specific transcription factor in epithelial cells. Oncogene 2002; 10: 1485-92 PMID:11896576; http://dx.doi.org/10.1038/sj/onc/1205211
- Deniaud E, Baguet J, Chalard R, Blanquier B, Brinza L, Meunier J, Michallet MC, Laugraud AL, Ah-Soon C, Wierinckx A, er al. Overexpression of transcription factor Sp1 leads to gene expression perturbations and cell cycle inhibition. PLoS One 2009; 4: e7035 PMID:19753117; http://dx.doi.org/10.1371/journal
- Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz, M, Bucher P. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem 2001; 276: 6675-88 PMID:11053426; http://dx.doi.org/10.1074/jbc.M001748200
- Hsu TI, Wang MC, Chen SY, Yeh YM, Su WC, Chang WC, Hung JJ. Sp1 expression regulates lung tumor progression. Oncogene 31, 3973-3988 PMID:22158040; http://dx.doi.org/10.1038/onc.2011.568
- Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res 2003; 1: 993-1000 PMID:14707282;
- Tan NY, Khachigian LM. Sp1 phosphorylation and its regulation of gene transcription. Mol Cell Biol 2009; 29, 2483-8 PMID:19273606; http://dx.doi.org/10.1128/MCB.01828-08
- He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev 2006; 20: 2949-54 PMID:17079684; http://dx.doi.org/10.1101/gad.1483206
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 2006; 443: 590-3 PMID:16964240; http://dx.doi.org/10.1038/nature05175
- Parsons LJ, Khoronenkova SV, Dianova II, Ternette N, Kessler BM, Datta PK, Dianov GL. Phosphorylation of PNKP by ATM prevents its proteasomal degradation and enhances resistance to oxidative stress. Nucleic Acids Res 2012; 40: 11404-15 PMID:23042680; http://dx.doi.org/10.1093/nar/gks909
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 1999; 13: 1501-12 PMID:10385618; http://dx.doi.org/10.1101/gad.13.12.1501
- Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov 2002; 1: 287-99 PMID:12120280; http://dx.doi.org/10.1038/nrd772
- Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurter V, Brosch G, Wintersberger E, Seiser C. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol 1999; 19: 5504-11 PMID:10409740
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 2004; 116: 499-509 PMID:14980218; http://dx.doi.org/10.1016/S0092-8674(04)00127-8
