Abstract
Posttranscriptional modification of the uridine located at the wobble position (U34) of tRNAs is crucial for optimization of translation. Defects in the U34 modification of mitochondrial-tRNAs are associated with a group of rare diseases collectively characterized by the impairment of the oxidative phosphorylation system. Retrograde signaling pathways from mitochondria to nucleus are involved in the pathophysiology of these diseases. These pathways may be triggered by not only the disturbance of the mitochondrial (mt) translation caused by hypomodification of tRNAs, but also as a result of nonconventional roles of mt-tRNAs and mt-tRNA-modifying enzymes. The evolutionary conservation of these enzymes supports their importance for cell and organismal functions. Interestingly, bacterial and eukaryotic cells respond to stress by altering the expression or activity of these tRNA-modifying enzymes, which leads to changes in the modification status of tRNAs. This review summarizes recent findings about these enzymes and sets them within the previous data context.
General Information
Over the last decade, accumulating data have shown the association of several rare human diseases with defects in the posttranscriptional modification of mitochondrial (mt) tRNAs.Citation1-6 However, the underlying pathophysiological mechanisms remain unclear.Citation2,4,7-9 In some of these diseases, lack of the modifications that are normally present in the uridine located at the wobble position (U34) of the anticodon is an indirect consequence of mutations in tRNA genes encoded by mt-DNA.Citation2 In other cases, disease is caused directly by defects in nuclear-encoded proteins responsible for U34 modifications of mt-tRNAs.Citation3,4 These proteins are evolutionary conserved from bacteria to humans,Citation10-13 but important aspects concerning their regulation and biochemical activities are not well-known. By exploring the precise cellular functions of the mt-tRNA modification enzymes and their regulatory mechanisms, it may be possible to uncover new aspects of the pathophysiology of the aforementioned diseases, and to design specific therapeutic approaches.
tRNAs are by far the most extensively and diversely modified of all cellular RNAs with about 10% of modified nucleotides per molecule.Citation14 Modifications are introduced posttranscriptionally by enzymes that are often highly specific for tRNA substrates and position, with some exceptions within pseudouridine synthases and dihydrouridine synthases.Citation15-17 A few methyltransferases have also been shown to act at several positions on an RNA molecule or different RNA types. Thus, Bacillus subtilis methyltransferase RlmCD acts at 2 positions of 23 S rRNA,Citation18 and Escherichia coli methyltransferase RlmN has dual specificity and recognizes both rRNA and tRNA as substrates,Citation19 whereas in eukaryotes, Trm4 is a multisite methyltansferase and methylthiotransferase CDK5RAP1 (with homology to the bacterial MiaB protein) acts on tRNAs and nuclear polyadenylated RNAs.Citation20,21
Modifications do not generally appear essential for cell viability, yet their importance may be revealed under stress or other specific conditions. Some play a critical role in the fine tuning of the function of tRNAs in translation or in other processes like cell signaling.Citation22-29
Modifications cluster in 2 main regions of the tRNA molecule: the structural core and the anticodon stem loop. Modifications in the structural core are relatively simple (e.g., methylations, pseudouridylations and dihydrouridylations) and often contribute to stabilize the L-shaped structure,Citation30,31 although thiolation of the uridine at position 8 (s4U) of E. coli tRNAs can serve as a cellular sensor.Citation27 Modifications within the anticodon stem loop include methylations and pseudouridylations together with more complex additions, which collectively optimize the efficiency of tRNAs in the mRNA decoding process, specially modifications at positions 34 (the wobble position) and purine 37, 3´-adjacent to the anticodon.Citation22,24,32 Positions 34 and 37 in the anticodon loop present the widest variety of modifications found among all RNAs, which reinforces the idea that the modified nucleosides at both positions play crucial roles in tRNA functions.Citation22,24,26
In this review, we highlight the modifications that occur at the wobble uridine of the tRNAs reading NNA/NNG codons in 2-codon boxes.
Function of Modifications at the Wobble Uridine
Modifications at wobble uridines are classified into 2 groups according to their chemical structures: 5-hydroxyuridine derivatives (xo5U), with an oxygen atom bonded directly to the C5 atom of the uracil base; 5-methyluridine derivatives (xm5U), with a methylene carbon bonded directly to the C5 atom. xo5U-type modifications (where x symbolizes any of several different groups and o5 stands for the oxygen bonded to uracil) are present in tRNAs reading family codon boxes and expand recognition to 3 or 4 synonymous codons.Citation33,34 xm5U-type modifications, where m5 stands for methylene carbon, are usually found in tRNAs that decode 2-family box codons ending in A or G. These nucleosides can also carry an additional 2-thio (xm5s2U) or a 2'-O-methyl group (xm5Um). It has been proposed that modified nucleosides of xm5s2U-type restrict the wobble capacity of uridine, thereby strengthening recognition of purine-ending codons and preventing misrecognition of the near-cognate codons ending in pyrimidines.Citation35 However, genetic and structural data suggest that the inability to pair with pyrimidine-ending codons might not be due to such modifications.Citation36-38 Instead both s2 and xm5 seem to be important for modulating geometry of the codon:anticodon pairs at the wobble position and thus the relative efficiency of anticodons in reading cognate codons.Citation38-41 Lack of s2 and xm5 causes translational frameshifting, reduces the read through at nonsense codons and the efficiency of suppressor tRNAs, and produces a pleitropic phenotype in both bacteria and eukaryotes.Citation24,32,42
The Escherichia coli Modification Pathways of the Wobble Uridine
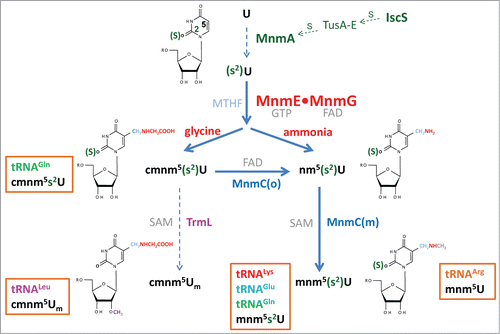
In E. coli, the wobble uridine (U34) of tRNALysmnm5s2UUU, tRNAGlumnm5s2UUC, tRNAGln(c)mnm5s2UUG, tRNALeucmnm5UmAA, and tRNAArgmnm5UCU is hypermodified through the action of the MnmEG pathway at position 5, the MnmA pathway at position 2, and SPOUT methyltransferase TrmL (previously named YibK), which is responsible for the methylation of the 2´-hydroxyl group of the ribose. MnmEG is the only pathway that can act on all 5 tRNAs, whereas MnmA functions on tRNALysmnm5s2UUU, tRNAGlumnm5s2UUC and tRNAGln(c)mnm5s2UUG, and TrmL works only on tRNALeucmnm5UmAA (). It is noteworthy that TrmL also modifies tRNALeuCmAA, but this tRNA is not a substrate in the MnmEG and MnmA pathways.Citation43
Figure 1. Synthesis of xm5(s2)U(m)-type nucleosides in E. coli. The MnmEG complex acts on position 5 of U34 in tRNALysmnm5s2UUU, tRNAGlumnm5s2UUC, tRNAGln(c)mnm5s2UUG, tRNALeucmnm5UmAA and tRNAArgmnm5UCU. MnmE binds GTP and MTHF, while MnmG is a FAD- and NADH-binding protein. In vitro, the MnmEG complex uses glycine and ammonium to respectively incorporate cmnm or nm at position 5 of U34 in the tRNA substrates. Thiolation at position 2 of U34 is catalyzed by MnmA on tRNALysmnm5s2UUU, tRNAGlumnm5s2UUC, tRNAGln(c)mnm5s2UUG, whereas the SAM-dependent TrmL enzyme methylates the 2´-OH group of the U-ribose in tRNALeucmnm5UmAA. MnmEG- and MnmA-catalyzed modifications occur independently of each other; thus thiolation may precede or follow the synthesis of the side chain at position 5. The FAD-dependent activity MnmC(o) of the bifunctional enzyme MnmC transforms cmnm5(s2)U into nm5(s2)U, whereas the MnmC(m) activity of MnmC transforms nm5(s2)U into mnm5s2U using SAM as the methyl donor.
TrmL requires a correct anticodon loop sequence and modification pattern. By using a chimera version of tRNALeuCAA, it has been shown that TrmL specifically recognizes a pyrimidine nucleoside at position 34, clearly prefers adenosine at position 35, and fails to methylate without prior addition of the ms2i6A modification at position 37.Citation43 This finding suggests that methylation by TrmL occurs as a late step in tRNA maturation. In vitro, TrmL catalyzes the 2´-O-methylation of the tRNALeuCAA chimera without the help of other proteins, which indicates that it functions independently despite being one of the smallest SPOUT enzymes. These enzymes exhibit an unusual α/β fold with a deep topological knot in the C-terminal half, and most harbor C-terminal or N-terminal extensions, which serve to bind the tRNA substrate. However, TrmL belongs to the group of minimalist SPOUT enzymes, which contain only the catalytic SPOUT domain and lack extensions.Citation44 A recent report has demonstrated that TrmL can efficiently methylate native tRNALeu isoacceptors.Citation45 In the same work, the crystal structures of TrmL in the apo form and in complex with S-adenosyl-homocysteine (the by-product of the methyl transfer reaction) were solved, revealing the cofactor binding site and a possible active site. Finally, a mutational analysis suggested that TrmL functions as a homodimer by using the C-terminal half of the SPOUT domain for catalysis and residues of the less-conserved N-terminal half of the other subunit for tRNA recognition.Citation45 It is important to note that loss of TrmL methylation reduces the efficiency of codon−wobble base interaction, which has a biological cost as a yibK-null mutant is out-competed by the wild-type strain in multiple-round growth experiments.Citation43
In the MnmA pathway (), cysteine desulfurase IscS transfers the persulfide moiety to MnmA through the sulfur relay chain formed by TusA/TusBCD/TusD.Citation46 IscS also provides other modification pathways with sulfur, which leads to the formation of s4U8, s2C32, and ms2i6A37.Citation47,48 The ability of IscS to interact with several different acceptor proteins is due to the conformational plasticity of a long loop where the catalytic Cys is located.Citation49 IscA and MnmA are evolutionary conserved,Citation24,50 but sulfur transfer mediators are not,Citation51 and the intermediate sulfur carriers in eukaryotic mitochondria remain to be identified. MnmA is a member of the ATP-pyrophosphatase family which exhibits a PP-loop as a signature motif. This enzyme recognizes nucleotides U34 and U35, which are present in the anticodon of tRNALysmnm5s2UUU, tRNAGlumnm5s2UUC and tRNAGln(c)mnm5s2UUG, and it uses a 2-step mechanism to sulfurate U34 through an adenylated U34 intermediate.Citation52 Further information on the bacterial IscS-MnmA pathway and similar eukaryotic pathways can be obtained from recent reviews.Citation24,32,50
It should be pointed out that the isolation of E. coli mutants containing mnm5 or s2 suggests that these modifications occur independently of each other.Citation53,54 In fact, both reactions have been performed in vitro using in vitro transcribed tRNA; i.e. an unmodified tRNA.Citation46,55 Notwithstanding, the possibility that the presence of the s2 group may facilitate the modification of position 5 by modulating the electron distribution in the uridine ring has not been studied.
The MnmEG pathway initiates the modification of U34 at position 5 with the action of the MnmEG complex, formed by multidomain proteins MnmE and MnmG. MnmE (formerly TrmE) is a GTP- and tetrahydrofolate- (THF-) binding protein, whereas MnmG (formerly GidA) is a FAD- and NADH-binding protein ().Citation32 MnmE is a dimeric protein with each monomer (50 kDa) consisting of 3 domains: an N-terminal domain responsible for constitutive dimerization and the binding of THF; a middle helical domain; a G-domain located far away from the THF-domain. MnmG is also a dimeric protein with each monomer (69 kDa) composed of a FAD-binding domain, an insertion domain and a helical C-terminal domain required for the interaction with MnmE. MnmE and MnmG form an α2β2 complex in which both proteins appear to function interdependently.Citation55,56 The complex catalyzes in vitro the addition of the aminomethyl (nm) and carboxymethylaminomethyl (cmnm) groups at position 5 of U34 using ammonium and glycine, respectively.Citation55 Both reactions require GTP, FAD, and a tetrahydrofolate (THF) derivative, likely methylene-THF, which serves as the donor of the methylene carbon bonded directly to the C5 atom. Since the reactions function in the absence of any THF-derivative, we thought that the recombinant MnmE protein copurifies with the one-carbon unit donor. To avoid this problem and identify the donor, MnmE and MnmG were purified from a folE::cat mutant, in which GTP cyclohydrolase I, the first enzyme of the de novo THF pathway, is lacking. Therefore, only some folate-related salvage pathways may be active. In this way, it was possible to observe a more efficient modification reaction when methylene-THF (MTHF) was used as the one-carbon unit donor. This finding, together with genetic data, has led to the proposal that MTHF, and not the more reactive formyl-THF, is the substrate for the modification reaction.Citation55
The MnmEG reaction also requires NADH if the FAD concentration is low (∼2 μM), which suggests that FAD undergoes an oxidation-reduction cycle during the modification reaction.Citation55 In the model proposed by our group, FAD would receive electrons from methylene-THF and would subsequently donate them to some reaction intermediate.Citation32,55 A high concentration of FAD (>50 μM) would guarantee the NADH-independent progress of the reaction, whereas a low FAD concentration (∼2 μM, a value close to the Kd for FAD binding to MnmG) would facilitate the FADH2 release from the enzyme, which could be reoxidized under the aerobic conditions used in the in vitro reaction. Oxidized FAD could once again occupy the binding site on MnmG, and would thus require the participation of NADH to be reduced and used in a later reaction step. The model, still awaiting experimental confirmation, requires the THF-binding site of MnmE and the FAD-binding site of MnmG being close enough in the complex so that FAD can receive electrons from MTHF.
The MnmE GTPase Cycle as a Regulator of the tRNA Modifying Function of the MnmEG Enzyme
GTP hydrolysis by MnmE is essential for tRNA modification,Citation57-60 yet the precise role of GTP hydrolysis remains unsolved. MnmE does not follow the prototypical Ras−GTPase cycle because its biochemical properties and activation mechanism render the participation of GTPase-activating proteins (GAPs) and guanine nucleotide-exchange factors (GEFs) unnecessary, at least under the in vitro assay conditions.Citation61,62 MnmE GTPase activity is stimulated by a cys, nucleotide- and potassium-dependent dimerization of its G-domains.Citation63 This dimerization leads to a reciprocal complementation of the G-domains at their active sites, which allows the GTPase machinery to achieve the catalytically competent conformation. Recently, the kinetics of the MnmE GTPase cycle has been studied under single-turnover conditions with stopped- and quench-flow techniques.Citation60 The data obtained indicate that the MnmE GTPase cycle is a sequential process consisting in GTP binding, G-domain dimerization, GTP hydrolysis, G-domain dissociation and the release of reaction products GDP and Pi, with G-domain dissociation being the rate-limiting step of the GTPase reaction (). The releases of GDP and Pi might occur instantaneously when the dimer is undone after GTP hydrolysis, although the conformational rearrangements leading to dimer dissociation appear relatively slow.
Figure 2. The GTPase cycle of MnmE. (A) Sequential process of MnmE conformational changes during the GTPase cycle. MnmE is a dimeric protein with each monomer consisting of 3 domains: an N-terminal domain responsible for constitutive dimerization and the binding of THF (smaller circle); a middle helical domain (oval); a G-domain (bigger circle) located far away from the THF domain. On the left of the schematic, a dimeric MnmE protein is represented with its G-domains in the apo monomeric state. Transition state and GTP-hydrolysis are rapidly reached by conformational reorganization after GTP binding. Dissociation of G-domains is the rate-limiting step of the GTPase cycle and likely the driving force for the functional activation ('ON' state) of MnmE. The releases of Pi and GDP may occur instantaneously during the G-domain dissociation. (B) The 'ON' and 'OFF' state of the MnmEG complex. A tetrameric MnmEG complex is represented, with the dimeric MnmE protein in the upper part of the complex and the MnmG dimeric protein in the lower part. The FAD-binding and insertion domains of each MnmG monomer are represented by a bigger oval, while the helical domain required for the interaction with MnmE is represented by a smaller oval. G-domain dissociation is directly responsible for the "ON" state of MnmE, in contrast to other GTPases like Ras-type proteins. At GDP and Pi physiological concentrations, the MnmE GTPase cycle is inhibited. This feedback mechanism may prevent useless GTP hydrolysis in vivo.
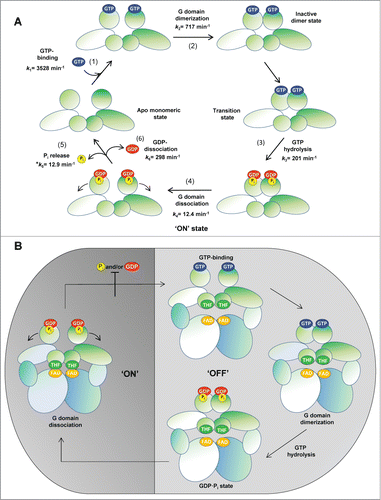
Mutational analyses have gradually revealed that GTP binding, GTP hydrolysis and, finally, post-hydrolysis G-domain dissociation are required for MnmE to be functionally active.Citation57,58,60,63 The GTPase-switch paradigm, in which a GTPase switches between an active GTP-bound state and an inactive GDP-bound state, has been used to interpret the regulatory mechanism of Ras-like GTPases. Strikingly, the MnmE GTPase cycle differs extensively from the Ras model as not only GTP hydrolysis, but also appropriate conformational changes during the G-domain dissociation, are required for MnmE to achieve the functionally active state. Therefore, we proposed that G-domain dissociation is directly responsible for the "ON" state of MnmE and, accordingly, that MnmE provides a new paradigm of how the ON/OFF cycling of GTPases can regulate a cellular process (). However, if we consider that molecular rearrangements occur in MnmE during the GTPase cycle (), the possibility of rearrangements associated with each cycle stage being control points to drive the complex modification reaction (multimodal switch model) cannot be ruled out. A mechanism based on multiple conformational switches, instead of the classical bimodal ON/OFF mechanism of Ras, has been previously proposed to control the functioning of the GTPase pair formed by the signal recognition particle (SRP) and the SRP receptor.Citation64
Interestingly, the MnmE GTPase cycle is controlled negatively by the hydrolysis products GDP and Pi, as it is inhibited at GDP and Pi physiological concentrations ().Citation60 This feedback mechanism may prevent useless GTP hydrolysis in vivo, but then, a conformational change might be required to remove product inhibition and to initiate a new GTPase/tRNA-modification cycle. This change could be mediated by the binding of a new unmodified tRNA molecule, but other options cannot be ruled out.
Recently, Versées and col. used small-angle X-ray scattering (SAXS) to characterize the nucleotide-induced conformational changes of MnmE and the mode of interaction among MnmE, MnmG and tRNA.Citation65 Models of the MnmEG complex were generated by docking MnmG to MnmE, and then validated and ranked using SAXS. The selected model was further supported by the molecular weight obtained by size exclusion chromatography (SEC) coupled to multiangle light scattering (SEC-MALS) measurements. In this model, one MnmE dimer was bound via the N-terminal domain and the helical domain of one subunit to the C-terminal domain of one subunit of the MnmG dimer in a nonsymmetric manner, leaving one protein-binding site vacant both on MnmE and MnmG, which could be used for further oligomerization. Notably, in the proposed MnmE-MnmG interface, the THF-binding site of the MnmE subunit and the FAD-binding site of the MnmG subunit are oriented toward each other, which may facilitate collaboration between both active sites, as required in the current model for the modification reaction.Citation55
Versées and col. also found that the binding of GDP and aluminum fluoride (GDP−AlFx), a transition-state mimic, by MnmE induces the formation of an MnmEG complex that contains 2 dimers of MnmE and one dimer of MnmG (α4β2).Citation65 A model was then built by placing a second MnmE dimer at the vacant binding site of MnmG in the α2β2 model, thus generating an “α2β2α2” form, which was supported by the coincidence of the theoretical and experimental scattering curves. Moreover by using an MnmE variant (E282A) with slow-hydrolase activity, a correlation between α4β2-complex disassembly and GTP hydrolysis was observed. Thus, the authors proposed a model in which GTP binding to MnmE in the α2β2 complex induced allosteric changes on MnmG, leading to the binding of a second MnmE dimer on the opposite side of MnmG, which resulted in an α2β2α2 (i.e., α4β2) complex, which would dissociate again to an α2β2 form after GTP hydrolysis. Given that SAXS experiments have also suggested that only one subunit of MnmG is bound to a tRNA molecule, Versees and col proposed that each MnmG monomer in the α4β2 complex could bind one tRNA, which would be modified prior to or during GTP hydrolysis, and would be released during the dissociation of the large complex.Citation65 Further research is still needed to solve both the role of the GTPase cycle in the modification reaction and the biological meaning of the oligomerization states of the MnmEG complex.
The MnmEG-MnmC Pathway. Reprogramming the Modification at Position 5 of U34.
The wobble uridine of the tRNA species modified by MnmEG may be further modified by the 2-domain, bifunctional enzyme MnmC, which transforms MnmEG products (nm5U and cmnm5U) into mnm5U (). The C-terminal domain of MnmC, named MnmC(o), is a FAD-dependent oxidoreductase that catalyzes the deacetylation of cmnm5U to produce nm5U, whereas the N-terminal domain of MnmC, designated MnmC(m), is a SAM-dependent methylase that transforms nm5U into mnm5U. In bulk tRNA purified from null mnmC mutants, the products of MnmEG cmnm5U and nm5U were also observed, indicating that the so-called glycine and ammonium pathways of MnmEG are functional in vivo.Citation55 However, cmnm5U had been previously identified as the final modification in tRNAs decoding Gln and Leu, suggesting that the ammonium pathway does not function on both tRNAs. In a recent study, we found that a small fraction of tRNA decoding Gln contains nm5U (when purified from a null mnmC mutant).Citation66 These results led us to propose that the nomenclature of this tRNA should be changed to tRNAGln(c)mnm5s2UUG. Strikingly, we were unable to observe activity of the ammonium pathway on tRNALeucmnm5UmAA in vivo despite the MnmEG complex modifying this tRNA substrate in vitro via the ammonium pathway.Citation66
The use of strains carrying the ΔmnmC or ΔmnmC(o) mutation allowed us to study the ability of MnmEG to use the glycine or the ammonium pathway under different growth conditions and, in this way, to detect the reprogramming of the U34 base modification in both bulk and specific tRNAs.Citation66 An analysis of total tRNA revealed that when strains were grown in the relatively rich medium LBT (LB with thymine), the glycine pathway prevailed over the ammonium pathway in the exponential phase as cmnm5s2U was the most abundant nucleoside, whereas the ammonium pathway appeared to be preferably used during entry into the stationary phase ().
Figure 3. Synthesis of cmnm5s2U and nm5s2U is depending on the growth medium, growth phase and tRNA species. HPLC analysis of total tRNA (A), tRNALysmnm5s2UUU (B), tRNAGln(c)mnm5s2UUG (C), and tRNALeucmnm5UmAA (D), purified from a strain carrying a ΔmnmC (A–C) or a null trmL mutation (D), was carried out as described.Citation66 The strains were grown in LBT. In (A), the percentage of nucleosides along the growth curve represents the distribution of the peak area of each nucleoside compared to the sum of the peak areas of the 2 nucleosides. In (B), (C) and (D), the specific tRNAs were purified at logarithmic (LogP) and stationary (StatP) phase.
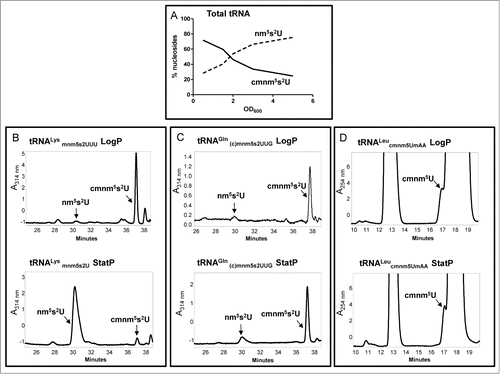
The tRNA species also determines the final output of the MnmEG pathways.Citation66 Thus, whereas tRNALysmnm5s2UUU follows the pattern of the bulk tRNA and accumulates nm5s2U in stationary cultures, the less abundant tRNAGln(c)mnm5s2UUG maintains the cmnm5U levels along the growth curve (). Notably, the ammonium pathway appears ineffective to modify tRNALeucmnm5UmAA in any growth phase (). The molecular bases of this behavior remain unknown. A preference by MnmEG for the ammonium or glycine pathway may depend on the availability of these substrates which, in turn, may depend on growth medium and cell metabolism. In fact, when cells are grown in minimal medium, the glycine pathway is used mostly in any growth phase, as observed in bulk tRNA, tRNALysmnm5s2UUU, and tRNAGln(c)mnm5s2UUG.Citation66 We cannot, however, rule out that the use of glycine or ammonium by MnmEG might be regulated by interactions of this complex with its tRNA substrates and other factors in vivo. Alternatively, tRNAGln(c)mnm5s2UUG and tRNALeucmnm5UmAA might be poor substrates for the ammonium pathway, and are thus out-competed by other tRNA species in vivo, or are unstable when carrying nm5(s2)U or mnm5(s2)U. Further research is required to investigate these hypotheses and to unravel the mechanisms underlying the selection of glycine and ammonium pathways by MnmEG.
The crystal structure of MnmC consists in 2 globular domains, MnmC(o) and MnmC(m), which interact through a rather hydrophilic interface.Citation67,68 The catalytic centers of MnmC(o) and MnmC(m) face opposite sides of the protein, thus favoring a model in which the 2 domains can function in a relatively independent manner. However, given the nature of their interface, the possibility that conformational changes within the entire MnmC protein may occur in vivo and facilitate the functional cooperation of the domains could not be excluded. The recent cloning and separate expression of both domains has facilitated the study of their biochemical activities and tRNA substrate specificity.Citation66 MnmC(o) and MnmC(m) can operate independently of each other (with a catalytic efficiency similar to that of the full protein) and differ in terms of their specificity for tRNAs. MnmC(o) cannot modify tRNAGln(c)mnm5s2UUG and tRNALeucmnm5UmAA, whereas MnmC(m) modifies both tRNAs in vitro, and also in vivo in the case of tRNAGln(c)mnm5s2UUG. Putative orthologs of the E. coli bifunctional MnmC protein are conserved only in γ-proteobacteria, but potential orthologs of a single domain have been identified in several genomes.Citation69,70 The phylogenetic analysis did not clear up whether the independent orthologs represent the ancestral or derived versions of the bifunctional MnmC enzyme. Nevertheless, the ability of the E. coli MnmC domains to function independently and to recognize different substrates suggests that the origin of the full MnmC protein present in γ-proteobacteria likely happened by domain fusion.Citation66
Phenotypes of E. coli Associated with U34 Modification Defects
Lack of modifications at U34 due to mnmE or mnmG mutations leads to impaired growth, high sensitivity to acidic pH and defects in translational fidelity, whereas MnmC impairment is less detrimental since it neither reduces the growth rate nor affects resistance to acidic pH.Citation32,56,66,71 Nevertheless, mnmC mutations have been shown to diminish not only the ability of cells to compete, but also the efficiency of a suppressor tRNA to read stop codons.Citation66,72 So it follows that adaptation of MnmEG to use ammonium and glycine, as well as the incorporation of MnmC activities, confers E. coli significant advantages to synthesize mnm5U and to survive under different conditions.
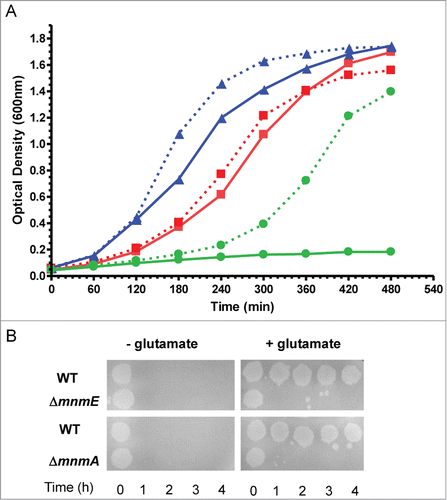
We constructed a double null mutant mnmE/mnmA by placing an additional chromosomal copy of mnmE under the control of the AraC-PBAD system (λaraC-PBAD::mnmE). We used this approach because it was not possible to obtain the double mutant by P1-transduction without having this extra copy of mnmE. These findings suggest that the combination of null mnmA and mnmE mutations produces synthetic lethality. Strikingly, the double mutant mnmE/mnmA (λaraC-PBAD::mnmE) was able to grow in minimal medium without the arabinose inducer, be it very slowly. This feature allowed us to grow the strain overnight in minimal medium (so all tRNA substrates of MnmEG and MnmA were finally unmodified) and, then, to study its growth rate in both LBT and minimal medium in the presence or absence of arabinose. No significant growth of strain mnmE/mnmA (λaraC-PBAD::mnmE) was observed in LBT without arabinose whereas induction of the MnmE synthesis from the extra copy of mnmE allow the double mutant to grow (). Altogether these results suggest that at least one of the 2 modifications at the U34 nucleobase, s2 or (c)mnm5, should be present for E. coli viability in rich medium. Notably, mutations mnmA and mnmE separately confer extreme sensitivity to acidic pH (), suggesting that, in this case, both genes are required for survival. The modification defect at both position 2 and 5 has been shown to promote translational frameshifting,Citation73-75 and this could affect growth. Considering that the level of affectation appears to be dependent on the growth conditions, it is tempting to speculate that translation of certain proteins required for growth under specific conditions is especially sensitive to the absence of s2 and/or (c)mnm5 modifications.
Figure 4. Phenotypic traits caused by mnmE and mnmA null mutations in E. coli. (A) Combination of mnmE and mnmA mutations confers synthetic lethality in LBT medium. The mutants were recovered in a DEV16 background through P1 procedures.Citation56 Overnight cultures were grown in YM9 buffer supplemented with 0.05% casamino acids and 0.4% glycerol; stationary-phase cultures were washed twice with YM9 and then were diluted 1:20 into LBT medium with or without 0.2% L-Arabinose. Cultures were incubated with shaking at 37 ºC. Growth was monitored by measuring the optical density at 600 nm. Mutants mnmE, mnmA and mnmA/mnmE (λaraC-PBAD::mnmE) are represented by triangles (blue lines), squares (red lines), and circles (green lines), respectively. Growth in the presence or absence of arabinose is represented by dashed and solid lines, respectively. (B) mnmE and mnmA mutations confer extreme sensitivity to acidic pH. Assays were carried out as described.Citation71 Briefly, cells were grown in LBT containing 0.4% glucose; stationary-phase cultures were diluted 1:1000 into EG pH 2 medium (minimal E medium containing 0.4% glucose) with or without 0.7 mM glutamate. Cultures were spotted on LBT plates at 0, 1, 2, 3 and 4 hours post acid challenge. Plates were incubated at 37ºC during 16 hours.
Mitochondrial-tRNA Modification Pathways
Mitochondria are pivotal organelles that play a critical role in ATP production via the oxidative phosphorylation (OXPHOS) system, but also in the generation of intermediary metabolites, biosynthesis, apoptotic cell death and intracellular signaling. The human mitochondrial genome encodes 13 key OXPHOS proteins and the 22 tRNAs and 2 rRNAs used for intra-mitochondrial protein synthesis. However, the vast majority of the OXPHOS components and proteins required for the synthesis, expression and regulation of mitochondrial DNA are encoded by the cell nucleus, including mitochondrial (mt) ribosomal proteins, aminoacyl-tRNA synthetases and tRNA-modifying enzymes.Citation76
Integration of mitochondrial functions within the cell depends on anterograde and retrograde signaling pathways.Citation77,78 Anterograde regulation arranges signals from external and internal stimuli at the nucleus to activate genetic programs in order to adjust the mitochondrial function accordingly. Retrograde regulation includes diverse, poorly known communication pathways from mitochondria to the nucleus that influence cellular activities in response to changes in the functional state of mitochondria.
Several human diseases have been related to defects in mt-tRNA modification.Citation6,42,79,80 Mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes (MELAS) and myoclonus epilepsy associated with ragged-red fibers (MERRF) are mostly due to mutations in genes mt-tRNALeu(UUR) and mt-tRNALys, respectively, with A3243G in mt-tRNALeu(UUR), and A8343G in mt-tRNALys, being the most frequent.Citation81 Patient cells exhibit mitochondrial translation defects, oxidative stress, and diminished respiratory enzyme activity and oxygen consumption.Citation82-85
mt-tRNAs carrying MERRF and MELAS mutations lack the U34 modifications that are normally present in nonmutated tRNAs.Citation42 These mutations appear to act as negative identity determinants which prevent tRNA recognition by the modifying enzymes. TRMU (also named MTU1 and MTO2) is homologous to bacterial MnmA and is thus responsible for the synthesis of s2U at U34 of mt-tRNAs decoding for Lys, Glu and Gln, whereas GTPBP3 and MTO1 are homologous to MnmE and MnmG, respectively. Notably, the human mt-tRNAs decoding for Lys, Glu, Gln, Leu(UUR), and Trp contain the taurinomethyl (τm) group at position 5 of U34.Citation86,87 So it is assumed that GTPBP3 and MTO1 use taurine instead of glycine to modify mt-tRNAs through a similar reaction to that occurring in E. coli;Citation55,87 however, no direct evidence in support of this proposal has been provided so far.
Lack of τm5U in tRNA-Leu carrying a MELAS mutation or τm5s2U in tRNA-Lys bearing a MERRF mutation has been postulated to produce a decoding defect, thus playing a crucial role in molecular pathogenesis.Citation42 Nevertheless, lack of the U34 modifications produced by mutations in genes MTO1 and TRMU, which are respectively associated with hypertrophic cardiomyopathy and acute infantile liver failure, does not appear to consistently affect mitochondrial protein synthesis in patient cells.Citation3,4,8 This is a surprising result if we consider the role of U34 modifications in protein translation optimization.Citation42 In fact, we previously reported that the transient knocking-down of GTPBP3 in HEK-293 with siRNAs diminished the incorporation of [3H]leucine into mtDNA-encoded proteins by about 20%.Citation13 If some compensatory mechanism(s) lower(s) this level of affectation, then the nondetection of a significant alteration at the overall mitochondrial translation rate might be possible. Compensatory mechanisms of translational defects can include the presence of mutant tRNAs and the overexpression of either specific tRNAs or tRNA synthetases.Citation73,88-94 Notably, acute infantile liver failure associated with TRMU mutations is reversible in some patients through a yet unknown mechanism.Citation3,95 Even though a compensatory mechanism of the translation defect may exist, the OXPHOS system function may be impaired, as observed in patient cells carrying MTO1 and TRMU mutations, and in TRMU-knocked-down cells.Citation4,8,96 This suggests that other mechanisms, in addition to altered translation, might contribute to mitochondrial dysfunction. In fact, it has been reported that MELAS mutation A3243G induces a retrograde signaling pathway involving ROS, kinase JNK, retinoid X receptor α and transcriptional coactivator PGC1α.Citation97 This pathway contributes to diminish the mRNA abundances of nuclear-encoded OXPHOS enzymes via transcriptional regulation, thereby aggravating the mitochondrial dysfunction (). Moreover, we recently found that ROS also induces the expression of microRNA 9/9* in MELAS cells, which reduces the abundance of proteins TRMU, GTPBP3 and MTO1 via mRNA destabilization given that TRMU, GTPBP3 and MTO1 mRNAs are direct targets for the microRNA ().Citation9 As far as we know, these results reveal for the first time that an mtDNA disorder directly affects miRNA expression and that cells may respond to stress by lowering the abundance of tRNA modification enzymes. Considering that the microRNA-9/9* overexpression has similar effects to knocking-down the TRMU, GTPBP3 and MTO1 expressions with siRNAs, both leading to mitochondrial dysfunction, we propose that the ROS-dependent induction of microRNA-9/9* in MELAS cells also contributes to the pathological mechanism by down-regulating mt-tRNA modification enzymes.Citation9
Figure 5. New retrograde and anterograde pathways between mitochondria and nucleus in MELAS cells. A MELAS mutation in mt-DNA affecting tRNALeu(UUR) prevents modification of U34 in tRNALeu(UUR) molecules, which impairs mitochondrial translation (step 1) and, consequently, leads to dysfunction of the OXPHOS system and production of ROS. Increased ROS levels have been proposed to activate kinase JNK and reduce the abundance of retinoid X receptor α (RXRA), which would decrease the formation of the transcriptional complex RXRA-coactivator PGC1α (step 2) and reduce the expression of nuclear-encoded OXPHOS genes, thus aggravating mitochondrial dysfunction.Citation97 We have recently found a new retrograde pathway based on the microRNA 9/9* induction by ROS in a NF-kB-dependent manner (step 3).Citation9 Increased levels of microRNA 9/9* reduce the abundance of the mt-tRNA modifying proteins TRMU, GTPBP3 and MTO1 by destabilizing the corresponding mRNAs (step 4), which also contributes to aggravate the mitochondrial dysfunction.
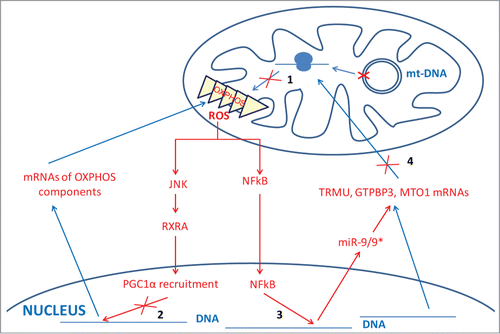
Interestingly, we have observed that microRNA-9/9* overexpression causes a slight, but significant, drop in the thiolation level of mt-tRNALys and tRNAGlu, likely as a result of the reduced amount of TRMU.Citation9 Future work should investigate whether the mitochondrial dysfunction associated with the induction of microRNA-9/9* and down-regulation of mt-tRNA-modifying enzymes is due to hypomodification of mt-tRNAs and/or to nonconventional functions of mt-tRNA-modifying enzymes.
In yeast, the TRMU homolog (named MTU1, MTO2 or SLM3) is also responsible for the 2-thiolation of tRNA substrates at U34, while MTO1 and MSS1 (the homologous protein of bacterial MnmE and human GTPBP3) promote the incorporation of the cmnm group at position 5.Citation12 Therefore, yeast proteins MTO1 and MSS1, like their bacterial homologs MnmG and MnmE, use glycine as a substrate in the modification reaction. Currently, it is not known whether the ammonium pathway can be used by the eukaryotic MnmG and MnmE homologs.
Interestingly, deletion of the TRMU and MTO1 homologs in yeast leads to a marked reduction in mitochondrial translation, whereas deletion of MSS1 results in a normal pattern of mitochondrial translation products.Citation12,98-100 Some data suggest that the regulation of mitochondrial translation in yeast involves other strategies than in humans.Citation76,101 Moreover in yeast, unlike humans, some mt-DNA genes (those for 21 S rRNA, CYTB and COX1) contain introns that are removed by intron-encoded maturases with the collaboration of a number of nuclear-encoded splicing factors.Citation101,102 Proteins TRMU, MTO1 and MSS1 are required for splicing, likely because they are needed for the translation of maturases.Citation99,100,103,104 Strikingly, effects of MTO1 inactivation are more severe than those of MSS1 inactivation on splicing and accumulation of certain mt-tRNAs,Citation100 which might explain why mitochondrial translation is affected more in MTO1 than in MSS1 mutants. These observations suggest that MTO1 and MSS1, apart from their shared role in mt-tRNA modification, may play independent roles in other cellular functions.
The possibility of human proteins TRMU, GTPBP3 and MTO1 performing additional roles to mt-tRNA modification cannot be ruled out. Accumulating evidence indicates that other mt-tRNA-modifying proteins are involved in different cellular functions. Thus human TRIT1 protein, which is homologous to bacterial MiaA and yeast Mod5, is responsible for the synthesis of N6-isopentenyladenosine (i6A) at position 37 of cytoplasmic and mitochondrial tRNAs, but is also involved in the tRNA gene-mediated silencing of RNA polymerase II promoters and in tumor suppression.Citation87,105,106 Protein CDK5RAP1, which is homologous to bacterial MiaB and responsible for the conversion of i6A into 2-methylthio-N6-isopentenyladenosine (ms2i6A37) in both cytoplasmic and mitochondrial tRNAs, has been initially identified as a repressor of the cyclin-dependent protein kinase 5.Citation21,87,107 Moreover, the methyltransferase for m1A9 or m1G9 in human mt-tRNAs (Trm10) is a component of mitochondrial RNase P,Citation108 whereas protein PusI, which modifies uridine to pseudouridine in several RNA types, including cytoplasmic and mitochondrial tRNAs, cooperates with the retinoic acid receptor to enhance transcription at target promoters.Citation87,109 In line with this, TRMU, which carries the sulfur moiety that is finally transferred to mt-tRNAs, has been suggested to be involved in the assembly of enzyme complexes containing iron-sulfur clusters, since an intermediate of the OXPHOS Complex II accumulates in TRMU-depleted cells.Citation8,95 However, this hypothesis does not explain why the accumulation of subunits of complex IV, which does not contain an iron-sulfur center, is also affected in the depleted cells.Citation95 Hence further work is required to clarify the hypothetical role of TRMU in sulfur trafficking.
Lack of U34 modifications can also affect cellular functions if hypomodified mt-tRNAs perform a signaling function. Loss of conserved wobble uridine modification in yeast cytosolic tRNAs has been recently reported to affect gene expression by perturbing cell signaling in a translation-independent manner.Citation29 In fact, the obtained data indicate that improperly modified tRNA elicits a noncanonical stress response by an unknown mechanism. This finding suggests a nonconventional role for tRNA modifications in regulating gene expression, which is in line with emerging evidence of signaling pathways involving tRNA and cleavage fragments.Citation110-112
Finally, special attention should be paid to synergic effects when studying the role(s) of tRNA modifications and tRNA-modifying enzymes. For instance, clinical outcome has been observed to be highly variable in patients harboring an identical homozygous MTO1 mutation, which suggests that genetic, epigenetic and environmental factors may play a crucial role in modulating the phenotype.Citation96 Interestingly, the simultaneous inactivation of MTO1, or MSS1 (the GTPBP3 homolog), and the TRMU homolog (named MTU1 or MTO2) in yeast has a dramatic synergic effect on respiratory activity as a double mutant MTO1/MTU1 or MSS1/MTU1 is unable to grow on nonfermentable medium.Citation12,100 Notably, translation of the COX1 subunit of complex IV, but not of other mtDNA-encoded proteins, was decreased by the combination of an MSS1 mutation with the paromomycin-resistance mitochondrial 15S rRNA C1409G mutation in a yeast strain devoid of mitochondrial introns.Citation99 This result suggests that the MSS1-dependent modification of U34 is required for translation of specific mRNAs by ribosomes with an altered decoding center. Moreover, knocking-down of TRMU in human cells carrying a homoplasmic mt-tRNAGlu mutation (m.14674T>C/G) associated with reversible infantile respiratory chain deficiency (RISCD) impairs mitochondrial translation despite translation is normal in both RISCD and TRMU patient cells.Citation95 Furthermore, the simultaneous inactivation in Caenorhabditis elegans of the homologs to the human TRMU and MTO1 genes causes severe developmental dysfunctions (our own unpublished results). All these synergic effects produced by combination of TRMU (MTU1), MTO1, and GTPBP3 (MSS1) mutations among themselves or with mutations affecting other translation players are reminiscent of those observed in E. coli () and highlight the importance of protein families TRMU/MTU1/MnmA, GTPBP3/MSS1/MnmE and MTO1/MnmG for bacterial and eukaryotic cells.
Conclusions and Future Prospects
Evolutionary conservation of proteins acting on the wobble uridine of tRNAs supports their crucial role for cell and organismal functions. Inactivation of these proteins often leads to impaired growth in microorganisms and diseases in humans. These effects are likely due to the role played by modifications in protein translation, but the possibility of tRNA-modifying enzymes and/or unmodified tRNAs performing functions that are unrelated with translation should be explored in future research. Accumulating evidence indicates that tRNA modifications are dynamic and that cells respond to stress by modulating the activity and/or levels of tRNA modification enzymes. However, a great deal of work needs to be done to unravel the underlying regulatory mechanisms. Major goals for future research also include determining the role of the GTPase cycle of MnmE and its homologous proteins in tRNA modification, as well as the structure, conformational and oligomeric dynamics, and the functioning of the MnmE-MnmG complex.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness [BFU2010–19737], and the Generalitat Valenciana [ACOMP/2012/065; and PROMETEO/2012/061] to M.-E.A.
References
- Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel-Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am J Hum Genet 2004; 74:1303-8; PMID:15108122; http://dx.doi.org/10.1086/421530
- Yasukawa T, Kirino Y, Ishii N, Holt IJ, Jacobs HT, Makifuchi T, Fukuhara N, Ohta S, Suzuki T, Watanabe K. Wobble modification deficiency in mutant tRNAs in patients with mitochondrial diseases. FEBS Lett 2005; 579:2948-52; PMID:15893315; http://dx.doi.org/10.1016/j.febslet.2005.04.038
- Zeharia A, Shaag A, Pappo O, Mager-Heckel AM, Saada A, Beinat M, Karicheva O, Mandel H, Ofek N, Segel R, et al. Acute infantile liver failure due to mutations in the TRMU gene. Am J Hum Genet 2009; 85:401-7; PMID:19732863; http://dx.doi.org/10.1016/j.ajhg.2009.08.004
- Ghezzi D, Baruffini E, Haack TB, Invernizzi F, Melchionda L, Dallabona C, Strom TM, Parini R, Burlina AB, Meitinger T, et al. Mutations of the mitochondrial-tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am J Hum Genet 2012; 90:1079-87; PMID:22608499; http://dx.doi.org/10.1016/j.ajhg.2012.04.011
- Tucker EJ, Hershman SG, Kohrer C, Belcher-Timme CA, Patel J, Goldberger OA, Christodoulou J, Silberstein JM, McKenzie M, Ryan MT, et al. Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell Metab 2011; 14:428-34; PMID:21907147; http://dx.doi.org/10.1016/j.cmet.2011.07.010
- Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med 2014; 20:306-14; PMID:24581449; http://dx.doi.org/10.1016/j.molmed.2014.01.008
- Kemp JP, Smith PM, Pyle A, Neeve VC, Tuppen HA, Schara U, Talim B, Topaloglu H, Holinski-Feder E, Abicht A, et al. Nuclear factors involved in mitochondrial translation cause a subgroup of combined respiratory chain deficiency. Brain 2011; 134:183-95; PMID:21169334; http://dx.doi.org/10.1093/brain/awq320
- Sasarman F, Antonicka H, Horvath R, Shoubridge EA. The 2-thiouridylase function of the human MTU1 (TRMU) enzyme is dispensable for mitochondrial translation. Hum Mol Genet 2011; 20:4634-43; PMID:21890497; http://dx.doi.org/10.1093/hmg/ddr397
- Meseguer S, Martinez-Zamora A, Garcia-Arumi E, Andreu AL, Armengod ME. The ROS-sensitive microRNA-9/9* controls the expression of mitochondrial-tRNA modifying enzymes and is involved in the molecular mechanism of MELAS syndrome. Hum Mol Genet 2014; 24:167–184; doi:10.1093/hmg/ddu427.
- Li X, Guan MX. A human mitochondrial GTP binding protein related to tRNA modification may modulate phenotypic expression of the deafness-associated mitochondrial 12S rRNA mutation. Mol Cell Biol 2002; 22:7701-11; PMID:12370316; http://dx.doi.org/10.1128/MCB.22.21.7701-7711.2002
- Li X, Li R, Lin X, Guan MX. Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of deafness-associated mitochondrial 12 S rRNA A1555G mutation. J Biol Chem 2002; 277:27256-64; PMID:12011058; http://dx.doi.org/10.1074/jbc.M203267200
- Umeda N, Suzuki T, Yukawa M, Ohya Y, Shindo H, Watanabe K. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J Biol Chem 2005; 280:1613-24; PMID:15509579; http://dx.doi.org/10.1074/jbc.M409306200
- Villarroya M, Prado S, Esteve JM, Soriano MA, Aguado C, Perez-Martinez D, Martinez-Ferrandis JI, Yim L, Victor VM, Cebolla E, et al. Characterization of human GTPBP3, a GTP-binding protein involved in mitochondrial tRNA modification. Mol Cell Biol 2008; 28:7514-31; PMID:18852288; http://dx.doi.org/10.1128/MCB.00946-08
- Grosjean H. Nucleic acids are not boring long polymers of only four types of nucleotides: a guided tour. In: Grosjean H, ed. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin, Texas: Landes Bioscience, 2009:1–18.
- Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 2000; 49:341-51; PMID:10902565; http://dx.doi.org/10.1080/152165400410182
- Behm-Ansmant I, Massenet S, Immel F, Patton JR, Motorin Y, Branlant C. A previously unidentified activity of yeast and mouse RNA:pseudouridine synthases 1 (Pus1p) on tRNAs. RNA 2006; 12:1583-93; PMID:16804160; http://dx.doi.org/10.1261/rna.100806
- Kasprzak JM, Czerwoniec A, Bujnicki JM. Molecular evolution of dihydrouridine synthases. BMC Bioinformatics 2012; 13:153; PMID:22741570; http://dx.doi.org/10.1186/1471-2105-13-153
- Desmolaize B, Fabret C, Bregeon D, Rose S, Grosjean H, Douthwaite S. A single methyltransferase YefA (RlmCD) catalyses both m5U747 and m5U1939 modifications in Bacillus subtilis 23S rRNA. Nucleic Acids Res 2011; 39:9368-75; PMID:21824914; http://dx.doi.org/10.1093/nar/gkr626
- Benitez-Paez A, Villarroya M, Armengod ME. The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA 2012; 18:1783-95; PMID:22891362; http://dx.doi.org/10.1261/rna.033266.112
- Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA 1999; 5:1105-18; PMID:10445884; http://dx.doi.org/10.1017/S1355838299982201
- Reiter V, Matschkal DM, Wagner M, Globisch D, Kneuttinger AC, Muller M, Carell T. The CDK5 repressor CDK5RAP1 is a methylthiotransferase acting on nuclear and mitochondrial RNA. Nucleic Acids Res 2012; 40:6235-40; PMID:22422838; http://dx.doi.org/10.1093/nar/gks240
- Björk GR, Hagervall TG. Transfer RNA Modification. EcoSal Plus, 2005; doi: 10.1128/ecosalplus.4.6.2
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 2010; 6:e1001247; PMID:21187895; http://dx.doi.org/10.1371/journal.pgen.1001247
- El Yacoubi B, Bailly M, de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet 2012; 46:69-95; PMID:22905870; http://dx.doi.org/10.1146/annurev-genet-110711-155641
- Gehrig S, Eberle ME, Botschen F, Rimbach K, Eberle F, Eigenbrod T, Kaiser S, Holmes WM, Erdmann VA, Sprinzl M, et al. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J Exp Med 2012; 209:225-33; PMID:22312113; http://dx.doi.org/10.1084/jem.20111044
- Helm M, Alfonzo JD. Posttranscriptional RNA Modifications: playing metabolic games in a cell's chemical Legoland. Chem Biol 2014; 21:174-85; PMID:24315934; http://dx.doi.org/10.1016/j.chembiol.2013.10.015
- Kramer GF, Baker JC, Ames BN. Near-UV stress in Salmonella typhimurium: 4-thiouridine in tRNA, ppGpp, and ApppGpp as components of an adaptive response. J Bacteriol 1988; 170:2344-51; PMID:3283108
- Nallagatla SR, Jones CN, Ghosh SK, Sharma SD, Cameron CE, Spremulli LL, Bevilacqua PC. Native tertiary structure and nucleoside modifications suppress tRNA's intrinsic ability to activate the innate immune sensor PKR. PLoS One 2013; 8:e57905; PMID:23483938; http://dx.doi.org/10.1371/journal.pone.0057905
- Zinshteyn B, Gilbert WV. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet 2013; 9:e1003675; PMID:23935536; http://dx.doi.org/10.1371/journal.pgen.1003675
- Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry 2010; 49:4934-44; PMID:20459084; http://dx.doi.org/10.1021/bi100408z
- Urbonavicius J, Durand JM, Bjork GR. Three modifications in the D and T arms of tRNA influence translation in Escherichia coli and expression of virulence genes in Shigella flexneri. J Bacteriol 2002; 184:5348-57; PMID:12218021; http://dx.doi.org/10.1128/JB.184.19.5348-5357.2002
- Armengod ME, Moukadiri I, Prado S, Ruiz-Partida R, Benitez-Paez A, Villarroya M, Lomas R, Garzon MJ, Martinez-Zamora A, Meseguer S, et al. Enzymology of tRNA modification in the bacterial MnmEG pathway. Biochimie 2012; 94:1510-20; PMID:22386868; http://dx.doi.org/10.1016/j.biochi.2012.02.019
- Nasvall SJ, Chen P, Bjork GR. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA 2004; 10:1662-73; PMID:15383682; http://dx.doi.org/10.1261/rna.7106404
- Nasvall SJ, Chen P, Bjork GR. The wobble hypothesis revisited: uridine-5-oxyacetic acid is critical for reading of G-ending codons. RNA 2007; 13:2151-64; PMID:17942742; http://dx.doi.org/10.1261/rna.731007
- Yokoyama S, Watanabe T, Murao K, Ishikura H, Yamaizumi Z, Nishimura S, Miyazawa T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc Natl Acad Sci U S A 1985; 82:4905-9; PMID:3860833; http://dx.doi.org/10.1073/pnas.82.15.4905
- Hagervall TG, Pomerantz SC, McCloskey JA. Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J Mol Biol 1998; 284:33-42; PMID:9811540; http://dx.doi.org/10.1006/jmbi.1998.2162
- Murphy FVt, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat Struct Mol Biol 2004; 11:1186-91; PMID:15558052; http://dx.doi.org/10.1038/nsmb861
- Westhof E, Yusupov M, Yusupova G. Recognition of Watson-Crick base pairs: constraints and limits due to geometric selection and tautomerism. F1000Prime Rep 2014; 6:19; PMID:24765524; http://dx.doi.org/10.12703/P6-19
- Bjork GR, Huang B, Persson OP, Bystrom AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 2007; 13:1245-55; PMID:17592039; http://dx.doi.org/10.1261/rna.558707
- Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol 2008; 28:3301-12; PMID:18332122; http://dx.doi.org/10.1128/MCB.01542-07
- Kruger MK, Pedersen S, Hagervall TG, Sorensen MA. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J Mol Biol 1998; 284:621-31; PMID:9826503; http://dx.doi.org/10.1006/jmbi.1998.2196
- Suzuki T, Nagao A. Human mitochondrial diseases caused by lack of taurine modification in mitochondrial tRNAs. Wiley Interdiscip Rev RNA 2011; 2:376-86; PMID:21957023; http://dx.doi.org/10.1002/wrna.65
- Benitez-Paez A, Villarroya M, Douthwaite S, Gabaldon T, Armengod ME. YibK is the 2'-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNA(Leu) isoacceptors. RNA 2010; 16:2131-43; PMID:20855540; http://dx.doi.org/10.1261/rna.2245910
- Tkaczuk KL, Dunin-Horkawicz S, Purta E, Bujnicki JM. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics 2007; 8:73; PMID:17338813; http://dx.doi.org/10.1186/1471-2105-8-73
- Liu RJ, Zhou M, Fang ZP, Wang M, Zhou XL, Wang ED. The tRNA recognition mechanism of the minimalist SPOUT methyltransferase, TrmL. Nucleic Acids Res 2013; 41:7828-42; PMID:23804755; http://dx.doi.org/10.1093/nar/gkt568
- Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol Cell 2006; 21:97-108; PMID:16387657; http://dx.doi.org/10.1016/j.molcel.2005.11.001
- Lauhon CT. Requirement for IscS in biosynthesis of all thionucleosides in Escherichia coli. J Bacteriol 2002; 184:6820-9; PMID:12446632; http://dx.doi.org/10.1128/JB.184.24.6820-6829.2002
- Lundgren HK, Bjork GR. Structural alterations of the cysteine desulfurase IscS of Salmonella enterica serovar Typhimurium reveal substrate specificity of IscS in tRNA thiolation. J Bacteriol 2006; 188:3052-62; PMID:16585765; http://dx.doi.org/10.1128/JB.188.8.3052-3062.2006
- Shi R, Proteau A, Villarroya M, Moukadiri I, Zhang L, Trempe JF, Matte A, Armengod ME, Cygler M. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol 2010; 8:e1000354; PMID:20404999; http://dx.doi.org/10.1371/journal.pbio.1000354
- Shigi N. Biosynthesis and functions of sulfur modifications in tRNA. Front Genet 2014; 5:67; PMID:24765101; http://dx.doi.org/10.3389/fgene.2014.00067
- Kotera M, Bayashi T, Hattori M, Tokimatsu T, Goto S, Mihara H, Kanehisa M. Comprehensive genomic analysis of sulfur-relay pathway genes. Genome Inform 2010; 24:104-15; PMID:22081593; http://dx.doi.org/10.1142/9781848166585_0009
- Numata T, Ikeuchi Y, Fukai S, Suzuki T, Nureki O. Snapshots of tRNA sulphuration via an adenylated intermediate. Nature 2006; 442:419-24; PMID:16871210; http://dx.doi.org/10.1038/nature04896
- Elseviers D, Petrullo LA, Gallagher PJ. Novel E. coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res 1984; 12:3521-34; PMID:6427754; http://dx.doi.org/10.1093/nar/12.8.3521
- Sullivan MA, Cannon JF, Webb FH, Bock RM. Antisuppressor mutation in Escherichia coli defective in biosynthesis of 5-methylaminomethyl-2-thiouridine. J Bacteriol 1985; 161:368-76; PMID:3881393
- Moukadiri I, Prado S, Piera J, Velazquez-Campoy A, Bjork GR, Armengod ME. Evolutionarily conserved proteins MnmE and GidA catalyze the formation of two methyluridine derivatives at tRNA wobble positions. Nucleic Acids Res 2009; 37:7177-93; PMID:19767610; http://dx.doi.org/10.1093/nar/gkp762
- Yim L, Moukadiri I, Bjork GR, Armengod ME. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli. Nucleic Acids Res 2006; 34:5892-905; PMID:17062623; http://dx.doi.org/10.1093/nar/gkl752
- Yim L, Martinez-Vicente M, Villarroya M, Aguado C, Knecht E, Armengod ME. The GTPase activity and C-terminal cysteine of the Escherichia coli MnmE protein are essential for its tRNA modifying function. J Biol Chem 2003; 278:28378-87; PMID:12730230; http://dx.doi.org/10.1074/jbc.M301381200
- Martinez-Vicente M, Yim L, Villarroya M, Mellado M, Perez-Paya E, Bjork GR, Armengod ME. Effects of mutagenesis in the switch I region and conserved arginines of Escherichia coli MnmE protein, a GTPase involved in tRNA modification. J Biol Chem 2005; 280:30660-70; PMID:15983041; http://dx.doi.org/10.1074/jbc.M503223200
- Meyer S, Wittinghofer A, Versees W. G-domain dimerization orchestrates the tRNA wobble modification reaction in the MnmE/GidA complex. J Mol Biol 2009; 392:910-22; PMID:19591841; http://dx.doi.org/10.1016/j.jmb.2009.07.004
- Prado S, Villarroya M, Medina M, Armengod ME. The tRNA-modifying function of MnmE is controlled by post-hydrolysis steps of its GTPase cycle. Nucleic Acids Res 2013; 41:6190-208; PMID:23630314; http://dx.doi.org/10.1093/nar/gkt320
- Cabedo H, Macian F, Villarroya M, Escudero JC, Martinez-Vicente M, Knecht E, Armengod ME. The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. EMBO J 1999; 18:7063-76; PMID:10601028; http://dx.doi.org/10.1093/emboj/18.24.7063
- Scrima A, Vetter IR, Armengod ME, Wittinghofer A. The structure of the TrmE GTP-binding protein and its implications for tRNA modification. EMBO J 2005; 24:23-33; PMID:15616586; http://dx.doi.org/10.1038/sj.emboj.7600507
- Scrima A, Wittinghofer A. Dimerisation-dependent GTPase reaction of MnmE: how potassium acts as GTPase-activating element. EMBO J 2006; 25:2940-51; PMID:16763562; http://dx.doi.org/10.1038/sj.emboj.7601171
- Zhang X, Schaffitzel C, Ban N, Shan SO. Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc Natl Acad Sci U S A 2009; 106:1754-9; PMID:19174514; http://dx.doi.org/10.1073/pnas.0808573106
- Fislage M, Brosens E, Deyaert E, Spilotros A, Pardon E, Loris R, Steyaert J, Garcia-Pino A, Versees W. SAXS analysis of the tRNA-modifying enzyme complex MnmE/MnmG reveals a novel interaction mode and GTP-induced oligomerization. Nucleic Acids Res 2014; 42:5978-92; PMID:24634441; http://dx.doi.org/10.1093/nar/gku213
- Moukadiri I, Garzon MJ, Bjork GR, Armengod ME. The output of the tRNA modification pathways controlled by the Escherichia coli MnmEG and MnmC enzymes depends on the growth conditions and the tRNA species. Nucleic Acids Res 2014; 42:2602-23; PMID:24293650; http://dx.doi.org/10.1093/nar/gkt1228
- Kim J, Almo SC. Structural basis for hypermodification of the wobble uridine in tRNA by bifunctional enzyme MnmC. BMC Struct Biol 2013; 13:5; PMID:23617613; http://dx.doi.org/10.1186/1472-6807-13-5
- Kitamura A, Sengoku T, Nishimoto M, Yokoyama S, Bessho Y. Crystal structure of the bifunctional tRNA modification enzyme MnmC from Escherichia coli. Protein Sci 2011; 20:1105-13; PMID:21574198; http://dx.doi.org/10.1002/pro.659
- Bujnicki JM, Oudjama Y, Roovers M, Owczarek S, Caillet J, Droogmans L. Identification of a bifunctional enzyme MnmC involved in the biosynthesis of a hypermodified uridine in the wobble position of tRNA. RNA 2004; 10:1236-42; PMID:15247431; http://dx.doi.org/10.1261/rna.7470904
- Kitamura A, Nishimoto M, Sengoku T, Shibata R, Jager G, Bjork GR, Grosjean H, Yokoyama S, Bessho Y. Characterization and structure of the Aquifex aeolicus protein DUF752: a bacterial tRNA-methyltransferase (MnmC2) functioning without the usually fused oxidase domain (MnmC1). J Biol Chem 2012; 287:43950-60; PMID:23091054; http://dx.doi.org/10.1074/jbc.M112.409300
- Gong S, Ma Z, Foster JW. The Era-like GTPase TrmE conditionally activates gadE and glutamate-dependent acid resistance in Escherichia coli. Mol Microbiol 2004; 54:948-61; PMID:15522079; http://dx.doi.org/10.1111/j.1365-2958.2004.04312.x
- Hagervall TG, Bjork GR. Undermodification in the first position of the anticodon of supG-tRNA reduces translational efficiency. Mol Gen Genet 1984; 196:194-200; PMID:6387394; http://dx.doi.org/10.1007/BF00328050
- Bregeon D, Colot V, Radman M, Taddei F. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev 2001; 15:2295-306; PMID:11544186; http://dx.doi.org/10.1101/gad.207701
- Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J 2001; 20:4863-73; PMID:11532950; http://dx.doi.org/10.1093/emboj/20.17.4863
- Jager G, Nilsson K, Bjork GR. The phenotype of many independently isolated +1 frameshift suppressor mutants supports a pivotal role of the P-site in reading frame maintenance. PLoS One 2013; 8:e60246.
- Smits P, Smeitink J, van den Heuvel L. Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J Biomed Biotechnol 2010; 2010:737385; PMID:20396601; http://dx.doi.org/10.1155/2010/737385
- Reinecke F, Smeitink JA, van der Westhuizen FH. OXPHOS gene expression and control in mitochondrial disorders. Biochim Biophys Acta 2009; 1792:1113-21; PMID:19389473; http://dx.doi.org/10.1016/j.bbadis.2009.04.003
- Whelan SP, Zuckerbraun BS. Mitochondrial signaling: forwards, backwards, and in between. Oxid Med Cell Longev 2013; 2013:351613; PMID:23819011; http://dx.doi.org/10.1155/2013/351613
- Abbott JA, Francklyn CS, Robey-Bond SM. Transfer RNA and human disease. Front Genet 2014; 5:158; PMID:24917879; http://dx.doi.org/10.3389/fgene.2014.00158
- Boczonadi V, Horvath R. Mitochondria: impaired mitochondrial translation in human disease. Int J Biochem Cell Biol 2014; 48:77-84; PMID:24412566; http://dx.doi.org/10.1016/j.biocel.2013.12.011
- Cohen BH. Neuromuscular and systemic presentations in adults: diagnoses beyond MERRF and MELAS. Neurotherapeutics 2013; 10:227-42; PMID:23549648; http://dx.doi.org/10.1007/s13311-013-0188-3
- Antonicka H, Floryk D, Klement P, Stratilova L, Hermanska J, Houstkova H, Kalous M, Drahota Z, Zeman J, Houstek J. Defective kinetics of cytochrome c oxidase and alteration of mitochondrial membrane potential in fibroblasts and cytoplasmic hybrid cells with the mutation for myoclonus epilepsy with ragged-red fibres ('MERRF') at position 8344 nt. Biochem J 1999; 342 Pt 3:537-44; PMID:10477264; http://dx.doi.org/10.1042/0264-6021:3420537
- Dunbar DR, Moonie PA, Zeviani M, Holt IJ. Complex I deficiency is associated with 3243G:C mitochondrial DNA in osteosarcoma cell cybrids. Hum Mol Genet 1996; 5:123-29; PMID:8789449; http://dx.doi.org/10.1093/hmg/5.1.123
- Enriquez JA, Chomyn A, Attardi G. MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNA(Lys) and premature translation termination. Nat Genet 1995; 10:47-55; PMID:7647790; http://dx.doi.org/10.1038/ng0595-47
- Vives-Bauza C, Gonzalo R, Manfredi G, Garcia-Arumi E, Andreu AL. Enhanced ROS production and antioxidant defenses in cybrids harbouring mutations in mtDNA. Neurosci Lett 2006; 391:136-41; PMID:16165271; http://dx.doi.org/10.1016/j.neulet.2005.08.049
- Suzuki T, Wada T, Saigo K, Watanabe K. Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J 2002; 21:6581-9; PMID:12456664; http://dx.doi.org/10.1093/emboj/cdf656
- Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Research 2014.
- El Meziane A, Lehtinen SK, Hance N, Nijtmans LG, Dunbar D, Holt IJ, Jacobs HT. A tRNA suppressor mutation in human mitochondria. Nat Genet 1998; 18:350-3; PMID:9537417; http://dx.doi.org/10.1038/ng0498-350
- Esberg A, Huang B, Johansson MJ, Bystrom AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell 2006; 24:139-48; PMID:17018299; http://dx.doi.org/10.1016/j.molcel.2006.07.031
- Fernandez-Vazquez J, Vargas-Perez I, Sanso M, Buhne K, Carmona M, Paulo E, Hermand D, Rodriguez-Gabriel M, Ayte J, Leidel S, et al. Modification of tRNA(Lys) UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet 2013; 9:e1003647; PMID:23874237; http://dx.doi.org/10.1371/journal.pgen.1003647
- Guo M, Schimmel P. Essential nontranslational functions of tRNA synthetases. Nat Chem Biol 2013; 9:145-53; PMID:23416400; http://dx.doi.org/10.1038/nchembio.1158
- Kirino Y, Yasukawa T, Marjavaara SK, Jacobs HT, Holt IJ, Watanabe K, Suzuki T. Acquisition of the wobble modification in mitochondrial tRNALeu(CUN) bearing the G12300A mutation suppresses the MELAS molecular defect. Hum Mol Genet 2006; 15:897-904; PMID:16446307; http://dx.doi.org/10.1093/hmg/ddl007
- Perli E, Giordano C, Tuppen HA, Montopoli M, Montanari A, Orlandi M, Pisano A, Catanzaro D, Caparrotta L, Musumeci B, et al. Isoleucyl-tRNA synthetase levels modulate the penetrance of a homoplasmic m.4277T>C mitochondrial tRNA(Ile) mutation causing hypertrophic cardiomyopathy. Hum Mol Genet 2012; 21:85-100; PMID:21945886; http://dx.doi.org/10.1093/hmg/ddr440
- Tyynismaa H, Schon EA. Mixing and matching mitochondrial aminoacyl synthetases and their tRNAs: a new way to treat respiratory chain disorders? EMBO Mol Med 2014; 6:155-7; PMID:24473201; http://dx.doi.org/10.1002/emmm.201303586
- Boczonadi V, Smith PM, Pyle A, Gomez-Duran A, Schara U, Tulinius M, Chinnery PF, Horvath R. Altered 2-thiouridylation impairs mitochondrial translation in reversible infantile respiratory chain deficiency. Hum Mol Genet 2013; 22:4602-15; PMID:23814040; http://dx.doi.org/10.1093/hmg/ddt309
- Baruffini E, Dallabona C, Invernizzi F, Yarham JW, Melchionda L, Blakely EL, Lamantea E, Donnini C, Santra S, Vijayaraghavan S, et al. MTO1 mutations are associated with hypertrophic cardiomyopathy and lactic acidosis and cause respiratory chain deficiency in humans and yeast. Hum Mutat 2013; 34:1501-9; PMID:23929671; http://dx.doi.org/10.1002/humu.22393
- Chae S, Ahn BY, Byun K, Cho YM, Yu MH, Lee B, Hwang D, Park KS. A systems approach for decoding mitochondrial retrograde signaling pathways. Sci Signal 2013; 6:rs4; PMID:23443683; http://dx.doi.org/10.1126/scisignal.2003266
- Colby G, Wu M, Tzagoloff A. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J Biol Chem 1998; 273:27945-52; PMID:9774408; http://dx.doi.org/10.1074/jbc.273.43.27945
- Decoster E, Vassal A, Faye G. MSS1, a nuclear-encoded mitochondrial GTPase involved in the expression of COX1 subunit of cytochrome c oxidase. J Mol Biol 1993; 232:79-88; PMID:8392589; http://dx.doi.org/10.1006/jmbi.1993.1371
- Wang X, Yan Q, Guan MX. Combination of the loss of cmnm5U34 with the lack of s2U34 modifications of tRNALys, tRNAGlu, and tRNAGln altered mitochondrial biogenesis and respiration. J Mol Biol 2010; 395:1038-48; PMID:20004207; http://dx.doi.org/10.1016/j.jmb.2009.12.002
- Herrmann JM, Woellhaf MW, Bonnefoy N. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim Biophys Acta 2013; 1833:286-94; PMID:22450032; http://dx.doi.org/10.1016/j.bbamcr.2012.03.007
- Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM. Human mitochondrial mRNAs–like members of all families, similar but different. Biochim Biophys Acta 2010; 1797:1081-5; PMID:20211597; http://dx.doi.org/10.1016/j.bbabio.2010.02.036
- Wang X, Yan Q, Guan MX. Mutation in MTO1 involved in tRNA modification impairs mitochondrial RNA metabolism in the yeast Saccharomyces cerevisiae. Mitochondrion 2009; 9:180-5; PMID:19460296; http://dx.doi.org/10.1016/j.mito.2009.01.010
- Yan Q, Li X, Faye G, Guan MX. Mutations in MTO2 related to tRNA modification impair mitochondrial gene expression and protein synthesis in the presence of a paromomycin resistance mutation in mitochondrial 15 S rRNA. J Biol Chem 2005; 280:29151-7; PMID:15944150; http://dx.doi.org/10.1074/jbc.M504247200
- Pratt-Hyatt M, Pai DA, Haeusler RA, Wozniak GG, Good PD, Miller EL, McLeod IX, Yates JR, 3rd, Hopper AK, Engelke DR. Mod5 protein binds to tRNA gene complexes and affects local transcriptional silencing. Proc Natl Acad Sci U S A 2013; 110:E3081-9; PMID:23898186; http://dx.doi.org/10.1073/pnas.1219946110
- Spinola M, Galvan A, Pignatiello C, Conti B, Pastorino U, Nicander B, Paroni R, Dragani TA. Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene 2005; 24:5502-9; PMID:15870694; http://dx.doi.org/10.1038/sj.onc.1208687
- Pierrel F, Douki T, Fontecave M, Atta M. MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J Biol Chem 2004; 279:47555-63; PMID:15339930; http://dx.doi.org/10.1074/jbc.M408562200
- Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase–extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res 2012; 40:11583-93; PMID:23042678; http://dx.doi.org/10.1093/nar/gks910
- Zhao X, Patton JR, Davis SL, Florence B, Ames SJ, Spanjaard RA. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell 2004; 15:549-58; PMID:15327771; http://dx.doi.org/10.1016/j.molcel.2004.06.044
- Durdevic Z, Schaefer M. tRNA modifications: necessary for correct tRNA-derived fragments during the recovery from stress? Bioessays 2013; 35:323-7; PMID:23315679; http://dx.doi.org/10.1002/bies.201200158
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev 2010; 24:1832-60; PMID:20810645; http://dx.doi.org/10.1101/gad.1956510
- Hurto RL. Unexpected functions of tRNA and tRNA processing enzymes. Adv Exp Med Biol 2011; 722:137-55; PMID:21915787; http://dx.doi.org/10.1007/978-1-4614-0332-6_9
