 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Death domain-associated protein 6 (DAXX) is a histone chaperone, putative regulator of apoptosis and transcription, and candidate modulator of p53-mediated gene expression following DNA damage. DAXX becomes phosphorylated upon DNA damage, however regulation of this modification, and its relationship to p53 remain unclear. Here we show that in human cells exposed to ionizing radiation or genotoxic drugs etoposide and neocarzinostatin, DAXX became rapidly phosphorylated in an ATM kinase-dependent manner. Our deletion and site-directed mutagenesis experiments identified Serine 564 (S564) as the dominant ATM-targeted site of DAXX, and immunofluorescence experiments revealed localization of S564-phosphorylated DAXX to PML nuclear bodies. Furthermore, using a panel of human cell types, we identified the p53-regulated Wip1 protein phosphatase as a key negative regulator of DAXX phosphorylation at S564, both in vitro and in cells. Consistent with the emerging oncogenic role of Wip1, its DAXX-dephosphorylating impact was most apparent in cancer cell lines harboring gain-of-function mutant and/or overexpressed Wip1. Unexpectedly, while Wip1 depletion increased DAXX phosphorylation both before and after DNA damage and increased p53 stability and transcriptional activity, knock-down of DAXX impacted neither p53 stabilization nor p53-mediated expression of Gadd45a, Noxa, Mdm2, p21, Puma, Sesn2, Tigar or Wip1. Consistently, analyses of cells with genetic, TALEN-mediated DAXX deletion corroborated the notion that neither phosphorylated nor non-phosphorylated DAXX is required for p53-mediated gene expression upon DNA damage. Overall, we identify ATM kinase and Wip1 phosphatase as opposing regulators of DAXX-S564 phosphorylation, and propose that the role of DAXX phosphorylation and DAXX itself are independent of p53-mediated gene expression.
Keywords:
Abbreviations
| ATM | = | ataxia telangiectasia mutated |
| CHX | = | cycloheximide |
| DAXX | = | Death domain-associated protein 6 |
| IR | = | ionizing radiation |
| NCS | = | neocarzinostatin |
| p21 | = | p21WAF1/Cip1 cyclin-dependent kinase inhibitor 1 |
| PML | = | promyelocytic leukemia protein |
| VP16 | = | etoposide |
| Wip1 | = | Wild-type p53-induced phosphatase 1 |
Introduction
Death domain-associated protein 6 (DAXX) is a highly conserved multifunctional protein implicated in complex biological functions ranging from cell death to regulation of transcription. Initially described as a binding partner of Fas death domain, DAXX was shown to potentiate Fas-induced apoptosis and activates the Jun N-terminal kinase (JNK) pathway.Citation1 Subsequently, other studies suggested that under certain stress conditions DAXX promotes JNK-mediated apoptosis through activation of apoptosis signal-regulating kinase (ASK1).Citation2 In contrast to its pro-apoptotic role, it has been reported that DAXX depletion sensitizes cells to multiple pro-apoptotic stimuli in certain cellular contexts.Citation3,4 In addition, homozygous deletion of the Daxx gene in mice is lethal at day 9.5 of embryonic development and is accompanied by massive apoptosis in all tissues, indicating that DAXX functions as an anti-apoptotic molecule and is critical for organismal development.Citation5 Thus, the exact function of DAXX in regulation of cell death mechanisms remains unclear and it has become a controversial issue.
Arguably the best characterized function of DAXX is that of a transcriptional regulator that can repress or activate gene transcription. Reportedly, DAXX interacts with transcriptional co-regulators including CREB-binding protein (CBP) and histone deacetylase (HDAC) and directly with a number of DNA-binding transcription factors, including Pax3 and Pax5, ETS1, and p53 and its family members p73 and p63.Citation6-14 Moreover, recent studies have shown that DAXX is a specific histone H3.3/H4 chaperone and plays a role in chromatin remodeling and DNA methylation indicating that it may control gene expression also via epigenetic mechanisms.Citation8,15-21
Consistent with the involvement in transcriptional regulation, DAXX is primarily localized in subnuclear compartments including PML bodies, nucleoli, heterochromatin domains and nucleoplasm, however, it can translocate to the cytoplasm under certain stress conditions.Citation22-25
Interestingly, DAXX was also proposed to cooperate with other cellular factors to stimulate the multifaceted function of p53 as a tumor suppressor. In unstressed cells, the association of DAXX with HAUSP, a de-ubiquitylating enzyme originally reported to act on p53,Citation26 and Mdm2 (RING-finger E3 ligase) results in Mdm2-dependent p53 ubiquitylation and degradation. In response to DNA damage, dissociation of HAUSP, DAXX and p53 from Mdm2 occurs by an unknown mechanism and Mdm2 is self-ubiquitylated and degraded, which allows accumulation of p53 and its activation.Citation27 Another example of p53 activation has been shown in cells after UV treatment. Here, an Axin/DAXX/HIPK2/p53 complex is formed that was proposed to promote transcriptional activation of pro-apoptotic p53 target genes.Citation28 It is therefore suggested that DAXX exerts its anti-apoptotic function in unstressed primary cells (considering data in knock-out mice mentioned above), and promotes apoptosis in tumor cells or transformed cells exposed to various stresses. However, a precise function and better understanding of the biological roles played by DAXX and its interplay with p53 in apoptosis and other cellular mechanisms in different cell types under various conditions remain to be elucidated.
Cellular responses to DNA damageCitation29 are mediated by signaling through diverse protein post-translational modifications, particularly phosphorylation by several protein kinases including ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3-related) – the master regulators critical for the maintenance of genome integrity.Citation30 Recently, large numbers of candidate ATM/ATR substrates were identified in high-throughput screening projects, thereby raising a formidable challenge of their functional characterization.Citation31-33 Given the controversies and open questions surrounding the regulation of DAXX, its role(s) in modulation of apoptosis and DAXX's relationship with p53 in response to DNA damage, we have performed this study focused on DAXX phosphorylation and its regulation in response to diverse genotoxic insults. Among the key questions we have addressed are whether DAXX is phosphorylated, what are the key kinase(s) and phosphatase(s) involved in such potential phosphorylation-mediated modulation of DAXX and its regulatory balance, and to what extent might such regulatory mechanism impact p53 stability and expression of p53 target genes, particularly those implicated in triggering apoptosis.
Results
DAXX is rapidly phosphorylated on Serine 564 after DNA damage and localizes at PML nuclear bodies
To examine whether human DAXX is phosphorylated in response to DNA damage, various cell types were transiently transfected with a vector expressing wild-type FLAG-tagged DAXX (FLAG-DAXXWT) and were treated with the DNA damaging agents etoposide (VP16) or neocarzinostatin (NCS). Immunoprecipitation experiments revealed that FLAG-DAXXWT was phosphorylated after exposure to either VP16 or NCS, as measured using an antibody against peptide motifs (SQ/TQ) that are phosphorylated by ATM and/or ATR (). In agreement with a previous report,Citation34 deletion mapping and mutational analysis of 4 putative ATM/ATR phosphorylation motifs in the C-terminus of DAXX (S564, S707, S712 and T726) identified S564 as the predominant site of DNA damage induced DAXX phosphorylation ( and Fig. S3A).
Figure 1. (See previous page) DAXX is phosphorylated on S564 rapidly after DNA damage. (A) HEK 293T cells were transfected with a vector expressing FLAG-DAXXWT and treated or not, as indicated, with 8 nM NCS or10 μM VP16 for 1 hour. Cells were lysed and FLAG-tagged proteins immunoprecipitated using anti-FLAG M2 beads, prior to analysis by SDS-PAGE and western blotting with antibodies against phospho-(S/T) ATM/ATR substrate, FLAG M2, phospho-Chk2 (T68) and DAXX. (B) HEK 293T cells were transfected with expression constructs encoding FLAG-tagged wild-type DAXX, single (DAXXS564A), triple or quadruple mutant as indicated and treated with 10 μM VP16 for 1 hour. Cells were lysed, immunoprecipitated using anti-FLAG M2 beads and analyzed by protein gel blotting with indicated antibodies. (C) BJ fibroblasts were exposed to 10 Gy of IR, collected at the indicated time points and subjected to western blotting analysis using specific phospho-DAXX (S564), DAXX, phospho-p53 (S15) and phospho-Chk2 (T68) antibodies. GAPDH was used as a loading control. (D) BJ fibroblasts were transfected with control (siCTRL) or DAXX siRNA and 3 d later treated or not as indicated with 10 μM VP16 for 1 hour. Immunofluorescence analysis using α-PML (red) and α-P-DAXX (S564) (green) showed that upon DNA damage DAXX is preferentially phosphorylated at PML nuclear bodies.
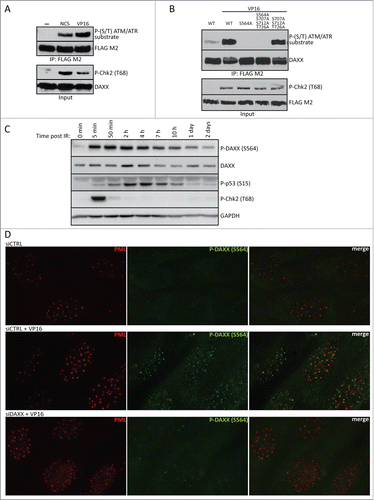
Next, we investigated the phosphorylation of endogenous DAXX upon DNA damage, using a rabbit polyclonal antibody raised against phosphorylated-DAXX (S564). Importantly, S564 was also phosphorylated in the context of endogenous DAXX protein in response to ionizing radiation (IR), appearing rapidly within minutes after irradiation and persisting for at least 24 hours following the 10 Gy radiation dose (). Moreover, phosphorylated DAXX was present in PML nuclear bodies, where DAXX normally resides (). Collectively, these experiments showed that DAXX is phosphorylated on S564 rapidly after DNA damage and localizes to PML nuclear bodies.
DAXX is not required for Mdm2/p53 stability or p53-mediated gene expression
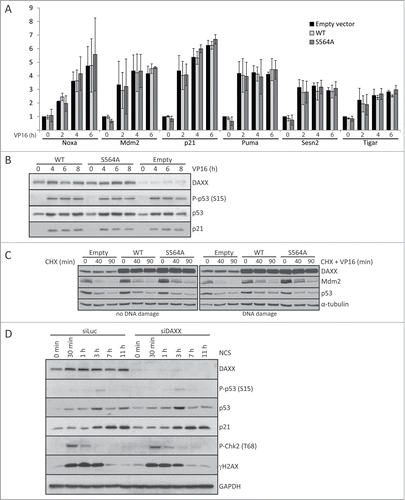
DAXX has been implicated in regulating wild-type p53,Citation27,35 however, the molecular basis of such link is poorly understood, and the data from diverse laboratories is controversial. For example, it has been shown that DAXX interacts with p53 and promotes p53-dependent apoptosis.Citation35 In contrast, the results of another study contradict this and suggest that DAXX only interacts with mutant p53.Citation14 Most recently, it was proposed that DAXX phosphorylation disrupts the interaction and stabilization of Mdm2 and thereby results in elevated p53 levels and induction of p53 target gene, p21WAF1/Cip1 (p21) encoding the CDK inhibitor p21.Citation34 To address these apparent contradictions and unanswered questions including impact of DAXX on p53-mediated induction of pro-apoptotic genes, we examined whether some central p53-dependent genes are regulated by S564 DAXX phosphorylation. First, we compared the expression of a series of 6 well-established p53-target genes in human cells expressing wt p53, before and after DNA damage at different time points. Surprisingly, damage-induced p21 transcription and protein expression, as well as transcription of 5 other important p53-dependent genes (Noxa, Mdm2, Puma, Sesn2 and Tigar) were not significantly altered in either human primary BJ fibroblasts ( and ) or U2OS sarcoma cells (Fig. S1A) stably over-expressing DAXXWT or DAXXS564A. Moreover, we failed to detect any effect of over-expressing either FLAG-DAXXWT or FLAG-DAXXS564A on Mdm2 and p53 protein stability, either before or after DNA damage ( and Fig. S1B). Finally, we did not observe any impact of DAXX depletion on both p53 stability and p21 expression in BJ cells before or after DNA damage ().
Figure 2. DAXX depletion or S564A mutation does not affect Mdm2/p53 stability or p53‑mediated gene expression. (A) BJ fibroblasts stably transduced with empty lentiviral pCDH vector or either pCDH-DAXXWT or pCDH-DAXXS564A were treated with 40 μM VP16 for 0, 2, 4 or 6 hours and RNA expression of the indicated p53-dependent genes was analyzed by quantitative RT‑PCR. Expression values were normalized to the average of 3 reference genes (β-actin, SDH and ALAS). (B) Transduced BJ fibroblast as in (A) were exposed to 40 μM VP16 for the indicated times and subjected to protein gel blotting analysis using add p53 - antibodies against DAXX, phospho-p53 (S15), p53 or p21. (C) U2OS cells transfected with pXJ41 Hdm2 (human Mdm2) together with empty FLAG-CMV, FLAG-DAXXWT or FLAG-DAXXS564A were treated with 50 μl/ml CHX alone or together with 10 μM VP16 for the specified time points. Cell were harvested and lysates separated by SDS–PAGE and probed with indicated antibodies. (D) BJ fibroblasts were depleted by control siRNA (siLuc) or siRNA against Wip1 and 3 d after transfection treated with 4 nM NCS. Cells were lysed at the indicated time points after DNA damage and analyzed by western blotting using labeled antibodies. GAPDH was used as a loading control.
In contrast to previously published results, our data suggested that DAXX does not regulate p53-dependent gene expression. However, the data described above and all previous reports have employed either cells over-expressing DAXX or cells in which DAXX was partially depleted, raising questions about the relevance of those findings. We therefore established U2OS cell lines in which DAXX was genetically deleted using TALEN-technology (Fig. S2A and B). To our knowledge, these are the first cellular experimental models in which human DAXX is completely absent.
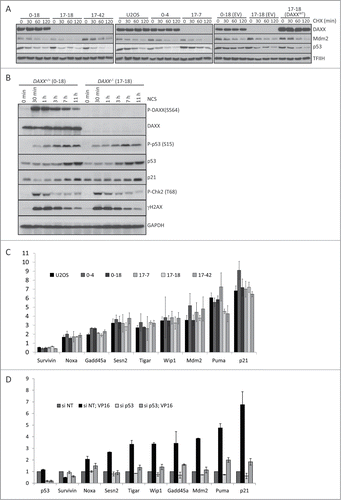
Next, we examined Mdm2 and p53 protein stability in control U2OS cells, 2 independent DAXX+/+single clonal isolates (clone 0–4 and clone 0–18) and 3 independent DAXX−/- clonal isolates (clones 17-7, 17–18, and 17–42, respectively). This experiment confirmed that DAXX deletion in U2OS cells does not significantly affect the stability of endogenous Mdm2 or p53 protein, in the absence of DNA damage (). Moreover, DAXX deletion also failed to impact DNA damage-induced increases in p53 or p21 protein levels in U2OS cells (). Finally, we examined whether DAXX deletion affected the activation of p53-dependent genes after NCS or VP16 treatment. Consistent with our results, obtained upon DAXX knock-down, we failed to detect any significant differences between DAXX+/+and DAXX−/- U2OS cells in expression of 9 established p53-dependent genes: Noxa, survivin, Gadd45a, Sesn2, Tigar, Wip1, Mdm2, Puma and p21 (). This negative result did not reflect the lack of p53 dependence for DNA damage-induced expression of these genes because apart from survivin, siRNA-mediated depletion of p53 prevented the VP16-induced expression of all the tested genes ().
Figure 3. DAXX deletion does not affect Mdm2/p53 stability or p53-mediated gene expression. (A) Mdm2 and p53 protein stability was examined in control U2OS cells, 2 independent DAXX+/+ clones (0–4, 0–18), 3 independent DAXX−/- clones (17-7, 17-18, 17–42) and in 17-7 DAXX−/- clone stably transduced with pCDH empty vector (EV) or pCDH-DAXXWT. Cells were collected after the treatment with 50 μl/ml CHX at the indicated time and subjected to protein gel blotting analysis using labeled antibodies. THIIF was used as a loading control. (B) DAXX+/+ (clone 0–18) and DAXX−/- (clones 17-18) cells were exposed to 4 nM NCS for the specified time points. Lysed cells were then separated by SDS–PAGE and immunoblotted with indicated antibodies. GAPDH was used as a loading control. (C) RNA expression of p53-dependent genes 8 hours after the treatment with 10 μM VP16 (expressed as fold change after VP16) in U2OS clones. RNA was analyzed by quantitative RT‑PCR and the expression values were normalized to the average of 3 reference genes (β-actin, SDH and ALAS). (D) U2OS cells transfected with control non-targeting siRNA and siRNA against p53 were treated with 10 μM VP16 for 8 hours and RNA expression of p53 and indicated p53-dependent genes was analyzed by quantitative RT‑PCR. The expression values were normalized to the average of 3 reference genes (β-actin, SDH and ALAS).
DAXX phosphorylation on Serine 564 is ATM-dependent
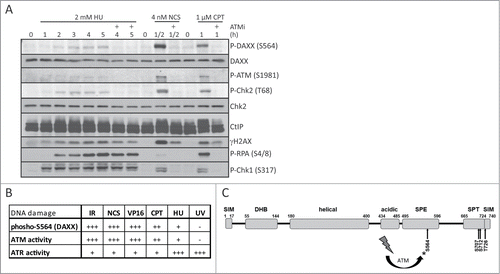
To gain further mechanistic insight into the observed phosphorylation of DAXX, we attempted to distinguish between the ATM and ATR kinase activity as plausible candidates responsible for phosphorylation of DAXX. First, we noted that S564 phosphorylation after NCS treatment causing mainly DNA double strand breaks was ablated by pretreatment of cells with either ATM inhibitor (KU-55933) or by shRNA-mediated depletion of ATM (Fig. S3A and B). Notably, we failed to detect significant phosphorylation of DAXX after exposure of cells to ultraviolet light (UV-C), a genotoxic insult that preferentially activates the ATR kinase (Fig. S3B). The fact that the cellular ATR response to UV irradiation was indeed activated in these experiments (yet did not lead to DAXX phosphorylation) is documented by the observed UV-induced phosphorylations of histone H2AX and Chk1 (Fig. S3B). In addition, treatment with a variety of DNA damaging agents revealed that the level of DAXX phosphorylation correlated with the extent of ATM activation, rather than with the extent of ATR activation ( and ). Intriguingly, our data therefore indicate, that DAXX may represent a very selective substrate of the ATM kinase that is not phosphorylated by ATR, despite both kinases share targeting the SQ/TQ motifs and the vast majority of the substrates identified to date are phosphorylated by both ATM and ATR.
Figure 4. Overview of ATM-dependent phosphorylation of DAXX at S564 after DNA damage. (A) U2OS cells were pretreated with DMSO or 10 μM ATM inhibitor KU-55933 (ATMi) for 30 min, exposed to 2 mM hydroxyurea (HU), 4 nM neocarzinostatin (NCS) or 1 μM camptothecin (CPT) for the indicated times and subjected to western blotting analysis using labeled antibodies. (B) Table showing the level of phosphorylated S564 on DAXX and the activity of ATM/ATR kinases after different types of DNA damage. (C) Sequence-based predicted modular organization of DAXX according to Escobar-Cabrera et al.Citation54 with indicated putative ATM/ATR phosphorylation motifs at C-terminus of DAXX (S564, S707, S712 and T726). SUMO-Interaction Motif (SIM), DAXX Helix Bundle (DHB) domain, segments rich in Ser/Pro/Glu residues (SPE) and in Ser/Pro/Thr (SPT) residues.
DAXX is a substrate of Wip1 phosphatase
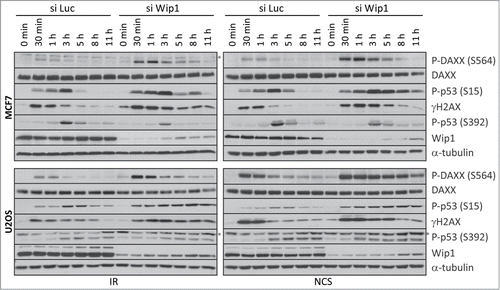
Given that DAXX is phosphorylated in response to a broad range of genotoxins, and is thus likely to be relevant for both physiologically occurring DNA damage and genotoxic treatments such as IR and chemotherapy, we addressed further how phosphorylation of S564 is regulated. Interestingly, we observed that DAXX S564 dephosphorylation coincides with an increased expression of the Wild-type p53-induced phosphatase 1 (Wip1 or PPM1D), the protein abundance of which was inversely correlated with DAXX S564 phosphorylation at 2–4 hours after IR (). Considering that Wip1 dephosphorylates multiple ATM substrates during the recovery phase of the DNA damage response,Citation36 we asked whether phospho-S564 of DAXX might also be a substrate of Wip1. To test this possibility, we first performed an in vitro phosphatase assay in which we incubated recombinant His-Wip1 with DAXX immunopurified from transfected U2OS cells pre-exposed to the DNA damaging VP16. Interestingly, wild-type Wip1 but not phosphatase-dead Wip1-D314A mutant was able to dephosphorylate DAXX protein (). To test whether Wip1 affects the level of phosphorylated DAXX in cells, we depleted Wip1 by siRNA in U2OS cells stably expressing FLAG-DAXXWT and assayed the level of phosphorylated DAXX after DNA damage. Western blotting showed that upon VP16-induced DNA damage, there was more phosphorylated DAXX in Wip1 siRNA-transfected cells compared to control GAPDH siRNA-transfected cells (). To determine whether Wip1 affects DNA damage-induced phosphorylation of endogenous DAXX, we examined S564 phosphorylation status in U2OS and MCF7 cells transfected with Wip1-targeting siRNA. Both IR-induced and NCS-induced phosphorylation of DAXX at S564 were greatly increased in U2OS and MCF7 cells in which Wip1 was depleted, compared to cells treated with control siRNA (). The effect of Wip1 downregulation on DAXX S564 phosphorylation was similar to established Wip1 substrates such as p53 (S15) and H2AX (S139).Citation37-39 In contrast, the phosphorylation of Ser392 in p53, an ATM-independent phosphorylation event, was not influenced by Wip1 depletion. Interestingly, in primary BJ cells, depletion of Wip1 impacted the phosphorylation of DAXX, p53 and H2AX to a lesser extent (Fig. S4A). This raises the intriguing possibility that the impact of Wip1 on DAXX regulation is more pronounced in cancer cells in which Wip1 is mutated and/or deregulated (Fig. S4B). Together, these results suggest that DAXX is a substrate of Wip1 phosphatase and that Wip1 regulates the level of DAXX phosphorylation before and after DNA damage in vivo.
Figure 5. DAXX is a substrate of Wip1 phosphatase. (A) BJ fibroblasts were exposed to 10 Gy or 2 Gy of IR and lysed at the indicated time points after DNA damage. Western blotting analysis using antibodies against phospho-DAXX (S564), DAXX, Wip1 or GAPDH showed that the DAXX S564 dephosphorylation coincides with increased expression of Wip1. (B) In vitro phosphatase assay was performed with recombinant wild-type Wip1 or phosphatase-dead Wip1-D314A mutant on FLAG-DAXXWT immunopurified from transfected U2OS cells exposed to DNA damage. Samples were separated by SDS–PAGE and probed with indicated antibodies. As control, phosphatase buffer without Mg2+ was used or treatment with lambda protein phosphatase (λPP). (C) Wip1 was depleted by siRNA in U2OS cells stably expressing FLAG-DAXXWT. Western blotting analysis using indicated antibodies showed that after DNA damage more phosphorylated DAXX is present in Wip1 siRNA treated cells compared to control GAPDH siRNA transfected cells.
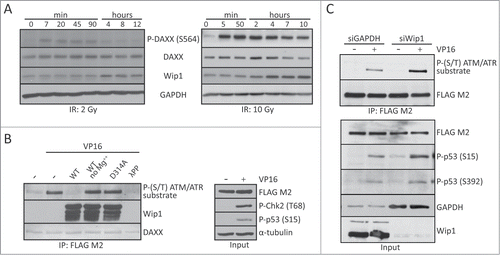
Figure 6. Wip1 affects DNA damage-induced phosphorylation of endogenous DAXX. MCF7 or U2OS cells were transfected with control (siLuc) or siRNA targeting Wip1 and 3 d later exposed to 5 Gy of IR or 4 nM NCS. Cells were lysed at the indicated time points after DNA damage and analyzed by protein gel blotting with the indicated antibodies (* unspecific band). α‑tubulin was used as a loading control.
In summary, in contrast to the currently prevailing concept, we find that neither DAXX phosphorylation nor DAXX itself functions to regulate p53 levels or p53-mediated gene expression. Mechanistically, we demonstrate that the phosphorylation status of DAXX at S564 is regulated following DNA damage by the opposing, yet coordinated, activities of ATM kinase and Wip1 phosphatase, respectively.
Discussion
DAXX is a multifunctional adaptor protein and putative histone chaperone that participates in numerous cellular processes, including transcriptional regulation, apoptosis and protein stability.Citation17,20,40-42 DAXX lacks any enzymatic activity and so most likely mediates its cellular function/s via direct interactions with its protein partners. Such biological functions of DAXX are likely fine-tuned by its interacting proteins and also by post-translational modifications of DAXX, such as phosphorylation. In this study we demonstrate that in human cells, both ectopic and endogenous DAXX is phosphorylated on S564 in an ATM-dependent manner following DNA damage. The apparently selective targeting of DAXX-S564 by ATM kinase is intriguing and could, if validated, serve as a surrogate biomarker for ATM activity in vivo. While we could exclude ATR as the S564 kinase, we did not directly examine a potential contribution of DNA-PK kinase to phosphorylation of DAXX on S564 in response to diverse genotoxic insults. On the other hand, the fact that the KU‑55933 inhibitor that we used in our present study is very selective in inhibiting ATM compared to DNA-PK (ATM is 200-fold more sensitive than DNA-PK), and that this inhibitor at a moderate concentration completely blocked the DNA damage-induced S564 phosphorylation suggests that DAXX is indeed a highly selective target of ATM kinase.
Another candidate phosphorylation site of DAXX, S712, was reported based on a proteomic screen for potential ATM/ATR-dependent phosphorylations.Citation32 We were unable to confirm phosphorylation of S712 in our multiple experiments, which suggested that S564 is the primary, if not the only phosphorylated SQ/TQ motif in human DAXX. Our finding that S564 is rapidly and reversibly phosphorylated by ATM in primary fibroblasts and also cancer cell lines following DNA damage is consistent with data from Tang et al.Citation34
The phosphorylation status of proteins is normally controlled by opposing activities of protein kinases and protein phosphatases. However, whether or how DAXX is dephosphorylated has so far been unknown. In mammalian cells, the serine/threonine phosphatase Wip1 acts on multiple substrates phosphorylated after DNA damage including p53, ATM, Mdm2, Chk1, Chk2 and p38 MAPK and is thus considered as a major homeostatic regulator of the DNA damage response.Citation37,43-47 In normal proliferating cells, Wip1 levels are relatively low and its expression increases after genotoxic stress.Citation48 We noticed in the current study that Wip1 protein levels increased during the same time period in which DAXX was dephosphorylated in vivo. Indeed, depletion of Wip1 resulted in persistent phosphorylation of DAXX, and this effect was apparent both before and after DNA damage, indicating that even the basal level of Wip1 expressed in the cells we examined contributes to a balanced steady-state level of DAXX phosphorylation. Moreover, phosphorylated DAXX was a substrate for recombinant Wip1 phosphatase activity, in vitro. These data identify, for the first time, DAXX as a novel substrate of Wip1 and reveal that Wip1 phosphatase can reverse DAXX phosphorylation at S564 within a few hours following DNA damage, similar to its previously identified role in dephosphorylating and reversing the activating phosphorylation of p53 at S15 and the key signaling phosphorylation of histone H2AX at Ser139.Citation37,38
Phosphorylation of several among the eighty-eight serines present within DAXX likely enables fine-tuning of its multiple functions, perhaps by regulating protein-protein interactions. For example, phosphorylation of S667 plays a role in DAXX interaction with the nuclear export carrier protein CRM1,Citation24 and CK2 kinase-mediated phosphorylation of S737/S739, within the Sumo-Interacting Motif (SIM), enhances binding of DAXX to SUMO-1-modified proteins.Citation49 In addition, phosphorylation of S669 by HIP kinases modulates the ability of DAXX to regulate transcriptionCitation50,51 and DAXX-mediated loading of histone H3.3 at gene promoters.Citation52 Finally, ASK1-mediated phosphorylation of DAXX at S176 and S184 regulates K63-linked polyubiquitylation of Lys122, a modification required for sustained activation of JNK.Citation53
In light of the above observations, the phosphorylation status of S564 might modulate DAXX binding to a known or some novel interacting partner(s), and/or impact on a posttranslational modification such as ubiquitylation or sumolyation. Interestingly, S564 is located in an unstructured SPE domain, the function of which is currently unknown (). The phosphorylation status of S564 might thus affect interaction with an as yet unidentified protein, or could modulate DAXX's spatial conformation and thereby potentially impact allosterically protein-protein binding through its N-terminal α-helical DHB domain or the C-terminal SPT and SIM domains. Intriguingly in this respect, the DAXX DHB domain (amino acids 55–144) was recently confirmed to be the site involved in interactions of DAXX with RassF1C, p53 and Mdm2 proteins.Citation54
Since the interaction of DAXX with Mdm2 or Rassf1C is disrupted following DNA damage,Citation27,55 perhaps ATM-mediated phosphorylation of S564 is involved in regulating this process, in order to regulate Mdm2 and p53 stability. Such a scenario would potentially be consistent with the reported co-association of DAXX with the Mdm2 deubiquitylating enzyme HAUSP, and the notion that HAUSP/DAXX- deubiquitylate and stabilize Mdm2 and thereby suppresses p53 activity, in the absence of DNA damage.Citation27,56 Consistent with this, it was reported by Tang et al.Citation34 that DAXX and Mdm2 no longer interact after DAXX is phosphorylated by ATM, thereby enabling Mdm2 degradation, p53 stabilization, and increased expression of p53-regulated genes in the presence of DNA damage. However, another group suggested that interaction between DAXX and wild-type p53 modulates expression of p53 target genes differentially, for example promoting the activation of Puma and inhibition of p21 and Mdm2.Citation35 It is clear from the above observations that the currently published data describing the role of DAXX in p53-mediated gene regulation are unclear and conflicting, perhaps reflecting differences in model systems employed to manipulate DAXX protein levels. Indeed, in our experiments, we failed to detect any impact of ectopically expressed wild-type DAXX or mutant DAXXS564A on Mdm2/p53 stability or on the efficacy of p53-mediated transactivation of any p53 target genes tested (e.g., p21, Puma, Tigar and other) either before or after DNA damage. The lack of impact observed in our experiments on either Mdm2 or p53 stability, or on expression of p53-regulated genes was not due to inefficient DAXX depletion, because we obtained similar results using a U2OS cell line in which DAXX was genetically deleted. To our knowledge our work represents the first time DAXX-deleted human cells have been employed to address the critical question of whether DAXX regulates p53-mediated gene expression, and unambiguously rules out a role for DAXX, and DAXX phosphorylation, in this process.
In summary, we show here that DAXX phosphorylated at S564 is regulated by opposing activities of ATM protein kinase and Wip1 protein phosphatase, both under standard growth conditions and particularly in response to DNA damage. While the interplay between the proteins examined here involves p53-induced Wip1 expression (and hence indirect impact of p53 on DAXX S564 phosphorylation status) and some form of interaction between DAXX and p53, our data demonstrate that DAXX regulates neither Mdm2 or p53 stabilization, nor p53-mediated gene expression. Our present findings may also be relevant to cancer, as ATM and p53 are established tumor suppressors and we and others have recently reported Wip1 activating mutations found in several types of tumors,Citation57,58 consistent with an oncogenic role of Wip1. Furthermore, recent studies have revealed coordinated roles of the DNA damage response and the ARF tumor suppressor pathway in their biological barrier functions against activated oncogenes and cancer progression.Citation59-61 Activated oncogenes evoke replication stress and endogenous DNA damage characterized by enhanced DNA double strand break formation (the DNA lesion to which ATM strongly responds) in transformed cells.Citation62 Considering that we find here ATM as the key kinase for DAXX phosphorylation after DNA damage, it will be important to find out to what extent might DAXX function, including its functional interplay with ATM, Wip1 and p53, and its interaction with ARF,Citation23 contribute to the DDR/ARF-mediated anti-cancer barrier, and whether and how is DAXX's role itself subverted in tumorigenesis.
Materials and Methods
Antibodies and reagents
The following antibodies were used for immunoblotting in this study: mouse monoclonal antibodies against Wip1 (sc-376257, Santa Cruz), DAXX (01, 02 + 03, Exbio), p53 (DO-1, sc-126, Santa Cruz), phospho-S139 of histone H2AX (05–636, Millipore), p21 (sc-56335, Santa Cruz), Mdm2 (AB-1, Calbiochem), α-tubulin (TU-01, Exbio), FLAG M2 (F1804, Sigma Aldrich), GAPDH (GTX30666, GeneTex), Chk2 (05–649, Millipore), ATM (ab31842, Abcam), phospho-S1981 of ATM (#4526, Cell Signaling); rabbit polyclonal antibodies against Wip1 (sc-20712, Santa Cruz), DAXX (M-112, sc-7152), phosho-(SQ/TQ) ATM/ATR substrate (#2851, Cell Signaling), phospho-S15 of p53 (#9284, Cell Signaling), phospho-S392 of p53 (#9281, Cell Signaling), phospho-S317 of Chk1 (#2344, Cell Signaling), phospho-T68 of Chk2 (#2661, Cell Signaling), p53 (FL‑393, sc-6243, Santa Cruz), HAUSP (sc-30164, Santa Cruz), TFIIH (sc-293, Santa Cruz), CtIP (A300–488A, Bethyl), phospho-S4/8 of RPA2 (A300-245A, Bethyl); rabbit monoclonal antibodies against DAXX (25C12, #4533, Cell Signaling), phospho-S139 of histone H2AX (#9718, Cell Signaling) and goat polyclonal antibody against β-actin (sc-1615, Santa Cruz). Rabbit polyclonal antibody specific to phosho-S564 of DAXX was made by Clonestar Peptide Services using EEESPV(Sp)QLFELE peptide. HRP-conjugated secondary antibodies goat anti-rabbit (170–6515) and goat anti-mouse (170–6516) were from Bio-Rad and rabbit anti-goat (sc2922) from Santa Cruz. Light chain specific antibodies mouse anti-rabbit (211-032-171) and goat anti-mouse (115-035-174) from Jackson ImmunoResearch Laboratories were used for protein detection after immunoprecipitation. Mouse monoclonal anti-PML (sc-966, Santa Cruz) and rabbit polyclonal anti-P-DAXX (S564) antibodies in combination with goat anti-mouse Alexa568 (A11036) and goat anti-rabbit Alexa488 (A11034) from Invitrogen were used for indirect immunofluorescence. Neocarzinostatin (NCS), etoposide (VP16), hydroxyurea (HU) and camptothecin (CPT) were purchased from Sigma and ATM kinase inhibitor (KU‑55933) and cycloheximide (CHX) were obtained from Calbiochem.
Plasmids and cloning
pFLAG-CMV-DAXXWT construct was generated by PCR and cloning into pFLAG-CMV-5a vector (Sigma Aldrich) and confirmed by sequencing. DAXX point mutations S564A, S707A, S712A, T726A (including their triple/quadruple combination) and S564E were prepared by site-directed mutagenesis. Briefly, PCR (using Phusion High Fidelity DNA polymerase from Thermo Scientific) was done with 30 nucleotides long primers containing desired point mutation. The PCR mixture was then incubated with DpnI restriction endonuclease to remove original plasmid and then transformed into competent E. coli strain TOP10. Lentiviral constructs expressing DAXX were prepared by subcloning the wild or site mutated forms of DAXX into pCDH-CMV-MCS-EF1-G418 vector (System Biosciences). pQE81L-Wip1WT and -Wip1D314A (phosphatase dead) constructs were described previously.Citation38
Cell lines
Human HEK 293T, MCF7 and U2OS cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Primary human BJ fibroblasts were grown in low glucose DMEM GlutaMAX (Gibco) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were grown at 37°C under 5% CO2 atmosphere and 95% humidity. Cell lines expressing stably integrated DAXX or control cells were prepared as follows: packaging lentiviral vector psPAX2 (Addgene, plasmid 12260) and pMD2.G (Addgene, plasmid 12259) were transfected using calcium phosphate into HEK 293T cells along with either empty pCDH-CMV or pCDH-CMV-DAXXWT or pCDH-CMV-DAXXS564A respectively. Medium containing lentiviral particles was harvested 36 hours after transfection and precipitated using PEG-it (System Biosciences) according to manufacturer´s instructions. Precipitated lentiviral particles were resuspended in 1xPBS and stored in -80°C. Target cells were infected with equivalent MOI for 24 hours. Transduced cells were selected using 2 μg/ml puromycine (Invitrogen) for 2–3 d or 1 mg/ml G418 (Invitrogen) for 4–5 d. Cells were irradiated by an X-ray generator T-200 (Wolf-Medizintechnik) as described.Citation63
Preparation of DAXX knockout U2OS cells
H6578 vectors for the expression of dimeric TALEN nuclease, targeting ACTGTTCTCAAG spacer present in exon 3 of DAXX gene, were purchased from TALEN Library Resource (Seoul National University). U2OS cells transfected with these constructs were subcloned into 96-wells plates and each of the obtained 150 clones was analyzed for the DAXX expression by SDS-PAGE. Down-regulation of DAXX mRNA, carried out by non-sense mediated decay, was tested by qRT-PCR in selected potential DAXX−/- clones. Three DAXX knock-out U2OS clones were finally obtained (clones 17-7, 17–18, 17–42). Clone 0–4 and clone 0–18 where DAXX deletion did not occur where chosen as control clones.
Transfection
HEK 293T and U2OS cells were transfected with plasmid DNA using FUGENE 6 (Roche) according to the manufacturer's instruction. Calcium phosphate transfection was performed under standard condition. Briefly, plasmids were mixed in 0.25 M CaCl2 solution followed by dropwise addition of BBS (BES buffered saline, pH 6.95) in 1:1 v/v ratio. Mixture was incubated 15 minutes at room temperature and added into medium supplemented with 25 μM chloroquine (C6628, Sigma) for 6 hours. siRNAs were transfected at final concentration 10 nM using RNAiMAX (Life Technologies). Oligonucleotides targeting DAXX (5′-CAGCCAAGCUCUAUGUCUA-3′) and Wip1 (5′‑CGAAAUGGCUUAAGUCGAA-3′) were purchased from Dharmacon. siRNA targeting LUC sequence (5′-CGUACGCGGAAUACUUCGA-3′) was from Microsynth and non-targeting control CTRL siRNA (Cat. Num. 4390843) and siRNA targeting GAPDH (Cat. Num. 4390849) were obtained from Ambion.
Immunoprecipitation and Western blotting
Collected cells were lysed in standard 1× Laemmli or ice cold IP lysis buffer (50 mM Tris-HCl pH 7.5; 150 mM NaCl; 0,5% NP40; 1 mM EDTA) supplemented with inhibitors of both proteases and phosphatases (Roche) followed by sonication and centrifugation. Extracts were incubated with anti-FLAG M2 (Sigma) beads for 2 h at 4°C. Beads were washed with IP buffer 3 times and bound proteins recovered by boiling in 2× Laemmli buffer. Samples were subjected to SDS PAGE, proteins transferred onto nitrocellulose membrane (Hybond ECL 0.45 μM; Amersham) and detected by specific antibodies combined with horseradish peroxidase-conjugated secondary antibodies. Peroxidase activity was detected by ECL (Amersham).
Phosphatase assay
His-Wip1WT and phosphatase dead His-Wip1D314A were purified from bacteria and in vitro phosphatase assay was performed as described.Citation38,64 Briefly, FLAG-DAXXWT was immunopurified from U2OS cells 1 hour after treatment with VP16 using FLAG M2 agarose (Sigma) and following extensive washing beads were further incubated (30°C for 15 min) with 100 ng of purified Wip1 in phosphatase buffer (40 mM HEPES pH 7.4, 100 mM NaCl, 50 mM KCl, 1 mM EGTA) supplemented or not with 50 mM MgCl2. Treatment with λ-phosphatase (New England Biolabs) was used as a positive control.
Quantitative Real Time PCR (qRT-PCR)
Total RNA was extracted from cells growing in 12‑wells plate using RNA Blue reagent (Top-BIO). 1 μg of extracted mRNA was reverse transcribed with RevertAid reverse transcriptase (Thermo Scientific) and prepared first-strand DNA was used for real-time qPCR with LightCycler 480 SYBR Green I master-mix, using LightCycler 480 instrument (Roche). Relative mRNA expression was calculated as according to Pfaffl,Citation65 normalization was done against ALAS, SDH, GAPDH and/or β-actin expression levels. Statistical analysis is based on the results of 3 independent experiments. Sequences of primers used for qRT-PCR:
β-actin (GGCATCCTCACCCTGAAGTA and AGGTG-TGGTGCCAGATTTTC),
SDH (AGATTGGCACCTAGTGGCTG and ACAAAGG-TAAGTGCCACGCT),
ALAS (GCGATGTACCCTCCAACACAACC and CCAC-TGGAAGAGCTGTGTGATGTG),
GAPDH (CACCACACTGAATCTCCCCT and CCCCTC-TTCAAGGGGTCTAC),
Mdm2 (TCGACCTAAAAATGGTTGCAT and GGCAGG-GCTTATTCCTTTTC),
p21WAF1/Cip1 (TGGAGACTCTCAGGGTCGAAA and GG-CGTTTGGAGTGGTAGAAATC),
Puma (TGAGCCAAACGTGACCACTA and GGCTGGC-TCAGGGAAGAT),
Noxa (GCTGGGGAGAAACAGTTCAG and AATGTGC-TGAGTTGGCACTG),
Sesn2 (GAGCGGAACCTCAAGGTCTA and AGTGCCT-CCAGAAGAGGTTG),
Tigar (ACCAGGTGAAAATGCGTGGA and AGTTGCT-TGGAGATCCTTGGG),
Wip1 (GGGAGTGATGGACTTTGGAA and CAAGATT-GTCCATGCTCACC),
Gadd45a (GAGCTCCTGCTCTTGGAGAC and TGTGG-ATTCGTCACCAGCAC),
DAXX (TCCCTCTGCATCCCTTCTC and CTTCTGGA-TCGCATTGTGTG),
p53 (GGCCCACTTCACCGTACTAA and TTCACAGA-TATGGGCCTTGA).
Fluorescence microscopy
Cells grown on coverslips were washed with PBS and fixed (4% formaldehyde in PBS) 10 min at room temperature. Cells were washed twice with PBS, permeabilized (10 min, 0.2% Triton X-100 in water), washed and blocked (20 min in 10% FBS in PBS). Incubation with the primary antibody (60 min, RT) was followed by wash (3 times in PBS) and incubation with appropriate fluorescently-labeled secondary antibody (60 min, room temperature). Coverslips were washed (3 times in PBS), stained with DAPI (1 μg/ml in water, 2 min) and mounted using VECTASHIELD (Vector Laboratories).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
988019_Supplementary_Materials.zip
Download Zip (919.1 KB)Acknowledgments
We thank K. Krejcikova for her excellent technical assistance.
Funding
This work was supported by the Grant Agency of the Czech Republic (Project P305/11/P683), the Danish Council for Independent Research and the Danish Cancer Society.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Yang X, Khosravi-Far R, Chang HY, Baltimore D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 1997; 89:1067-76; PMID:9215629; http://dx.doi.org/10.1016/S0092-8674(00)80294-9
- Chang HY, Nishitoh H, Yang X, Ichijo H, Baltimore D. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science 1998; 281:1860-3; PMID:9743501; http://dx.doi.org/10.1126/science.281.5384.1860
- Michaelson JS, Leder P. RNAi reveals anti-apoptotic and transcriptionally repressive activities of DAXX. J Cell Sci 2003; 116:345-52; PMID:12482920; http://dx.doi.org/10.1242/jcs.00234
- Chen LY, Chen JD. Daxx silencing sensitizes cells to multiple apoptotic pathways. Mol Cell Biol 2003; 23:7108-21; PMID:14517282; http://dx.doi.org/10.1128/MCB.23.20.7108-7121.2003
- Michaelson JS, Bader D, Kuo F, Kozak C, Leder P. Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev 1999; 13:1918-23; PMID:10444590; http://dx.doi.org/10.1101/gad.13.15.1918
- Huang YS, Chang CC, Huang TC, Hsieh YL, Shih HM. Daxx interacts with and modulates the activity of CREB. Cell Cycle 2012; 11:99-108; PMID:22185778; http://dx.doi.org/10.4161/cc.11.1.18430
- Kuo HY, Chang CC, Jeng JC, Hu HM, Lin DY, Maul GG, Kwok RP, Shih HM. SUMO modification negatively modulates the transcriptional activity of CREB-binding protein via the recruitment of Daxx. Proc Nat Acad Sci U S A 2005; 102:16973-8; PMID:16287980; http://dx.doi.org/10.1073/pnas.0504460102
- Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci 2002; 115:3319-30; PMID:12140263
- Hollenbach AD, Sublett JE, McPherson CJ, Grosveld G. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO J 1999; 18:3702-11; PMID:10393185; http://dx.doi.org/10.1093/emboj/18.13.3702
- Emelyanov AV, Kovac CR, Sepulveda MA, Birshtein BK. The interaction of Pax5 (BSAP) with Daxx can result in transcriptional activation in B cells. J Biol Chem 2002; 277:11156-64; PMID:11799127; http://dx.doi.org/10.1074/jbc.M111763200
- Li R, Pei H, Watson DK, Papas TS. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene 2000; 19:745-53; PMID:10698492; http://dx.doi.org/10.1038/sj.onc.1203385
- Zhao LY, Colosimo AL, Liu Y, Wan Y, Liao D. Adenovirus E1B 55-kilodalton oncoprotein binds to Daxx and eliminates enhancement of p53-dependent transcription by Daxx. J Virol 2003; 77:11809-21; PMID:14557665; http://dx.doi.org/10.1128/JVI.77.21.11809-11821.2003
- Kim EJ, Park JS, Um SJ. Identification of Daxx interacting with p73, one of the p53 family, and its regulation of p53 activity by competitive interaction with PML. Nucleic Acids Res 2003; 31:5356-67; PMID:12954772; http://dx.doi.org/10.1093/nar/gkg741
- Ohiro Y, Usheva A, Kobayashi S, Duffy SL, Nantz R, Gius D, Horikoshi N. Inhibition of stress-inducible kinase pathways by tumorigenic mutant p53. Mol Cell Biol 2003; 23:322-34; PMID:12482984; http://dx.doi.org/10.1128/MCB.23.1.322-334.2003
- Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Nat Acad Sci U S A 2003; 100:10635-40; PMID:12953102; http://dx.doi.org/10.1073/pnas.1937626100
- Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, Picketts DJ, Yang X. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J Biol Chem 2004; 279:20369-77; PMID:14990586; http://dx.doi.org/10.1074/jbc.M401321200
- Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev 2010; 24:1253-65; PMID:20504901; http://dx.doi.org/10.1101/gad.566910
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010; 140:678-91; PMID:20211137; http://dx.doi.org/10.1016/j.cell.2010.01.003
- Denizio JE, Elsasser SJ, Black BE. DAXX co-folds with H3.3/H4 using high local stability conferred by the H3.3 variant recognition residues. Nucleic Acids Res 2014; 42:4318-31; PMID:24493739; http://dx.doi.org/10.1093/nar/gku090
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Nat Acad Sci U S A 2010; 107:14075-80; PMID:20651253; http://dx.doi.org/10.1073/pnas.1008850107
- Corpet A, Olbrich T, Gwerder M, Fink D, Stucki M. Dynamics of histone H3.3 deposition in proliferating and senescent cells reveals a DAXX-dependent targeting to PML-NBs important for pericentromeric heterochromatin organization. Cell Cycle 2014; 13:249-67; PMID:24200965; http://dx.doi.org/10.4161/cc.26988
- Torii S, Egan DA, Evans RA, Reed JC. Human Daxx regulates Fas-induced apoptosis from nuclear PML oncogenic domains (PODs). EMBO J 1999; 18:6037-49; PMID:10545115; http://dx.doi.org/10.1093/emboj/18.21.6037
- Ivanchuk SM, Mondal S, Rutka JT. p14ARF interacts with DAXX: effects on HDM2 and p53. Cell Cycle 2008; 7:1836-50; PMID:18583933; http://dx.doi.org/10.4161/cc.7.12.6025
- Song JJ, Lee YJ. Tryptophan 621 and serine 667 residues of Daxx regulate its nuclear export during glucose deprivation. JJ Biol Chem 2004; 279:30573-8; PMID:15128734; http://dx.doi.org/10.1074/jbc.M404512200
- Ishov AM, Vladimirova OV, Maul GG. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J Cell Sci 2004; 117:3807-20; PMID:15252119; http://dx.doi.org/10.1242/jcs.01230
- Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 2004; 13:879-86; PMID:15053880; http://dx.doi.org/10.1016/S1097-2765(04)00157-1
- Tang J, Qu LK, Zhang J, Wang W, Michaelson JS, Degenhardt YY, El-Deiry WS, Yang X. Critical role for Daxx in regulating Mdm2. Nat Cell Biol 2006; 8:855-62; PMID:16845383; http://dx.doi.org/10.1038/ncb1442
- Li Q, Wang X, Wu X, Rui Y, Liu W, Wang J, Liou YC, Ye Z, Lin SC. Daxx cooperates with the Axin/HIPK2/p53 complex to induce cell death. Cancer Res 2007; 67:66-74; PMID:17210684; http://dx.doi.org/10.1158/0008-5472.CAN-06-1671
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009; 461:1071-8; PMID:19847258; http://dx.doi.org/10.1038/nature08467
- Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 2011; 13:1161-9; PMID:21968989; http://dx.doi.org/10.1038/ncb2344
- Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem 2007; 282:17330-4; PMID:17478428; http://dx.doi.org/10.1074/jbc.C700079200
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316:1160-6; PMID:17525332; http://dx.doi.org/10.1126/science.1140321
- Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics: MCP 2010; 9:1314-23; PMID:20164059; http://dx.doi.org/10.1074/mcp.M900616-MCP200
- Tang J, Agrawal T, Cheng Q, Qu L, Brewer MD, Chen J, Yang X. Phosphorylation of Daxx by ATM contributes to DNA damage-induced p53 activation. PloS One 2013; 8:e55813; PMID:23405218; http://dx.doi.org/10.1371/journal.pone.0055813
- Gostissa M, Morelli M, Mantovani F, Guida E, Piazza S, Collavin L, Brancolini C, Schneider C, Del Sal G. The transcriptional repressor hDaxx potentiates p53-dependent apoptosis. J Biol Chem 2004; 279:48013-23; PMID:15339933; http://dx.doi.org/10.1074/jbc.M310801200
- Yamaguchi H, Durell SR, Chatterjee DK, Anderson CW, Appella E. The Wip1 phosphatase PPM1D dephosphorylates SQ/TQ motifs in checkpoint substrates phosphorylated by PI3K-like kinases. Biochemistry 2007; 46:12594-603; PMID:17939684; http://dx.doi.org/10.1021/bi701096s
- Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev 2005; 19:1162-74; PMID:15870257; http://dx.doi.org/10.1101/gad.1291305
- Macurek L, Lindqvist A, Voets O, Kool J, Vos HR, Medema RH. Wip1 phosphatase is associated with chromatin and dephosphorylates gammaH2AX to promote checkpoint inhibition. Oncogene 2010; 29:2281-91; PMID:20101220; http://dx.doi.org/10.1038/onc.2009.501
- Moon SH, Nguyen TA, Darlington Y, Lu XB, Donehower LA. Dephosphorylation of gamma H2AX by WIP1 An important homeostatic regulatory event in DNA repair and cell cycle control. Cell Cycle 2010; 9:2092-6; PMID:20495376; http://dx.doi.org/10.4161/cc.9.11.11810
- Elsaesser SJ, Allis CD. HIRA and Daxx constitute two independent histone H3.3-containing predeposition complexes. Cold Spring Harbor Symp Quant Biol 2010; 75:27-34; PMID:21047901; http://dx.doi.org/10.1101/sqb.2010.75.008
- Elsasser SJ, Huang H, Lewis PW, Chin JW, Allis CD, Patel DJ. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature 2012; 491:560-5; PMID:23075851; http://dx.doi.org/10.1038/nature11608
- Liu CP, Xiong C, Wang M, Yu Z, Yang N, Chen P, Zhang Z, Li G, Xu RM. Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX. Nat Struct Mol Biol 2012; 19:1287-92; PMID:23142979; http://dx.doi.org/10.1038/nsmb.2439
- Lu X, Nguyen TA, Moon SH, Darlington Y, Sommer M, Donehower LA. The type 2C phosphatase Wip1: an oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metast Rev 2008; 27:123-35; PMID:18265945; http://dx.doi.org/10.1007/s10555-008-9127-x
- Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer cell 2007; 12:342-54; PMID:17936559; http://dx.doi.org/10.1016/j.ccr.2007.08.033
- Takekawa M, Adachi M, Nakahata A, Nakayama I, Itoh F, Tsukuda H, Taya Y, Imai K. p53-inducible wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. EMBO J 2000; 19:6517-26; PMID:11101524; http://dx.doi.org/10.1093/emboj/19.23.6517
- Fujimoto H, Onishi N, Kato N, Takekawa M, Xu XZ, Kosugi A, Kondo T, Imamura M, Oishi I, Yoda A, et al. Regulation of the antioncogenic Chk2 kinase by the oncogenic Wip1 phosphatase. Cell Death Differ 2006; 13:1170-80; PMID:16311512; http://dx.doi.org/10.1038/sj.cdd.4401801
- Shreeram S, Hee WK, Demidov ON, Kek C, Yamaguchi H, Fornace AJ Jr, Anderson CW, Appella E, Bulavin DV. Regulation of ATM/p53-dependent suppression of myc-induced lymphomas by Wip1 phosphatase. J Exp Med 2006; 203:2793-9; PMID:17158963; http://dx.doi.org/10.1084/jem.20061563
- Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, O'Connor PM, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Nat Acad Sci U S A 1997; 94:6048-53; PMID:9177166; http://dx.doi.org/10.1073/pnas.94.12.6048
- Chang CC, Naik MT, Huang YS, Jeng JC, Liao PH, Kuo HY, Ho CC, Hsieh YL, Lin CH, Huang NJ, et al. Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol Cell 2011; 42:62-74; PMID:21474068; http://dx.doi.org/10.1016/j.molcel.2011.02.022
- Ecsedy JA, Michaelson JS, Leder P. Homeodomain-interacting protein kinase 1 modulates Daxx localization, phosphorylation, and transcriptional activity. Mol Cell Biol 2003; 23:950-60; PMID:12529400; http://dx.doi.org/10.1128/MCB.23.3.950-960.2003
- Lan HC, Wu CF, Shih HM, Chung BC. Death-associated protein 6 (Daxx) mediates cAMP-dependent stimulation of Cyp11a1 (P450scc) transcription. J Biol Chem 2012; 287:5910-6; PMID:22199361; http://dx.doi.org/10.1074/jbc.M111.307603
- Michod D, Bartesaghi S, Khelifi A, Bellodi C, Berliocchi L, Nicotera P, Salomoni P. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron 2012; 74:122-35; PMID:22500635; http://dx.doi.org/10.1016/j.neuron.2012.02.021
- Fukuyo Y, Kitamura T, Inoue M, Horikoshi NT, Higashikubo R, Hunt CR, Usheva A, Horikoshi N. Phosphorylation-dependent Lys63-linked polyubiquitination of Daxx is essential for sustained TNF-{alpha}-induced ASK1 activation. Cancer Res 2009; 69:7512-7; PMID:19789334; http://dx.doi.org/10.1158/0008-5472.CAN-09-2148
- Escobar-Cabrera E, Lau DK, Giovinazzi S, Ishov AM, McIntosh LP. Structural characterization of the DAXX N-terminal helical bundle domain and its complex with Rassf1C. Structure 2010; 18:1642-53; PMID:21134643; http://dx.doi.org/10.1016/j.str.2010.09.016
- Kitagawa D, Kajiho H, Negishi T, Ura S, Watanabe T, Wada T, Ichijo H, Katada T, Nishina H. Release of RASSF1C from the nucleus by Daxx degradation links DNA damage and SAPK/JNK activation. EMBO J 2006; 25:3286-97; PMID:16810318; http://dx.doi.org/10.1038/sj.emboj.7601212
- Tang J, Qu L, Pang M, Yang X. Daxx is reciprocally regulated by Mdm2 and Hausp. Biochem Biophys Res Commun 2010; 393:542-5; PMID:20153724; http://dx.doi.org/10.1016/j.bbrc.2010.02.051
- Ruark E, Snape K, Humburg P, Loveday C, Bajrami I, Brough R, Rodrigues DN, Renwick A, Seal S, Ramsay E, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature 2013; 493:406-10; PMID:23242139; http://dx.doi.org/10.1038/nature11725
- Kleiblova P, Shaltiel IA, Benada J, Sevcik J, Pechackova S, Pohlreich P, Voest EE, Dundr P, Bartek J, Kleibl Z, et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J Cell Biol 2013; 201:511-21; PMID:23649806; http://dx.doi.org/10.1083/jcb.201210031
- Kotsinas A, Papanagnou P, Galanos P, Schramek D, Townsend P, Penninger JM, Bartek J, Gorgoulis VG. MKK7 and ARF: new players in the DNA damage response scenery. Cell Cycle 2014; 13:1227-36; PMID:24675893; http://dx.doi.org/10.4161/cc.28654
- Velimezi G, Liontos M, Vougas K, Roumeliotis T, Bartkova J, Sideridou M, Dereli-Oz A, Kocylowski M, Pateras IS, Evangelou K, et al. Functional interplay between the DNA-damage-response kinase ATM and ARF tumour suppressor protein in human cancer. Nat Cell Biol 2013; 15:967-77; PMID:23851489; http://dx.doi.org/10.1038/ncb2795
- Evangelou K, Bartkova J, Kotsinas A, Pateras IS, Liontos M, Velimezi G, Kosar M, Liloglou T, Trougakos IP, Dyrskjot L, et al. The DNA damage checkpoint precedes activation of ARF in response to escalating oncogenic stress during tumorigenesis. Cell Death Differ 2013; 20:1485-97; PMID:23852374; http://dx.doi.org/10.1038/cdd.2013.76
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science 2008; 319:1352-5; PMID:18323444; http://dx.doi.org/10.1126/science.1140735
- Moudry P, Lukas C, Macurek L, Hanzlikova H, Hodny Z, Lukas J, Bartek J. Ubiquitin-activating enzyme UBA1 is required for cellular response to DNA damage. Cell Cycle 2012; 11:1573-82; PMID:22456334; http://dx.doi.org/10.4161/cc.19978
- Macurek L, Benada J, Mullers E, Halim VA, Krejcikova K, Burdova K, Pechackova S, Hodny Z, Lindqvist A, Medema RH, et al. Downregulation of Wip1 phosphatase modulates the cellular threshold of DNA damage signaling in mitosis. Cell Cycle 2013; 12:251-62; PMID:23255129; http://dx.doi.org/10.4161/cc.23057
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45; PMID:11328886; http://dx.doi.org/10.1093/nar/29.9.e45
