Abstract
To date, a complete understanding of the molecular events leading to DNA replication origin activation in mammalian cells still remains elusive. In this work, we report the results of a high resolution chromatin immunoprecipitation study to detect proteins interacting with the human Lamin B2 replication origin. In addition to the pre-RC component ORC4 and to the transcription factors USF and HOXC13, we found that 2 components of the AP-1 transcription factor, c-Fos and c-Jun, are also associated with the origin DNA during the late G1 phase of the cell cycle and that these factors interact with ORC4. Both DNA replication and AP-1 factor binding to the origin region were perturbed by cell treatment with merbarone, a topoisomerase II inhibitor, suggesting that DNA topology is essential for determining origin function.
Introduction
Eukaryotic DNA replication is a highly organized process that initiates at multiple locations along each chromosome, the origins of DNA replication, and ensues in entire genome duplication once and only once during the S phase of the cell cycle. In the budding yeast Saccharomyces cerevisiae, origin selection is dictated by the sequence-specific recognition of the essential ARS consensus sequence (ACS) by the 6-subunit origin recognition complex (ORC). ORC then acts as a platform for the subsequent assembly, during the G1 phase, of other members of the pre-replicative complex (Pre-RC).Citation1 While the multistep process of pre-RC assembly is essentially conserved in all eukaryotes, as it is the identity of its essential protein components, the specification of origins along the chromosomal DNA sequence however varies. In Schizosaccharomyces pombe, ORC binds AT-rich origin sequences by virtue of an AT-hook DNA binding domain of its ORC4 subunit.Citation2 Metazoan ORCs, instead, have apparently lost sequence-specificity in origin DNA recognition.Citation3,4 In these organisms, accessory proteins and chromatin structure, rather than primary DNA sequence alone, appear essential in dictating origin choice.Citation5-7 The lack of a primary consensus sequence for origin recognition has significantly slowed down the comprehensive identification of metazoan origins and, in most instances, the determinants for this recognition have remained still elusive.
Among the essential proteins that assist metazoan ORC in origin binding are various cellular transcription factors.Citation8 This justifies both the presence of transcription factor binding sites in close proximity to origins and the well established connection between origin location and the transcriptional activity and chromatin conformation of the regions where origins are positioned.Citation9-12 Indeed, transcription factor-induced remodeling of chromatin toward an accessible state is believed to be a key regulatory element determining pre-RC assembly at origins.Citation13,14
The first human origin to be characterized in molecular detail was the Lamin B2 origin, located on chromosome 19p13.3 in the 3′ region of the lamin B2 gene and upstream another closely-position gene, TIMM13.Citation15 Work performed over the last several years have explored pre-RC assembly at this origin during the cell cycle,Citation16 have pinpointed the exact site of DNA replication initiationCitation17 and have defined the role of topology in origin regulation.Citation18 Of interest, downstream the ORC binding site, which also defines the DNA replication start site, the Lamin B2 origins contains an E-box binding site, which was shown to interact with the USF transcription factor.Citation12,19 In addition, the homeotic transcription factor HOXC-13 also binds this origin and regulate its function.Citation11
A transcription factor that mediates the cellular response to activating stimuli, including the induction of cell cycle progression, is AP-1. This is a heterodimeric factor composed of proteins belonging to the Fos, Jun, ATF and JDP families, which becomes rapidly induced in response to different stimuli to then activate a variety of cellular processes, among which cell proliferation.Citation20-22 Of interest, the c-Fos/c-Jun heterodimer binds a degenerate, conserved DNA sequence, the AP-1 binding site,Citation23 but also contributes to the regulation of genes that have no apparent AP-1 sites.Citation24,25 Multiple evidence indicates that the control of cell proliferation by AP-1 is mediated by the transcriptional induction of several cell cycle regulators.Citation22,26 However, the findings that AP-1 proteins constitute a control point for G1 progression and are required for initiation of DNA synthesis in response to serumCitation27 and that they enhance replication of the polyomavirus genome,Citation28,29 also render these factors potential candidates for a direct role in the regulation of DNA replication initiation.
In this work we investigate AP-1, ORC4, USF-1 and HOXC13 protein binding at the LaminB2 DNA replication origin. We found that binding of these proteins define 2 interacting zones in a range of 250 base pairs, involving the start site of DNA replication and the downstream TIMM13 gene promoter region. The c-Fos and c-Jun AP-1 components were detected both to bind the origin DNA region and to interact with the ORC4 pre-RC component. Of note, during G1 progression, DNA-binding of the 2 factors displayed a peculiar kinetics, by which, in mid-G1 cells, both c-Fos and c-Jun were detected at the TIMM13 promoter area, while, in late G1, binding of c-Fos was restricted at the DNA replication start site region. Both DNA replication and interaction of the 2 AP-1 factors with the origin area was perturbed by cell treatment with merbarone, a topoisomerase II inhibitor, suggesting that DNA topology is essential in determining origin function.
Results
Analysis of the Lamin B2 origin of DNA replication by high resolution ChIP
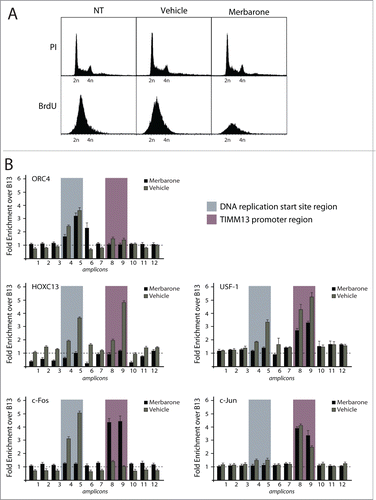
We investigated protein binding across the Lamin B2 origin sequence using the high resolution-chromatin immunoprecipitation (HR-ChIP) procedure, able to generate high resolution information over a relatively short genomic region. In brief, this method entails fomaldehyde-crosslinking of protein-protein and protein-DNA interactions on isolated nuclei, followed by micrococcal nuclease (MNase) digestion to mononucleosomal size before immunoprecipitation with specific antibodies and detection of the co-precipitated DNA by real-time, quantitative PCR.Citation9,11 We focused on a 1.1 kb-long DNA region, encompassing the DNA replication start siteCitation17 and a downstream-located, 250 bp-DNA segment in the TIMM13 promoter region, which was scanned using 12 overlapping PCR segments to achieve resolution in the order of 100–200 bp (). Among the antibodies used for HR-ChIP were those against H2B and K14-acetylated H3, to detect chromatin modification, against 3 proteins already known to bind in proximity of the replication start site (ORC4Citation30 and HOXC13Citation9,11 or the region immediately downstream of it (USF-1Citation12) and against the AP-1 components c-Fos and c-Jun, which were identified as potential regulators of origin function.Citation8 The results of these experiments, which were performed in asynchronous T98G human cells, are shown in . Binding of H2B was detected throughout the investigated area, with the exception of the start site region (fragments 4 and 5) and the nucleosome-free region of the TIMM13 promoter, which is known to associate with transcription factors USF1, Sp1 and NRF1 (fragments 8 and 9). H3K14 acetylation was mostly detected immediately downstream this region (fragments 10 and 11). ORC4 was enriched at fragment 5, encompassing the DNA replication start site. Both this region and the TIMM13 promoter were bound by both USF1 and HOXC13. Of note, c-Fos was detected in correspondence to the start site region (enrichment in fragments 4, 5 and 6), while c-Jun to the nucleosome-free region of the TIMM13 promoter. No binding to any of the investigated fragments was detected for another member of the Jun family (Jun D), NF-κB p65, used as a negative control, and 2 known interactors of HOX and AP1 proteins, namely the 3 members of the PBX (pre-B-cell leukemia transcription factor) family and NFAT1 (nuclear factor of activated T cells), respectively.Citation31-34
Figure 1 (See previous page). High resolution chromatin immunoprecipitation analysis at the lamin B2 origin of DNA replication. (A) Schematic representation of the position of the 12 overlapping PCR segments, used for ChIP analysis, across the 1.1 kbp DNA region containing the start site of the Lamin B2 origin of DNA replication (highlighted in gray) and the TIMM13 promoter (purple) regions. B48 and B13 exemplify origin and the non-origin regions.Citation15 The promoter of the TIMM13 gene is located ∼250 bp downstream the LaminB2 start site and contains the binding sites for USF-1 (green box), SP-1 (red box) and NRF-1 (blue box) transcription factors. (B) Real time, quantitative PCR was used to assess the chromatin binding profiles of nucleosomal proteins (H2B and K14 acetylated H3), of 3 proteins known to interact with the start site and the TIMM13 promoter (ORC4, HOXC13 and USF-1), of the AP-1 proteins c-Fos, c-Jun and JunD, of NFAT-1 and NF-κB p65, and of the AP-1 cofactor PBX.
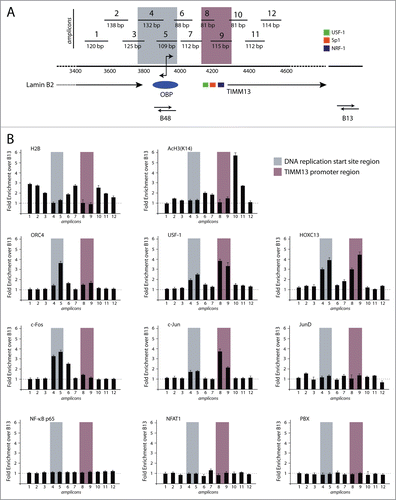
Taken these results together, the investigated proteins that bind the Lamin B2 origin region define 2 major areas, one corresponding to the actual start site for DNA replication and the second to the TIMM13 promoter. Quite unexpectedly, interaction with USF1 and HOXC13 was detected for both regions, while the 2 AP-1 components selectively bound either the start site (c-Fos) or the promoter (c-Jun).
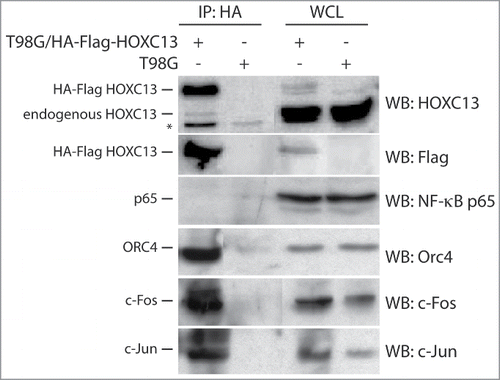
Finally, we also wanted to assess whether a specific interaction between the proteins binding the Lamin B2 origin could also be detected by co-immunoprecipitation. We took advantage of a stable cell clone derived from T98G cells and overexpressing a HOXC13 protein carrying a double HA-Flag tag at its N-terminus. After immunoprecipitation using an anti-HA antibody, ORC4, c-Jun and c-Fos were all found in the immunoprecipitate (). This observation is consistent with the conclusion that that these proteins are part of a multi-protein complex interacting with the Lamin B2 origin region.
Figure 2. Co-immunoprecipitation analysis of proteins interacting with HOXC13. HA immunoprecipitation was performed with lysates from asynchronous crosslinked T98G cells overexpressing an HA-Flag tag HOXC13 construct using a specific anti-HA antibody for immuno-precipitation and the indicated antibodies for blottings. Lysates from asynchronous T98G cells served as a control. The NF-κB p65 subunit was used as a negative control for immunoprecipitation, whereas whole cell lysates (WCL) of both cell lines were used as positive controls. The asterisk indicates an unspecific signal detected by the anti-HOXC13 antibody.
Spatial and temporal analysis of AP-1 binding onto the LaminB2 origin
To start evaluating a possible correlation between the association of c-Fos and c-Jun with the Lamin B2 origin region and with origin activity itself, we applied our HR-ChIP analysis to chromatin collected at different time points during cell cycle progression. T98G cells were synchronized in G0 by 72 hours of serum starvation, followed by serum re-addition. shows the flow cytometry profiles of cellular DNA content, detected after propidium iodide staining, at different times after serum addition (9, 14, 16 and 20 h for mid-G1, late-G1, G1-S border and early S phase, respectively, as already reported).Citation35 Analysis of the levels of expression of c-Fos and c-Jun at the same time points by western blotting indicated that both proteins were significantly induced in mid- and late-G1 compared to the G0 (starved) cells ().
Figure 3. Spatial and temporal analysis of the localization of c-Fos and c-Jun proteins onto the origin DNA. (A) Flow cytometry profiles of T98G cells synchronized by 72 h serum starvation followed by the re-addition of serum. Cells were collected at 9, 14, 16 h and 20 h for mid-G1, late-G1, G1-S border and early S respectively. (B) Western blotting analysis of whole cell lysates (WCL) from each time point to detect the levels of c-Fos, c-Jun and Cyclin A. (C) Real time PCR analysis of chromatin immunoprecipitated at the different time points using the indicated antibodies. (D) Immunoblot using an anti ORC4 antibody performed on the samples used for the c-Fos and c-Jun ChIPs. The red dotted square indicates the co-presence of ORC4, mostly in late G1, in the c-Jun and c-Fos immunoprecipitated protein-complexes.
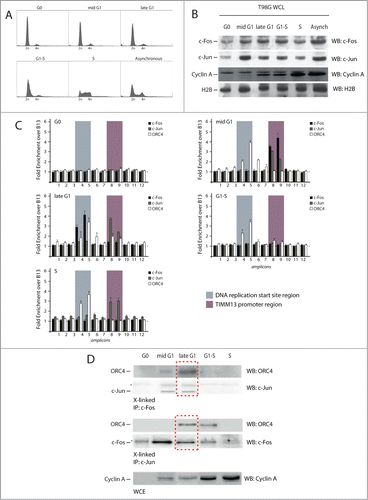
HR-ChIP was then performed over the Lamin B2 origin at different times of the cell cycle using antibodies against ORC4, c-Fos and c-Jun. The results of these experiments are shown in . None of the investigated proteins was found to interact with the origin region in G0. Beginning in mid G1 to S phase, ORC4 was detected at the DNA replication start site, consistent with its role as a member of the ORC complex and in agreement with our previous DNA footprinting and protein-DNA interaction results.Citation18,36 In contrast, the binding pattern of the 2 AP-1 members was peculiar. In mid-G1 cells, both c-Fos and c-Jun were detected at the TIMM13 promoter area. In late G1, however, while c-Jun was still present on the promoter, binding of c-Fos was detected at the start site region. At the G1/S transition, when the pre-RC complex becomes activated, neither c-Jun nor c-Fos were any longer detectable at the origin area, with c-Jun returning again during the S-phase. Efficacy of immunoprecipitation with each of the 3 antibodies was verified by protein gel blotting in all HR-ChIP immunoprecipitates before real time qPCR analysis (not shown).
We then wanted to understand whether a possible protein-protein interaction between the 2 AP-1 proteins and ORC4 could be detected in the different phases of the cell cycle. We analyzed the samples used for the c-Fos and c-Jun HR-ChIPs by western blotting, searching for the presence of ORC4. This was found in both the c-Fos and c-Jun immunoprecipitations in late G1 ().
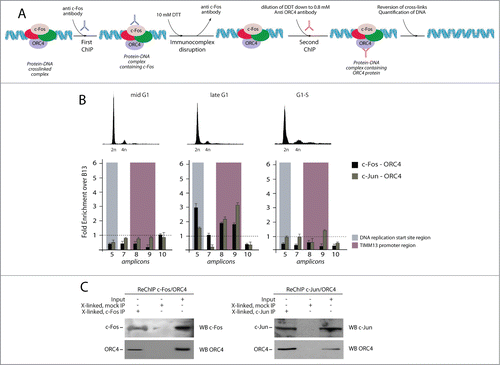
Since positive signals were also detected in mid G1 for ORC4/c-Fos and G1-S for ORC4/c-Jun, we wanted to re-probe these interactions with the Re-ChIP technology.Citation37,38 This method, which is applied to crosslinked, immunoprecipitated chromatin, allows one to detect the presence of a specific protein in a complex formed at a chosen genomic region (). The results of the Re-ChIP experiments revealed the presence of ORC4 in both the c-Fos and c-Jun immunoprecipitates exclusively obtained from late G1 cells, both at the DNA replication and TIMM13 promoter areas (). These results indicate that an interaction between ORC4 and the 2 AP-1 proteins occurs in vivo at the Lamin B2 in a specific window of the cell cycle that precedes the G1/S transition.
Figure 4. Sequential chromatin immunoprecipitation analysis (Re-ChIP) to detect interaction between AP-1 factors and ORC4 onto the Lamin B2 origin. (A) Schematic representation of the Re-ChIP procedure. Crosslinked chromatin, which was immunoprecipitated with anti-c-Fos or c-Jun antibodies, was subjected to a mild DTT treatment and re-probed using an anti ORC4 antibody. Finally, the immunocomplex was treated to reverse the crosslink and DNA analyzed by real time qPCR. (B) Upper part: flow cytometry profiles of T98G cells synchronized at different time points corresponding to different stages of the cell cycle, and subjected to Re-ChIP. Lower part: Results of real time qPCR analysis, showing that interaction between c-Fos, c-Jun and ORC4 occurs on the Lamin B2 origin only during the late G1 phase of the cell cycle. (C) Part of the material of each re-ChIP was subjected to immunoblot before qPCR to confirm the co-presence of c-Fos/ORC4 and c-Jun/ORC4.
Effects of the disruption of topological structure on protein binding across the LaminB2 origin region
Previous studies have demonstrated that topoisomerase II joins the pre-RC complex on the LaminB2 origin during the mid-G1 phase of the cell cycle, and that this interaction is highly sensitive to topological interference.Citation18 For this reason, we analyzed the influence of DNA topology on the pattern of protein-DNA interaction in the origin area by treating asynchronously growing cells with the reversible topoisomerase II DNA-binding inhibitor merbarone (5-(N-phenylcarbamoyl)-2-thiobarbituric acid).Citation39 The effect of the drug was confirmed by a drastic reduction in BrdU incorporation after 2 hours of treatment, despite no apparent variation in cell cycle profile, as detected by propidium iodide staining (). ORC4 binding to the start site region, an event occurring early in G1, was not affected by drug treatment, as it was not binding of USF1 to the TIMM promoter. In contrast, merbarone completely blocked origin interaction with HOXC13, a protein that we previously showed to bind the DNA replication start site in late G1,Citation9,11 in addition to USF1 binding with the start site region (). Of interest, we found that the binding profile of c-Fos was drastically altered by the drug, since the protein was found associated to the TIMM13 promoter instead of the start site, thus with a profile similar to the one observed in the mid-G1 phase of the cell cycle. Taken together, these results indicate that passage from the early to the late phases of the cell cycle is concomitant with a shift in the position of c-Fos binding and that this event requires a topological modification in the origin region.
Figure 5. Effect of the topoisomerase II inhibitor merbarone on multi-protein complex assembly. (A) Flow cytometry analysis of T98G cells treated with merbarone. The upper panels show propidium iodide (PI) staining; the lower panels show BrdU incorporation. (B) Results of high resolution ChIP experiments in T98G cells treated with merbarone.
Discussion
Application of a high-resolution ChIP technique to the analysis of protein-DNA interactions at the lamin B2 origin here led to the identification, in addition to the DNA replication start site, of another region that appears involved in the regulation of origin activity. This region, which encompasses the downstream positioned TIMM13 gene promoter area, is relatively devoid of histones and interacts with transcription factors USF-1 and HOXC13; our previous work has shown that the former protein acts as a transcription factor for TIMM13 gene expression [13] while the latter plays a role during the late G1 phase, as a pre-RC interactor.Citation9,11
In addition to these factors, we found that the AP-1 subunits c-Fos and c-Jun also interact with both the origin downstream region and the start site region itself. Participation of c-Jun and c-Fos to origin binding was consistent with their interaction, as detected by co-immunoprecipitation, with both ORC4, a canonical member of the pre-RC,Citation30 and with HOXC-13, which also participates in origin function.Citation9,11 That the interaction of c-Jun and c-Fos with ORC4 specifically occurs at the origin region was proven by the Re-ChIP technology, which allows one to investigate the protein-protein interactions specifically occurring at a given DNA region.
What might be the significance of c-Fos and c-Jun binding to an origin of DNA replication? Since mammalian origins lack a specific, primary DNA sequence similarity, there is a general consensus that additional factors should assist the pre-RC in defining chromosomal origins and participating in their function. This appears to be particularly true for transcription factors.Citation9-12 In particular, the c-Myc transcription factor was also found to bind early replication origins and to be involved in origin activity in human cellsCitation3,10; similar to c-Myc, also c-Fos and c-Jun have a well established role in promoting G1-S phase transition and favoring S-phase entry.Citation21,22,40 In addition, bioinformatics analysis has indicated that the presence of AP-1 binding sites might positively identify human DNA replication origins.Citation8
The observation that c-Jun and c-Fos bind the lamin B2 origin with a peculiar kinetics during the cell cycle is more puzzling. In particular, we found that both c-Fos and c-Jun start to selectively interact with the TIMM13 promoter area in mid-G1 after synchronization. While the cell cycle progressed, however, c-Jun continued to interact with this area, while c-Fos instead associated with the start site region. Further toward the G1/S transition, neither protein was detected at the origin. These observations point to the existence of specific and dynamic structural reorganizations of the complexes assembled at the origin region along with origin activation. This conclusion is also supported by our experiments using merbarone, a reversible inhibitor of topoisomerase II, which was already used to define the kinetics of protein-DNA interactions at other origins.Citation41 The drug did not prevent binding of neither ORC4 to the start site region nor USF to the TIMM promoter. However, it blocked both interaction of the origin with HOXC13 and, most notably, the transition of c-Fos binding from the TIMM13 promoter area to the start site. These observations indicate that the topological configuration of origin DNA during the cell cycle is an essential determinant to permit origin activation. In cells treated with merbarone, as expected, there was a dramatic block in DNA replication. Previous experiments have shown that the lamin B2 origin assumes non-canonical DNA structures,Citation42 and that these structures are important for proper association with pre-RC members.Citation4,43 We speculate that the detected shift in c-Fos binding region during the cell cycle and its inhibition upon merbarone treatment might be the consequence of the formation of such structures. Further experiments are clearly required to directly address this possibility.
Materials and Methods
Cell culture and treatments
T98G (ATCC CRL-1690), HeLa (ATCC CCL-2) and T98G stably expressing HA-FLAG HOXC13 (a kind gift of Dr. Ramiro Mendoza-Maldonado from ICGEB in Trieste) were cultured in 1 g/ml glucose, 1 mM pyruvate D-MEM with Glutamax (Invitrogen) supplemented with 10 U/L penicillin, 10 μg/L streptomycin and 10% fetal calf serum (Gibco) at 37°C and 5% CO2 according to standard conditions. Sub-confluent T98G cells were synchronized in serum free medium. After 72 h, serum free medium was replaced with complete serum and the cells were left in these conditions at 37°C and 5% CO2 for 9, 15, 16 and 20 h to synchronize them in mid G1, late G1, G1/S and S, respectively, as already reported.Citation35 To evaluate cell cycle profiles, cells were fixed with cold absolute ethanol and then stained with 500 μl of a solution containing 375 μl sodium citrate 0.1% w/v, 125 μl 1 mg/ml propidium iodide (Sigma P4864), 6.25 μl NP-40 0.1% v/v and 0.625 μl 10 mg/ml RNAse A (Roche) and analyzed with the FACScalibur flow cytometer (BD Bioscences). Topoisomerase II inhibitor merbarone (5-(N-Phenylcarbamoyl)-2-thiobarbituric acid, NSC-336628) (Sigma M2070) was dissolved in DMSO and added to complete medium at a final concentration of 100 μM; the cells were incubated at 37°C and 5% CO2 for 1 h.
Antibodies
The following primary antibodies were used for either ChIP or Western Blot (WB) experiments: mouse monoclonal anti FLAG M2 HRP conjugated A8592 from Sigma (WB 1:1000), rabbit polyclonal anti-HOXC13 (a kind gift of Dr. Ramiro Mendoza-Maldonado from ICGEB Trieste (WB 1:500), mouse monoclonal anti HA agarose conjugated Clone HA-7 A2095 from Sigma (ChIP 0.5 μg/mg proteins). rabbit polyclonal anti c-Fos sc-52 from Santa cruz (WB 1:400, ChIP 1 μg/mg proteins), rabbit polyclonal anti c-Jun sc-45 from Santa cruz (WB 1:500, ChIP 1 μg/mg proteins), rabbit polyclonal anti NF-kB p50 sc-7178 from Santa Cruz (WB 1:500), rabbit polyclonal anti NF-kB p65 sc-372 from Santa Cruz (ChIP 1 μg/mg proteins), mouse monoclonal anti NF-kB p65 MAB3026 Upstate (WB 1:500), mouse monoclonal anti ORC4 611170 from BD Biosciences (WB 1:1000), rabbit polyclonal anti ORC4 sc-20634 from Santa Cruz (ChIP 1 μg/mg proteins), rabbit polyclonal anti USF-1 sc-229 from Santa Cruz (WB 1:500, ChIP 1 μg/mg proteins), rabbit polyclonal anti H2B sc-10808 (WB1:500, ChIP 1 μg/mg proteins), rabbit polyclonal anti acetyl-Histone 3 (K14) Millipore 06–599 (WB 1:1000), rabbit polyclonal anti PBX 1-2-3 sc-888 from Santa Cruz (WB 1:500, ChIP 1 μg/mg proteins), rabbit polyclonal anti JunD sc-74 from Santa Cruz (WB 1:500, ChIP 1 μg/mg proteins) rabbit polyclonal anti NFAT1 sc-13034 (WB 1:500, ChIP 1 μg/mg proteins), mouse monoclonal anti Cyclin A sc-239 from Santa Cruz (WB1:500). As negative controls, pre-immune mouse or rabbit from Sigma (R9133 and M5905 respectively) were used. Secondary antibodies used in Western Blot experiments were monoclonal mouse anti rabbit light chain specific (211-032-171) and goat anti mouse light chain specific (115-035-174), both from Jackson immunoResearch.
Chromatin immuno precipitation (ChIP) and Sequential chromatin immuno precipitation (Re-ChIP)
Chromatin was obtained by biochemical fractionation. Briefly, cells were lysed by Dounce homogenization in hypothonic buffer (10 mM Hepes pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 5 mM Sodium Butyrate). Nuclei were collected by centrifugation, resuspended in buffer S1 (20 mM Hepes pH 7.9, 250 mM Sucrose, 10 mM MgCl2), gently overlaid on buffer S2 (20 mM Hepes pH 7.9, 350 mM Sucrose, 0.5 mM MgCl2) and recovered by centrifugation. Purified nuclei were washed with buffer B (20 mM Hepes pH 7.9, 1.5 mM MgCl2, 20 mM Kcl, 5 mM Sodium Butyrate, 10% glycerol) and incubated at 4°C in lysis buffer (20 mM Hepes pH 7.9, 750 mM ϵ-aminocaproic acid, 0.5% Triton-X 100). Chromatin was recovered by centrifugation and resuspended in crosslink buffer (1% formaldehyde in 20 mM Hepes pH 7.9). After 10 min, the reaction was quenched by adding 125 mM glycine. Unbound material was removed performing one wash in NaCl 1 M in buffer B; after 2 more washing steps in buffer B, chromatin was resuspended in buffer C (20 mM Hepes pH 7.9, 3 mM CaCl2) and subjected to micrococcal nuclease (Roche) digestion. The reaction was stopped by adding 4 mM EDTA, the material was sonicated 3 times in ice for 5 sec in order to further shear the DNA (∼200 bp) and then, after quantification by the Bradford assay, subject to immunoprecipitation. The immunocomplexes were washed with low salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 7.9, 150 mM NaCl), high salt buffer (500 mM NaCl), LiCl buffer (250 mM LiCl, 1% IGEPAL, 1% deoxycholic acid, 1 mM EDTA, 10 mM Tris-HCl, pH 8) and TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8). The washed precipitates were divided for protein gel blot and DNA extraction. DNA was recovered by phenol:chloroform:isoamyl alcohol (24:25:1) from Sigma after RNAseA (Roche) and Proteinase K digestion of the immunocomplexes. Finally, DNA was precipitated with absolute ethanol and washed once with 70% ethanol and quantified. For Re-ChIP, chromatin was obtained and DNA:protein complexes were immuno-precipitated as described. Immuno-complexes bound to the agarose beads were incubated with an equal volume (∼50 μl) of 10 mM DTT for 30 min at 37°C. After centrifugation, the supernatant was transferred into a new tube and the immuno-complexes were subjected to a second round of 10 mM DTT treatment. The eluates were combined and diluted 100 times; 10% of the sample was kept for input, while the rest was processed by adding the second antibody. The immunoprecipitated material was washed and DNA was purified as described above.
High resolution LaminB2 PCR
A 1.1 Kbp region encompassing both the DNA replication start site of the LaminB2 origin and the downstream TIMM13 promoter was divided into 12 partially overlapping fragments by designing primer pairs spanning 80–120 bp each. All primers were chosen to have similar annealing temperatures, with the exception of those encompassing the start site region, which is particularly A/T rich. All primers sets were analyzed by real time PCR, amplifying 4 scalar concentrations of genomic DNA, obtained from 3 independent purifications, to assess their efficiency using the slope analysis approach. Real time PCR experiments were carried out and the enrichments for all the investigated fragments were calculated using the ΔΔCt method. Primers are listed in .
Table 1. List of primers and amplification conditions used for the analysis of the Lamin B2 origin
Acknowledgments
This work is dedicated to the memory of Arturo Falaschi, in whose laboratory this research project was conceived and took shape. Prof. Falaschi passed away on 2nd June 2010 but his memory and enthusiasm has remained present throughout. The authors are grateful to Suzanne Kerbavcic for precious editorial work.
Funding
This work was supported by a grant from the ICGEB intramural program.
References
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem 2002; 71:333-74; PMID:12045100; http://dx.doi.org/10.1146/annurev.biochem.71.110601.135425
- Kong D, DePamphilis ML. Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol Cell Biol 2001; 21:8095-103; PMID:11689699; http://dx.doi.org/10.1128/MCB.21.23.8095-8103.2001
- DePamphilis ML. The ‘ORC cycle’: a novel pathway for regulating eukaryotic DNA replication. Gene 2003; 310:1-15; PMID:12801628; http://dx.doi.org/10.1016/S0378-1119(03)00546-8
- Mendez J, Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays 2003; 25:1158-67; PMID:14635251; http://dx.doi.org/10.1002/bies.10370
- Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature 2004; 430:372-6; PMID:15254542; http://dx.doi.org/10.1038/nature02694
- Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J 2004; 23:897-907; PMID:14765124; http://dx.doi.org/10.1038/sj.emboj.7600077
- Zhou J, Chau CM, Deng Z, Shiekhattar R, Spindler MP, Schepers A, Lieberman PM. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J 2005; 24:1406-17; PMID:15775975; http://dx.doi.org/10.1038/sj.emboj.7600609
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci U S A 2008; 105:15837-42; PMID:18838675; http://dx.doi.org/10.1073/pnas.0805208105
- Comelli L, Marchetti L, Arosio D, Riva S, Abdurashidova G, Beltram F, Falaschi A. The homeotic protein HOXC13 is a member of human DNA replication complexes. Cell Cycle 2009; 8:454-9; PMID:19182517; http://dx.doi.org/10.4161/cc.8.3.7649
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature 2007; 448:445-51; PMID:17597761; http://dx.doi.org/10.1038/nature05953
- Marchetti L, Comelli L, D’Innocenzo B, Puzzi L, Luin S, Arosio D, Calvello M, Mendoza-Maldonado R, Peverali F, Trovato F, et al. Homeotic proteins participate in the function of human-DNA replication origins. Nucleic Acids Res 2010; 38:8105-19; PMID:20693533; http://dx.doi.org/10.1093/nar/gkq688
- Toth EC, Marusic L, Ochem A, Patthy A, Pongor S, Giacca M, Falaschi A. Interactions of USF and Ku antigen with a human DNA region containing a replication origin. Nucleic Acids Res 1993; 21:3257-63; PMID:8341600; http://dx.doi.org/10.1093/nar/21.14.3257
- Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Mechali M. Specification of a DNA replication origin by a transcription complex. Nat Cell Biol 2004; 6:721-30; PMID:15247921; http://dx.doi.org/10.1038/ncb1149
- Hu YF, Hao ZL, Li R. Chromatin remodeling and activation of chromosomal DNA replication by an acidic transcriptional activation domain from BRCA1. Genes Dev 1999; 13:637-42; PMID:10090719; http://dx.doi.org/10.1101/gad.13.6.637
- Giacca M, Zentilin L, Norio P, Diviacco S, Dimitrova D, Contreas G, Biamonti G, Perini G, Weighardt F, Riva S, et al. Fine mapping of a replication origin of human DNA. Proc Natl Acad Sci U S A 1994; 91:7119-23; PMID:8041756; http://dx.doi.org/10.1073/pnas.91.15.7119
- Abdurashidova G, Riva S, Biamonti G, Giacca M, Falaschi A. Cell cycle modulation of protein-DNA interactions at a human replication origin. EMBO J 1998; 17:2961-9; PMID:9582289; http://dx.doi.org/10.1093/emboj/17.10.2961
- Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 2000; 287:2023-6; PMID:10720330; http://dx.doi.org/10.1126/science.287.5460.2023
- Abdurashidova G, Radulescu S, Sandoval O, Zahariev S, Danailov MB, Demidovich A, Santamaria L, Biamonti G, Riva S, Falaschi A. Functional interactions of DNA topoisomerases with a human replication origin. EMBO J 2007; 26:998-1009; PMID:17290216; http://dx.doi.org/10.1038/sj.emboj.7601578
- Falaschi A, Biamonti G, Cobianchi F, Csordas-Toth E, Faulkner G, Giacca M, Pedacchia D, Perini G, Riva S, Tribioli C. Presence of transcription signals in two putative DNA replication origins of human cells. Biochim Biophys Acta 1988; 951:430-42; PMID:3145020; http://dx.doi.org/10.1016/0167-4781(88)90117-0
- Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 2001; 20:2438-52; PMID: 11402339; http://dx.doi.org/10.1038/sj.onc.1204385
- Kovary K, Bravo R. The jun and fos protein families are both required for cell cycle progression in fibroblasts. Mol Cell Biol 1991; 11:4466-72; PMID:1908553
- Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene 2001; 20:2390-400; PMID:11402335; http://dx.doi.org/10.1038/sj.onc.1204383
- Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophy Acta 1991; 1072:129-57; PMID:1751545
- Zhou H, Zarubin T, Ji Z, Min Z, Zhu W, Downey JS, Lin S, Han J. Frequency and distribution of AP-1 sites in the human genome. DNA Res 2005; 12:139-50; PMID:16303745; http://dx.doi.org/10.1093/dnares/12.2.139
- Li M, Ge Q, Wang W, Wang J, Lu Z. c-Jun binding site identification in K562 cells. J Genet Genomics 2011; 38:235-42; PMID:21703547; http://dx.doi.org/10.1016/j.jgg.2011.05.004
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol 2002; 4:E131-6; PMID:11988758; http://dx.doi.org/10.1038/ncb0502-e131
- Riabowol K, Schiff J, Gilman MZ. Transcription factor AP-1 activity is required for initiation of DNA synthesis and is lost during cellular aging. Proc Natl Acad Sci U S A 1992; 89:157-61; PMID:1729683; http://dx.doi.org/10.1073/pnas.89.1.157
- Asano M, Ishikawa H, Ito Y. Possible involvement of nuclear oncoproteins in regulation of DNA replication. Tohoku J Exp Med 1992; 168:183-7; PMID:1339100; http://dx.doi.org/10.1620/tjem.168.183
- Ito K, Asano M, Hughes P, Kohzaki H, Masutani C, Hanaoka F, Kerppola T, Curran T, Murakami Y, Ito Y. c-Jun stimulates origin-dependent DNA unwinding by polyomavirus large Tantigen. EMBO J 1996; 15:5636-46; PMID:8896457
- Quintana DG, Hou Z, Thome KC, Hendricks M, Saha P, Dutta A. Identification of HsORC4, a member of the human origin of replication recognition complex. J Biol Chem 1997; 272:28247-51; PMID:9353276; http://dx.doi.org/10.1074/jbc.272.45.28247
- Laurent A, Bihan R, Omilli F, Deschamps S, Pellerin I. PBX proteins: much more than Hox cofactors. Int J Dev Biol 2008; 52:9-20; PMID:18033668; http://dx.doi.org/10.1387/ijdb.072304al
- Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene 2001; 20:2476-89; PMID:11402342; http://dx.doi.org/10.1038/sj.onc.1204386
- Ramirez-Carrozzi VR, Kerppola TK. Dynamics of Fos-Jun-NFAT1 complexes. Proc Natl Acad Sci U S A 2001; 98:4893-8; PMID:11320240; http://dx.doi.org/10.1073/pnas.091095998
- Wang WM, Lee AY, Chiang CM. One-step affinity tag purification of full-length recombinant human AP-1 complexes from bacterial inclusion bodies using a polycistronic expression system. Protein Expr Purif 2008; 59:144-52; PMID:18329890; http://dx.doi.org/10.1016/j.pep.2008.01.016
- Paolinelli R, Mendoza-Maldonado R, Cereseto A, Giacca M. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat Struct Mol Biol 2009; 16:412-20; PMID:19343071; http://dx.doi.org/10.1038/nsmb.1583
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 2000; 20:8602-12; PMID:11046155; http://dx.doi.org/10.1128/MCB.20.22.8602-8612.2000
- Hatzis P, Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol Cell 2002; 10:1467-77; PMID:12504020; http://dx.doi.org/10.1016/S1097-2765(02)00786-4
- Kouskouti A, Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J 2005; 24:347-57; PMID:15616580; http://dx.doi.org/10.1038/sj.emboj.7600516
- Drake FH, Hofmann GA, Mong SM, Bartus JO, Hertzberg RP, Johnson RK, Mattern MR, Mirabelli CK. In vitro and intracellular inhibition of topoisomerase II by the antitumor agent merbarone. Cancer Res 1989; 49:2578-83; PMID:2540903
- Cosenza SC, Yumet G, Soprano DR, Soprano KJ. Induction of c-fos and c-jun mRNA at the MG1 border is required for cell cycle progression. J Cell Biochem 1994; 55:503-12; PMID:7962180; http://dx.doi.org/10.1002/jcb.240550410
- Rampakakis E, Zannis-Hadjopoulos M. Transient dsDNA breaks during pre-replication complex assembly. Nucleic Acids Res 2009; 37:5714-24; PMID:19638425; http://dx.doi.org/10.1093/nar/gkp617
- Kusic J, Kojic S, Divac A, Stefanovic D. Noncanonical DNA elements in the lamin B2 origin of DNA replication. J Biol Chem 2005; 280:9848-54; PMID:15611042; http://dx.doi.org/10.1074/jbc.M408310200
- Kusic J, Tomic B, Divac A, Kojic S. Human initiation protein Orc4 prefers triple stranded DNA. Mol Biol Rep 2010; 37:2317-22; PMID:19690980; http://dx.doi.org/10.1007/s11033-009-9735-8
