Abstract
Given the possible critical importance of placental gene imprinting and random monoallelic expression on fetal and infant health, most of those genes must be identified, in order to understand the risks that the baby might meet during pregnancy and after birth. Therefore, the aim of the current study was to introduce a workflow and tools for analyzing imprinted and random monoallelic gene expression in human placenta, by applying whole-transcriptome (WT) RNA sequencing of placental tissue and genotyping of coding DNA variants in family trios. Ten family trios, each with a healthy spontaneous single-term pregnancy, were recruited. Total RNA was extracted for WT analysis, providing the full sequence information for the placental transcriptome. Parental and child blood DNA genotypes were analyzed by exome SNP genotyping microarrays, mapping the inheritance and estimating the abundance of parental expressed alleles. Imprinted genes showed consistent expression from either parental allele, as demonstrated by the SNP content of sequenced transcripts, while monoallelically expressed genes had random activity of parental alleles. We revealed 4 novel possible imprinted genes (LGALS8, LGALS14, PAPPA2 and SPTLC3) and confirmed the imprinting of 4 genes (AIM1, PEG10, RHOBTB3 and ZFAT-AS1) in human placenta. The major finding was the identification of 4 genes (ABP1, BCLAF1, IFI30 and ZFAT) with random allelic bias, expressing one of the parental alleles preferentially. The main functions of the imprinted and monoallelically expressed genes included: i) mediating cellular apoptosis and tissue development; ii) regulating inflammation and immune system; iii) facilitating metabolic processes; and iv) regulating cell cycle.
Abbreviations
| WT | = | whole-transcriptome |
| IUGR | = | intrauterine growth restriction |
| RNA-Seq | = | RNA-sequencing |
| UCSC | = | University of California Santa Cruz |
| FDR | = | false discovery rate |
| ASE | = | allele-specific expression |
| MHC | = | major histocompatibility complex |
| NK cells | = | natural killer cells |
| GEO | = | Gene Expression Omnibus |
| RPKM | = | reads per kilobase per million |
| MAF | = | minor allele frequency |
Introduction
The placenta is essential for fetal growth and survival, by producing specific hormones, transporting nutrients and waste between the fetal and maternal vascular systems and protecting the fetus from the maternal immune system. Placenta is formed mostly by fetal chorion and maternal decidual tissue. The gene expression patterns of the fetal and maternal components of the placenta differ from each otherCitation1 and exhibit temporal changes during placental development and growth.Citation2,3 The correct regulation of gene expression is crucial for a healthy pregnancy, as deviations in gene expression are associated with pathologies, such as intrauterine growth restriction (IUGR)Citation4 and preeclampsia.Citation5
Genomic imprinting is a unique epigenetic feature, characterized by parent-specific monoallelic gene expression. It involves DNA methylation and histone modifications, mostly at gene promoter areas, without altering the genetic sequence. Most imprinted genes are expressed in the placenta, where they contribute to pregnancy development. Postnatal genomic imprinting may also occur, predominantly in the brain.Citation6 The fact that imprinting has been mostly found in placental mammals has resulted in several hypotheses to explain its evolutionary origin. According to the most accepted theory, the “parental conflict hypothesis,”Citation7 paternally expressed genes maximize the resources received by an individual offspring, whereas maternally expressed genes conserve and distribute resources equally among all offspring. Thus, imprinting includes 2 opposing parental interests that are balanced during the development of a normal healthy pregnancy, while dysregulation could result in growth restriction of the newborn and, in more extreme cases, rejection of the pregnancy.
Well-known examples of imprinted genes that play critical roles in fetal growth and placental development include the paternally expressed insulin-like growth factor 2 (IGF2) and paternally expressed gene 10 (PEG10), and the maternally expressed pleckstrin homology-like domain, family A, member 2 (PHLDA2) and cyclin-dependent kinase inhibitor 1C (CDKN1C). IGF2 promotes the proliferation of cells in diverse fetal and placental tissues. The importance of paternally inherited IGF2 in allocating maternal resources for fetal benefit has been confirmed by the suppressed gene activity in IUGR placentas versus those from normal pregnancies.Citation4 In contrast, PHLDA2 and CDKN1C restrict feto-placental growth to the energetic advantage of the mother, as evidenced by their elevated gene expression levels in the placentas of sporadic IUGR pregnancies.Citation4 The embryonic lethality of Peg10 knock-out mice due to placental defectsCitation8 revealed the indispensability of this paternally inherited gene for placental biology.
Although more than 50 placental imprinted genes have been detected in the human genome (Genomic Imprinting Website: www.geneimprint.com, Table S1), the list is not complete. The earliest studies on gene expression in mouse embryos and the characterization of genes involved in imprinting disorders such as Beckwith-Wiedemann syndrome, including both IGF2 and CDKN1C, provided the first evidence on parentally biased gene expression. Thereafter, genome-wide attempts to identify novel imprinted genes exploited genotyping microarrays,Citation9,10 mapping of differentially methylated DNA regionsCitation11 and computational prediction.Citation12 More recently, high-throughput RNA sequencing (RNA-Seq) was used to provide the most representative genome-wide list of imprinted genes in the placenta of mice,Citation13 as well as horse and donkey hybrids.Citation14
Gene imprinting is just one particular manifestation of monoallelic expression. The remaining 2 classes are characterized by random monoallelic expression and include autosomal monoallelic expression and X-inactivated genes (as reviewed in ref. 15). These 2 other mechanisms contrast the imprinted genes as the choice to express either of the parental autosomal allele or silencing of most genes in one X-chromosome is taken randomly and autonomously at the cellular level. Still common to all manifestations of monoallelic expressions, they are stably inherited during cell divisions, since the epigenetic marks are established in precursor cells. Although, X-chromosome inactivation in placental tissue is a matter of active research, there is only one systematic report on random monoallelic autosomal gene expression in placental tissue,Citation16 and its functional consequences on fetal development and survival have still remained unknown.
Given the possible critical importance of placental imprinting and monoallelic gene expression in general on fetal and infant health, most of those genes active during embryonal and fetal development must be identified, in order to understand the risks that the baby might meet during pregnancy and after birth. Therefore, our objective was to develop a more comprehensive method for updating the list of developmentally important imprinted and randomly monoallelically expressed placental genes by applying whole-transcriptome (WT) RNA-Seq of placental tissue and genotyping of coding variants in family trios.
Results
Identification of candidate imprinted genes
Ten family trios (mother, father and child) were recruited after the delivery of a healthy newborn from an uncomplicated term pregnancy. DNA from the umbilical cord and peripheral blood samples were used to obtain single nucleotide polymorphism (SNP) genotypes from newborns and parents, respectively (). Exome SNP microarrays (Illumina SNP HumanExome BeadChip v1.1) were utilized to genotype the coding SNPs of all participants. This microarray contains SNPs for 18,474 genes, representing 78% of the known genes in the University of California Santa Cruz (UCSC) databaseCitation17 (Fig. S1). On average, 13.5 SNPs were included per gene, with most genes represented by >1 SNP (17,246 out of 18,474; 93.4%) and 818 genes (4.4%) harboring >40 SNPs (distribution of the SNP content of all genes is shown in Fig. S2).
Figure 1. Schematic view to identify imprinted and monoallelically expressed genes. The pipeline shows the steps involved in analyzing and detecting imprinted and monoallelically expressed genes using RNA-Seq data and R scripts. Arrows show the data flow. Data sources (including RNA-Seq, SNP array data and external sources) are labeled by light gray boxes, while dark gray box shows the final result about imprinted and monoallelically expressed genes. UCSC: University of California Santa Cruz database.Citation17
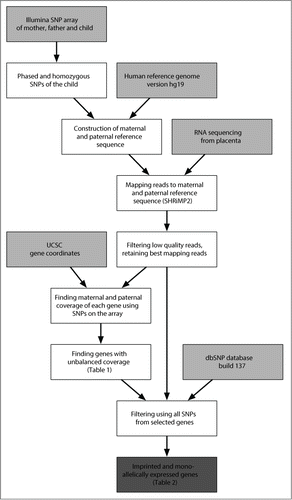
The genotyping of all family members resulted in a list of genes eligible for analysis by our methodology. Only genes containing coding SNPs for which a newborn was heterozygous and at least one of the parents was homozygous were considered informative for further analysis. This list of genes was generated by phasing each SNP for which the child was heterozygous to the parental genotypes. In this way, we were able to define which of the child's 2 alleles was inherited from the mother and which was from the father. We identified at least one phased SNP for at least one family trio in 55.0% (n = 10,167) of genes in the array (Fig. S1). As a result, an average of 8,019 SNPs per trio (referred to as phased SNPs) among all genotyped SNPs were considered suitable for further analysis (Table S2 provides information for every family trio).
Next, parental genomes were constructed in relation to the reference genome (hg19).Citation17 These genomes were used to build the child's diploid genome, consisting of the inherited paternal and maternal haplotypes. SNP positions from the microarray annotation file were used to define the same SNPs in the RNA-Seq reads () and were compared to the UCSC gene coordinatesCitation17 to identify genes that harbored the studied SNPs.
Placenta samples from all family trios were analyzed by RNA-Seq, and the reads were mapped to both parental genomes. Phasing information was used to find maternal and paternal read counts of each phased SNP, which reflect the expression levels of maternally and paternally inherited alleles. All trios and all SNPs in a gene were summed to obtain gene-level read counts representing the abundance of the parental alleles, assuming that the imprinted genes show consistent predominant expression from either parental allele, as evidenced by the SNP content of the sequenced transcripts. A binomial test was used to detect statistically significant deviations between maternal and paternal read counts. Multiple testing-corrected p-values (q-values) were calculated by the false discovery rate (FDR) method, to limit the number of false-positive results. The fraction of maternal and paternal reads was significantly unbalanced for 33 genes (q < 0.05), which were reported as having allele-specific expression (ASE) and as being potentially imprinted ().
Table 1. Candidate imprinted genes after RNA-Seq and exome genotyping
Selection of the final list of imprinted and randomly monoallelically expressed genes
To select the final list of imprinted and monoallelically expressed genes, another bioinformatic filtering step was introduced. Besides the SNPs on the microarray, all SNPs revealed by RNA-Seq and verified with the dbSNP databaseCitation18 were used to confirm the expression mode of the genes. We expect that all of the heterozygous SNPs in imprinted and randomly monoallelically expressed genes of the placental transcriptome, regardless of parental inheritance, will show a greater imbalance in the allelic ratio of the SNPs, with a higher proportion of statistically significant ASE (referred to as ASE-SNPs).
We reused a similar approach as in the original analysis pipeline to detect ASE-SNPs, with the exception that trios and SNPs were not aggregated to the gene level (). Genes with ≥75% of the child's heterozygous SNPs showing significant ASE were selected, because this arbitrary level exceeded, by >3 times, the proportion of ASE-SNPs (22.6%) among approximately 10,000 randomly picked genome-wide SNPs. The more stringent analysis resulted in 12 genes listed in and with the full names and functions given in .
Table 2. Imprinted and monoallelically expressed genes found in the current study
Table 3. Summary of detected imprinted and monoallelically expressed genes
As the initial approach of summing up the information from all family trios including all SNPs may mask and bias the results in identifying the imprinted regulation of gene expression in individual families, we subsequently analyzed the parental specific gene expression of these 12 genes in individual families. This way of analyzing the results eliminates the risk that the gene imprinting ascertained is mostly caused by the biased ratio of gene activity in one or a few of the families analyzed, while in rest of the trios the gene is expressed in biallelic mode. Furthermore, by using this filtering step, we were further able to restrict the list of potentially imprinted genes and were capable to discriminate between the imprinted genes and those having monoallelic expression, but showing random activity of the parental alleles.
In trio-specific approach we confirmed the imprinting and paternal expression of PEG10 gene. In addition, we corroborated the imprinted regulation of recently established AIM1Citation19, and RHOBTB3Citation11. For all these genes, we were able to identify one (PEG10) or 3-4 (RHOBTB3 and AIM1, respectively) phased SNPs for showing the parental specific expression of the genes among the 10 families studied ().
Figure 2. Allele-specific expression in different placental samples. Each grid shows the allele-specific expression for each of the 12 imprinted and monoallelically expressed genes in different placental samples. The X-axis is the genomic position of the SNP and the Y-axis shows the percentage of maternal reads, with 100% refers to complete maternal expression, 0% corresponds to complete paternal expression and 50% refers to perfect biallelic expression. Statistically significant results are shown in red (maternal expression) and blue (paternal expression). From all non-significant results (depicted in black), only those are shown where the total number of RNA-Seq reads was at least 10. P001 - P014 are family trios analyzed.
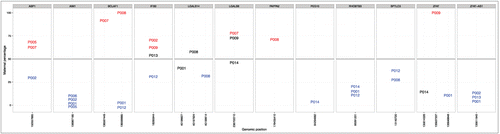
Lately, the study of Barbaux et al.Citation9 showed the imprinting and paternal expression of ZFAT locus harboring also a non-coding antisense RNA overlapping ZFAT gene named ZFAT-AS1. Although the number of phased SNPs was limited for ZFAT and ZFAT-AS1 locus in our study, we were able to re-establish the consistent paternal expression of ZFAT-AS1 by SNP rs135611945 in 3 family trios (). However, we could not repeat the results for ZFAT imprinting as we demonstrated the monoallelic expression of ZFAT of 2 different SNPs in 2 different family trios, revealing monoallelic expression, but random activity of either of the parental allele ().
In addition to ZFAT gene, ABP1, BCLAF1 and IFI30 demonstrated random monoallelic expression as all these 3 genes showed consistent and statistically significant monoallelic expression for 1-2 SNPs in 3–4 family trios (). The monoallelic expression of these 4 genes seems to be convincing as almost all informative SNPs identified in these genes in our family trios showed monoallelic, but mixed paternal and maternal activity of the corresponding gene. Surprisingly, we found biased parental allele usage for those 4 genes if we calculated the aggregated coverage for RNA-Seq reads summarised for all SNPs and families analyzed (). This could mean that the silencing of parental alleles could be different and may result in some variations in the relative amount of parental alleles in individual families. We were also unable to identify additional randomly monoallelically expressed genes neither among all genes nor the 33 candidate genes shown in .
According to our results, we showed possible imprinted regulation for 4 novel genes (LGALS8, LGALS14, PAPPA2 and SPTLC3). However, future studies are required in order to clearly show the imprinted regulation of these 4 genes as the number of SNPs and trios remained too small to draw the final conclusion. Furthermore, as for galectin genes (LGALS8 and LGALS14), we found SNPs with both statistically significant ASE and biallelic expression, the activity of these genes is most likely regulated by mosaic imprinting not present in all family trios. However, this finding should also be confirmed by subsequent studies including the higher number of families.
Among the 8 imprinted genes found in the current study only 2 genes (PAPPA2 and LGALS8) were expressed predominantly from the maternal allele, while in rest of the genes the paternal allele was more active. The proportions of maternal reads for the imprinted genes are shown in , ranging from 3.5% of maternally expressed alleles for the paternally active PEG10 to 70% of maternally expressed alleles in PAPPA2.
Expression levels and genomic locations of imprinted and monoallelically expressed genes
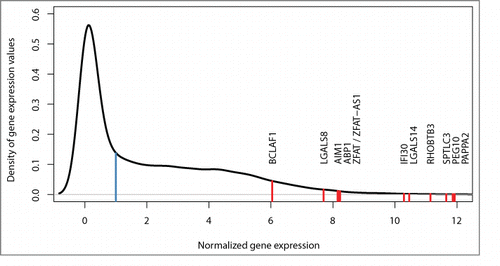
All 12 imprinted and randomly monoallelically active genes were expressed at levels above the median expression level of all genes transcribed in human placental tissue. The lowest and highest expressions were observed for BCLAF1 and PAPPA2, respectively. The density curve depicted in shows the distribution of the expression values for all genes.
Figure 3. Density curve for gene expression. Red lines show the distribution of expression levels for all 12 imprinted and monoallelically expressed genes detected in the current study (see for details). Median expression of all genes is normalized to 1 (blue line).
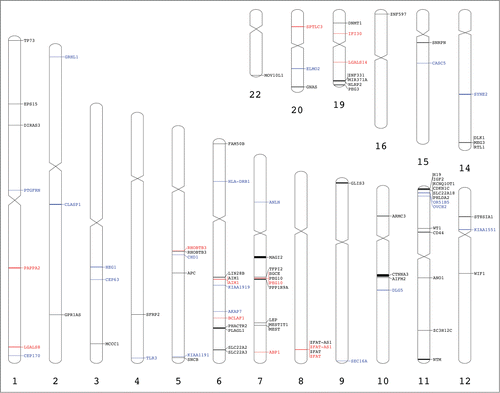
The ideogram in highlights the locations of the candidate imprinted genes (), the positions of the imprinted and randomly monoallelically expressed genes (), and the chromosomal coordinates of the previously known imprinted genes (Table S1). Some of the imprinted genes found in the current study belonged to well-known clusters of imprinted genes, such as AIM1 at 6q21 and PEG10 at 7q21. Still, most of the imprinted and monoallelically active genes were randomly scattered across the entire human genome.
Figure 4. Genome visualization of the imprinted and monoallelically expressed genes. Previously reported placental imprinted genes are in black (Table S1), candidate imprinted genes are marked in blue () and imprinted and monoallelically expressed genes found in our analysis are marked in red (). This image was generated using Idiographica web service.Citation49
Validation of imprinted genes
High-throughput screening methods are useful for genome-wide scans, but can sometimes cause false-positive results. To confirm the validity of our approach, 3 imprinted genes - PEG10, Rho-related BTB domain containing 3 (RHOBTB3) and pappalysin 2 (PAPPA2), were chosen for further validation. PEG10 is the most studied and evolutionarily conserved maternally imprinted and paternally expressed gene.Citation20 In contrast, RHOBTB3 was only very recently shown to be imprinted based on DNA methylation profilingCitation11 and requires additional proof from gene expression analysis. Potential imprinting of PAPPA2 in human placenta has not been reported previously.
The 500-bp genomic windows with the densest SNP concentration were selected and genotyped by Sanger sequencing (Table S3). Genotyping results for the SNPs were obtained for maternal, paternal and newborn DNA and placental cDNA. If a gene has monoallelic expression in placenta according to its cDNA profile (so called “transcriptome-derived genotype”), then the genotype assigned for the placental cDNA will be homozygous. In case the child's blood DNA genotype is heterozygous, the silencing of one allele in placental transcriptome is proven.
We genotyped 2 SNPs (rs13073 and rs13226637) within PEG10 in 7 families. In 4 families, we were able to confirm heterozygosity in the newborn and the monoallelic placental cDNA genotype. In all cases, the placental alleles were paternally inherited, confirming the exclusive expression from the paternal allele. In the other 3 tested families, all obtained genotypes were homozygous; therefore, it was not possible to gather information about imprinting. For RHOBTB3, 7 SNPs (rs6815, rs7622, rs3184188, rs6697, rs12351, rs34898 and rs34899) were tested in 8 family trios. There were no informative genotypes for one SNP (rs3184188). For the other 6 SNPs, we were able to see the same effect as for PEG10 in 5 families, namely, the placental cDNA had the homozygous genotype, and the child's blood DNA was heterozygous in the same position. The allele detected in the placental cDNA was inherited from the father in all cases, indicating imprinted status for the paternally expressed RHOBTB3 (Fig. S3). Three potentially informative SNPs (rs12137448, rs12137517 and rs726252) were genotyped for PAPPA2 to confirm the RNA-Seq results shown in . We were able to confirm heterozygous genotypes for all 3 SNPs in the newborn. Only maternal alleles were detected in the same positions in the placental cDNA, proposing imprinting of the paternal allele and expression of the maternal allele of PAPPA2.
Contribution of maternal decidua to the study of fetal genes
As placenta comprises both fetal and maternal tissues, it is possible that the maternal part of the placenta could provide more sequencing reads. To estimate this potential bias, the fraction of maternal alleles was assessed by using phased SNPs from all autosomes. The fraction of maternal reads was equal to paternal reads, being close to 50% (46.5–51.2%) in all studied placentas (Fig. S4), supporting the conclusion that genes active in maternal decidua do not interfere with the identification of imprinted and monoallelically expressed genes in placental tissue.
Discussion
We used WT RNA-Seq and genotyping of coding SNPs to study genomic imprinting and monoallelic autosomal expression in general in human placenta. We confirmed imprinting of well-established PEG10 and recently shown AIM1, RHOBTB3 and ZFAT-AS1. All these 4 genes are expressed from the paternal allele. In addition, we provided the evidence of possible imprinting of 4 additional genes, exhibiting either paternal (LGALS14 and SPTLC3) or maternal (LGALS8 and PAPPA2) expression. However, these findings should be taken with the caution and require confirmation by future studies including higher number of family trios. Even more interesting, we were able to demonstrate the random monoallelic expression of 4 genes (ABP1, BCLAF1, IFI30 and ZFAT), showing opposite expression of parental alleles in placental tissue in individual families.
We used full-thickness placental samples to study placental genomic activity. This approach raises the concern that a maternal contribution might distort the study of fetal genes in placental tissue. Furthermore, the differences in invasive behavior of feto-placental cells could be a possible reason for predominance of maternally or paternally expressed imprinted genes in placental tissue samples in different families. However, these 2 alarming precautions were considered unlikely as the contribution of maternal tissues to the total amount of transcribed alleles in placenta was found to be minimal because the proportions of maternally and paternally expressed alleles were nearly equal in all of our analyzed samples (Fig. S4). This finding adds confidence about our study design, in terms of its ability to highlight imprinting and monoallelic expression in the fetal part of the placenta. Furthermore, by mapping the inheritance of the fetal alleles, we were able to focus only on the maternal alleles that were passed from the mother to her child.
The main functions of the proteins encoded by the imprinted and monoallelically expressed genes identified in the current study can be grouped as follows: i) mediating cellular apoptosis and tissue development; ii) regulating inflammation and the immune system; iii) facilitating metabolic processes; and iv) regulating the cell cycle (). Among our detected genes, PEG10, AIM1, BCLAF1 and PAPPA2 all participate in the regulation of cellular apoptosis and tissue development.
BCL2-associated transcription factor 1 (BCLAF1) encodes a protein that interacts with anti-apoptotic members of the BCL2 family, thereby acting as an inducer of apoptosis.Citation21 The functional BCLAF1 protein prevents carcinogenic processes and is suppressed in various carcinoma cells.Citation22 Nephroblastoma, also known as Wilms tumor, is a kidney cancer that commonly affects children with Beckwith-Wiedemann syndrome. In approximately 20% of nephroblastomas, the cancer is caused by mutations in the Wilms tumor 1 gene (WT1),Citation23 which exhibits polymorphic imprinting in human placenta.Citation24 Recently, the BCLAF1 protein was identified as a member of the WT1-associating protein complex required for cell-cycle progression.Citation25 Here, we provide the first evidence that placental BCLAF1 is an epigenetically regulated gene with random monoallelic expression in human placenta.
One of the most intriguing findings of our study is the imprinting status of the pregnancy-associated plasma protein 2 gene (PAPPA2; also known as pappalysin-2). The proteases PAPPA2 and its paralog PAPPA are involved in normal placental development, are produced by human trophoblasts,Citation26,27 and have complementary functions. Both proteins degrade maternal decidua-produced insulin-like growth factor-binding proteins (IGFBPs), thereby releasing IGF2.Citation28,29 PAPPA is a known marker of fetal genetic disorders, such as Down syndrome,Citation30 and PAPPA2 is upregulated in the placenta in preeclampsia (discussed by ref. 31).
We found preliminary evidence that PAPPA2 was maternally expressed in the human full-term placenta. If this finding is supported by future studies, then the imprinting status of PAPPA2 would help to outline alternative scenarios through which 2 mammalian species, mouse and human, have uniquely solved the problem of balancing the growth-promoting effect of paternally expressed IGF2. In mice, the maternally expressed Igf2 receptor (Igf2r) binds and targets paternal Igf2 for lysosomal degradation, thereby reducing its bioavailability.Citation32 However, in humans, the bioavailability of IGF2 might be regulated by the selective expression of maternal PAPPA2, thereby limiting the cleavage rate of IGFBPs and the release of IGF2.
During pregnancy, controlling infections and inflammation while modulating the maternal immune system are essential elements for fetal survival and development within the mother. We found 5 genes, IFI30, LGALS8, LGALS14, ZFAT and ZFAT-AS1, which may be associated with those mechanisms.
Interferon gamma (INF-γ) inducible protein 30 (IFI30) encodes a lysosomal thiol reductase that reduces protein disulfide bonds at low pH to control intracellular proteases. IFI30 is expressed constitutively in antigen-presenting cells and induced by INF-γ in other cell types. IFI30 has an important role in MHC class II-restricted antigen processing, by regulating proteases essential for the degradation of endogenous and exogenous antigens in the antigen presentation process.Citation33 More importantly, IFI30 helps to shape central and peripheral CD4+ T-cell tolerance to self-antigens.Citation34,35 Here, we detected monoallelic expression of maternal IFI30 in term placentas. This is in concordance with the general view that monoallelic expression is more often seen among the genes crucial for immune response and adaptation and encoding for immunoglobulins, T cell receptors, interleukins and receptors on natural killer (NK) cells (as reviewed in ref. 15). Although remaining highly speculative, we are suggesting that monoallelic expression of IFI30 may regulate pregnancy-related changes in the maternal immune system, particularly by inducing and maintaining immune tolerance toward the fetus and protecting the fetus from infection.
Two imprinted genes, LGALS14 (lectin, galactoside-binding, soluble, 14) and LGALS8, encode members of the galectin family of proteins. Galectins are β-galactoside-binding animal lectins with conserved carbohydrate recognition domains. LGALS14 is predominantly expressed in placenta and hardly detectable in fetal organs.Citation36 Although there are limited research data on LGALS14, its membrane-bound counterparts are evolutionary conserved proteins that regulate immune function. For instance, the membrane-bound galectin-9 on T helper cells regulates immune responses toward immune tolerance.Citation37 Hence, we assume that the paternally expressed LGALS14 in term placenta aids in regulating the maternal immune system, to protect the fetus from maternal rejection.
LGALS8 is widely expressed in tumor tissues and seems to be involved in integrin-like cell interactions. We observed that LGALS8 was maternally expressed, unlike LGALS14, indicating that these galectins exert different roles in pregnancy. Galectin-8 is a cytosolic lectin that monitors endosomal and lysosomal integrity and detects bacterial invasion, thereby activating antibacterial defense.Citation38 Galectin-8 is expressed in villous and extravillous trophoblasts.Citation39 Together, these data suggest that galectin-8 most likely contributes to cellular proliferation, adhesion, invasion, and protection from microbial infection, all controlled by the maternal LGALS8 gene.
Using genome-wide mapping of differentially methylated regions, a recent study reported that RHOBTB3 has imprinted regulation.Citation11 However, this study largely challenged the earlier study of Gimelbrant et al.Citation16 showing biallelic expression for RHOBTB3 in human placenta. RHOBTB3 gene encodes a member of a family of small GTPases, which is involved in diverse cellular processes, such as endocytosis, vesicle trafficking, morphogenesis, cytokinesis, transcriptional activation, cell-cycle progression and apoptosis.Citation40 Our results seem to support the view of RHOBTB3 imprinting in human placenta as we demonstrated a strong bias toward expression of paternal allele, similar to PEG10 gene. However, the more depth understanding about the need for paternal expression of RHOBTB3 awaits for future studies.
All our imprinted and monoallelically expressed genes demonstrated high activity levels compared to the median level of all expressed genes (). This elevated activity could be because these genes are likely important for pregnancy development and are thus highly upregulated in term placenta. Alternatively, the detection of imprinted and monoallelically expressed genes might have been limited to those that were expressed at high levels, as in the case of insufficient expression, any method can lack statistical power to demonstrate the monoallelic mode of gene expression.
Previous studies suggested that imprinted genes tend to be found in clusters.Citation41 Thus, the search for novel imprinted genes is often confined to the proximity of already established clusters of imprinted genes. This approach can present a bias that is eliminated by using RNA-Seq, which provides the advantage of capturing an entire transcriptome at the gene level. Unexpectedly, our findings seem to violate this general rule, as none of the newly identified 4 imprinted genes matched any known cluster (). Among the 33 candidate imprinted genes, OR51B5 and OVCH2 were located in the IGF2 imprinted cluster at chromosomal locus 11p15, whereas KIAA1919 was positioned close to the imprinted AIM1 gene at 6q21. Still, the overall distribution of imprinted genes observed in our study is supportive of the view that imprinted genes tend to show a more disperse allocation throughout the human genome than initially thought. This possibility was demonstrated in another genome-wide RNA-Seq study of placental imprinting, as newly identified imprinted genes were randomly distributed across the entire equine genome.Citation14
Our report is among a very few studies that have systematically addressed bi- and monoallelic gene expression in placenta. Surprisingly, 5–10% of autosomal human genes undergo monoallelic expression, as shown in recent study.Citation16 The high proportion of monoallelically expressed genes in that previous study seems to contradict our results with only 4 genes (ABP1, BCLAF1, IFI30 and ZFAT) showing monoallelic expression. However, this can be easily explained by the study design and different nature of gene expression regulation. In the study of Gimelbrant et al.,Citation16 the authors analyzed clonal B-lymphoblastoid cell lines and very small patches (1 mmCitation3) of placental tissue. The latter most probably consisted of clonally expanded placental cells which had inherited the identical epigenetic pattern of monoallelic expression to all daughter cells. Instead, in our study, full-thickness placental blocks of around 2–3 cm were taken from 3 different regions of placenta thus eliminating the possibility that we analyzed only the cells having common origin.
If the clonally expanded cells are analyzed, including the small clonal patches of tissue samples, the analyzed cells might express the same parental allele. However, if non-clonal cells or larger tissue samples with cells of different origin are analyzed the monoallelic expressions of opposite alleles are usually compensated and masked, making the monoallelic expression difficult to follow. This explains that monoallelic gene expression can be identified in larger extent only when single cells are analyzed or alternatively, when clonal descendants of the single cells are examined all together. This makes our observation of 4 monoallelically expressed genes valuable as we focused on the placental tissue as a mixture of different cell-types and having diverse origin. Our finding also implies that the epigenetic decision to inactivate one of the parental alleles was made early in the placental development and this choice was stably transmitted to all placental cells.
Although there is a growing body of evidence that random monoallelic gene expression seems to be a common phenomenon that greatly contributes to phenotypic differences among individual cells, our findings also hint that it might have consequences on pregnancy development. Still, the roles of random monoallelic gene expression as well as the detailed mechanisms of coordinated allele silencing remain to be discovered by the future studies.
The major shortcoming of RNA-Seq is the need to map the short sequence reads to their correct genomic locations in a reference genome. The mapping of reads with heterozygous SNPs might create a bias toward favored mapping of the allele in the reference sequence, compared with the alternative allele,Citation42 thus leading to a possible false discovery of gene imprinting. To overcome this systemic bias toward the reference allele, we used exome SNP genotyping of a child and her/his parents. We adopted a computational pipeline methodCitation43 that integrated both maternal and paternal genome variation data into a diploid reference sequence. Mapping reads to both parental genomes and using the best mapping for further analysis helped to ensure that we were not reporting false-positive imprinted genes in our study.
We were able to confirm only 4 genes from previously known imprinted genes in placenta. The reason for this low number of genes may be due to the exome genotyping array that was used. The array did not include informative SNPs for identifying most of the previously known imprinted genes as only 78% of all annotated genes were represented in the microarray with at least one SNP (Fig. S1). This proportion is further reduced to 43% if we count only those genes that possessed at least one informative SNP in at least one family trio. Therefore, our methodological approach would likely miss more than half of all possibly imprinted genes. For example, the microarray analysis did not contain any phased SNPs in any of the trios for the analysis of PHLDA2 and CDKN1C. Only one phased SNP was found in 3 family trios in IGF2, but the low mapping coverage of the RNA-Seq reads to that specific locus was not sufficient to confirm its imprinting. However, when all heterozygous SNPs in the RNA-Seq data were analyzed in IGF2, almost 80% of them showed statistically significant ASE, thereby indicating the imprinted status for IGF2 in our study.
Owing to these limitations, our methodology to reveal the imprinted genes in placental tissue is biased toward the identification of highly expressed genes with denser sets of SNPs on the genotyping microarray, as summarized in Figure S5. In future studies, exome sequencing of all family members should be used to reveal the full genetic diversity and heritability. Still, we proposed the imprinting of 4 novel genes (LGALS8, LGALS14, PAPPA2 and SPTLC3). When compared to already known imprinted genes, it seems that all novel genes are characterized with much subtle parental allelic expression bias. For instance, the less active allele accounts, in average, for 9.6% and 33.1% of total gene expression for already known (AIM1, PEG10, RHOBTB3 and ZFAT-AS1) and novel imprinted genes, respectively. Furthermore, gene imprinting may depend on the developmental stage of the placenta and the health of the pregnancy. As such, the results of the current study must be considered in the context of full-term healthy pregnancies.
In conclusion, the aim of the current study was to introduce a workflow and tools for analyzing imprinted and random monoallelic autosomal gene expression in human placental transcriptome. We identified 4 novel possible imprinted genes (LGALS8, LGALS14, PAPPA2 and SPTLC3) and confirmed the imprinting of 4 genes (AIM1, PEG10, RHOBTB3 and ZFAT-AS1) in human term placenta. Still, the major finding is the identification of 4 genes that demonstrated random allelic bias with the expression of one of the parental alleles preferentially. However, instead of the randomness at the cellular or clonal level the monoallelic expression was observed in the entire placental tissue.
Materials and Methods
Description of participants and sample collection
The study was approved by the Research Ethics Committee of the University of Tartu. Written informed consent was obtained from each participant of the study (permission no. 213/T-21). Ten family trios (mother, father and newborn) of Caucasian descent were recruited from the Maternity Department of the Women's Clinic at Tartu University Hospital from April to June 2012. Participant characteristics are summarized in Table S4. The average (mean ± standard deviation [SD]) maternal and paternal ages were 26.7 ± 5.4 and 30.5 ± 6.4 years, respectively. Data regarding parental diseases, smoking status, somatometric data and maternal childbirth history were obtained from the medical records during the pregnancy and after birth. Cases of documented fetal or chromosomal abnormalities, families with a history of inherited diseases, and patients with diabetes mellitus, hypertension or renal diseases as well as with pregnancy complications were excluded.
Clinical and biological materials were collected from singleton pregnancies at term. The average gestational age at delivery was 40.5 ± 1.0 weeks (range, 38–42 weeks). Eight women had spontaneous vaginal deliveries. Two babies were born by elective and emergency caesarean section due to scarred uterus and psychosocial reasons; and obstructed labor, respectively. The newborn babies (6 boys and 4 girls) were healthy, with birth parameters appropriate for their gestational age of 10th to 90th percentile, according to the Estonian Medical Birth Registry.Citation44 The average birth weight was 3,629.6 ± 324.1 g. The average Apgar scores at 1 and 5 minutes were 8.6 ± 0.5 and 9.2 ± 0.6 points, respectively.
Placental biopsies were obtained within 1 hour after removal of placenta during the caesarean section or vaginal delivery. Full-thickness placental blocks of around 2–3 cm were taken from 3 different regions of placenta, placed immediately into RNAlater (Ambion®, Life Technologies™), stored 24 hours at 4°C, and stored at -80°C for subsequent RNA extraction. All samples were collected by the same medical personnel. The maturity and health of the term placenta were confirmed by histological examination. Postnatal umbilical cord and parental peripheral blood samples were collected for DNA genotyping.
DNA and RNA extraction
A total of 9 mL of peripheral venous blood from parents and 4 mL of umbilical cord blood were collected into ethylenediaminetetraacetic acid-containing tubes (BD Vacutainer, Becton, Dickinson and Co) for DNA extraction. DNA was isolated from blood by a salting-out protocol.Citation45 DNA samples were eluted with 1 mL of water and stored at -20°C until genotyping. NanoDrop 2000 (Thermo Fisher Scientific Inc.) was used to assess DNA concentration and purity.
Total RNA was extracted from frozen placental tissue with the mirVana™ miRNA Isolation Kit (Ambion®, Life Technologies™), according to the manufacturer's instructions. Extractions were performed from 3 samples collected from the same placenta, and the RNA samples were pooled together. Contaminating DNA was removed from RNA samples with the DNA-free™ Kit (Invitrogen™, Life Technologies™), according to the manufacturer's instructions. An Agilent 2100 Bioanalyzer and the RNA 6000 Nano Kit (Agilent Technologies) were used to assess the integrity and concentration of total RNA samples. RNA samples with an RNA integrity number of ≥5 were used for further analyses.
DNA SNP genotyping
Exome SNPs of all participants (mother, father and child) were genotyped with the Illumina HumanExome BeadChip v1.1 SNP microarray (Illumina®). A 95% call rate criterion was used for genotyped samples and markers. Calling was performed with GenomeStudio (2010.3). PLINK software (http://pngu.mgh.harvard.edu/purcell/plink/) was used for quality control filtering.Citation46 Samples were controlled for gender mismatches and larger deviations of heterozygosity to detect mislabelling and contamination. SNPs from the X and Y chromosomes, all insertions and deletions (indels), mutant SNPs (i.e., a child had a SNP allele that was missing in his/her parents, most likely representing genotyping errors) and SNPs with at least one missing allele (e.g., due to low-quality DNA and/or poor genotyping data) were excluded from further analysis (Table S2). Genotyping data is available in GEO (accession number: GSE56781).
RNA-Seq and gene expression analysis
To prepare the WT library for WT RNA-Seq, 45 ng of total RNA were used. RNA was amplified with the Ovation RNA-Seq System V2 Kit (NuGen). Output double-stranded cDNA was used to prepare the SOLiD 5500 System DNA fragment library, according to the manufacturer's protocols (Life Technologies™). Barcoding primers for library preparation and paired-end sequencing chemistry were applied (75 bp in forward and 35 bp in reverse directions). In the case of 10 pooled samples analyzed on 3 FlowChip lanes, approximately 40 million mappable paired-end reads were obtained per sample. The DESeq packageCitation47 in the R statistical language and environment (version 3.0.2) was used to calculate the reads per kilobase per million (RPKM). Log2 RPKM was used as a measure of gene expression. Background gene expression was calculated as the median value of all genes, with the gene expression scaled so that the background expression was considered equal to one. RNA-Seq data is available in GEO (accession number: GSE56781).
Identifying imprinted and monoallelically expressed genes
A custom pipeline, similar to the one used by Rozowsky et al.Citation43 was used (). This pipeline was mostly based on a collection of custom scripts (available on http://biit.cs.ut.ee/supplementary/placenta_imprinting/) written in R software (version 2.15). Differences from the Rozowsky et al. pipeline are mainly due to the data type analyzed. Only SNPs and not structural variants or indels, were used to construct the parental genomes.
WT RNA-Seq reads from placenta were mapped to both parental genomes with the SHRiMP2 mapping program.Citation48 For each read, the alignment with the best parental score was used in the downstream analysis. When the mapping quality was equal for the maternal and paternal genomes, the underlying sequence was most probably the same; therefore, either of the 2 mappings was chosen randomly. Reads with gapped alignment, or in which the paired-end reads were mapped too far away from each other (>10,000 bp) or to different chromosomes were excluded. After excluding low-quality mappings, approximately 50% of the reads remained for further analyses (Fig. S6). For each phased SNP, the number of maternal and paternal reads was counted. The deviation between the read numbers was analyzed with the binomial test in R software (version 2.15).
To select the final list of genes, further filtering was performed, including all RNA-Seq SNPs with at least 1% minor allele frequency (MAF), according to the dbSNP database,Citation18 and 3% MAF in the RNA-Seq data; in this way, possible sequencing errors were minimized. The 2 alleles were confirmed to be the same as those found in the dbSNP database. The analysis pipeline similar to microarray SNPs was repeated (), and the genes, which had ≥75% of ASE-SNPs from the total number of heterozygous SNPs, were selected.
As an additional filtering step, we subsequently analyzed the parental specific gene expression in individual families that helped to discriminate between the imprinted and randomly monoallelically expressed genes ().
Validation of gene imprinting and monoallelic expression
Selected regions of genomic DNA samples of PEG10, RHOBTB3 and PAPPA2 (Table S3) from all family members were amplified by PCR in 10 μL containing 1 × Hot FirePol® Buffer B1 (Solis BioDyne), 0.25 mM of each dNTP (Thermo Fisher Scientific Inc.), 2.5 mM MgCl2 (Thermo Fisher Scientific Inc.), 1 U of Hot FirePol® DNA polymerase (Solis BioDyne) and 0.8 μM of each primer (Metabion GmbH). Cycling was performed with the MyCycler Thermal Cycler (Bio-Rad Laboratories Inc.) under following conditions: 95°C for 15 minutes (initial denaturation), 35 cycles of 95°C for 30 seconds, 58°C for 30 seconds and 72°C for 50 seconds, followed by 72°C for 5 minutes.
Reverse transcription of placental RNA was performed with the RevertAid™ First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc.) with random hexamer primers. The acquired cDNA was amplified with the same specified primers (Table S3) and the aforementioned thermocycling conditions. For sequencing, the BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies™) was used in a 10-μL reaction volume containing 1 μL of purified PCR product. Electrophoresis was performed on an ABI Prism 3730 DNA Analyzer (Life Technologies™).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are grateful to all of the families who participated in this study. We also thank the midwives Sirje Kõvermägi and Kätlin Mark for collecting the clinical data and biological samples. Agne Velthut-Meikas is acknowledged for critical reading of the manuscript.
Supplementary_Tables_S1-S4.docx
Download MS Word (156.9 KB)Supplementary_Figures_S1-S6.docx
Download MS Word (1.2 MB)Funding
This study was supported by grants from the Center of Excellence in Genomics (grant EXCEGEN); Estonian Ministry of Education and Research (grants SF0180044s09, SF0180008s12, SF0180035s08 and by institutional research funding IUT20-46); Enterprise Estonia (grant EU30020); EU-FP7 Eurostars program (grant EU41564); EU-FP7 IAPP project (grant EU324509); Center of Translational Genomics, University of Tartu (SP1GVARENG); COST Action FA1201; European Regional Development Fund through Estonian eXcellence in Computer Science and through BioMedIT projects and through Center of Translational Medicine, University of Tartu and the Tiger University program of the Information Technology Foundation for Education.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Sood R, Zehnder JL, Druzin ML, Brown PO. Gene expression patterns in human placenta. Proc Nat Acad Sci U S A 2006; 103:5478-83; PMID:16567644; http://dx.doi.org/10.1073/pnas.0508035103
- Sitras V, Fenton C, Paulssen R, Vartun A, Acharya G. Differences in gene expression between first and third trimester human placenta: a microarray study. PloS One 2012; 7:e33294; PMID:22442682; http://dx.doi.org/10.1371/journal.pone.0033294
- Uuskula L, Mannik J, Rull K, Minajeva A, Koks S, Vaas P, Teesalu P, Reimand J, Laan M. Mid-gestational gene expression profile in placenta and link to pregnancy complications. PloS One 2012; 7:e49248; PMID:23145134; http://dx.doi.org/10.1371/journal.pone.0049248
- McMinn J, Wei M, Schupf N, Cusmai J, Johnson EB, Smith AC, Weksberg R, Thaker HM, Tycko B. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta 2006; 27:540-9; PMID:16125225; http://dx.doi.org/10.1016/j.placenta.2005.07.004
- Meng T, Chen H, Sun M, Wang H, Zhao G, Wang X. Identification of differential gene expression profiles in placentas from preeclamptic pregnancies versus normal pregnancies by DNA microarrays. Omics: A J Integr Biol 2012; 16:301-11; PMID:22702245; http://dx.doi.org/10.1089/omi.2011.0066
- Renfree MB, Suzuki S, Kaneko-Ishino T. The origin and evolution of genomic imprinting and viviparity in mammals. Philos T Roy Soc Lond Series B, Biol Sci 2013; 368:20120151; PMID:23166401; http://dx.doi.org/10.1098/rstb.2012.0151
- Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet: TIG 1991; 7:45-9; PMID:2035190; http://dx.doi.org/10.1016/0168-9525(91)90230-N
- Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, Hino T, Suzuki-Migishima R, Ogonuki N, Miki H, et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet 2006; 38:101-6; PMID:16341224; http://dx.doi.org/10.1038/ng1699
- Barbaux S, Gascoin-Lachambre G, Buffat C, Monnier P, Mondon F, Tonanny MB, Pinard A, Auer J, Bessieres B, Barlier A, et al. A genome-wide approach reveals novel imprinted genes expressed in the human placenta. Epigenetics: Off J DNA Methylation Soc 2012; 7:1079-90; PMID:22894909; http://dx.doi.org/10.4161/epi.21495
- Daelemans C, Ritchie ME, Smits G, Abu-Amero S, Sudbery IM, Forrest MS, Campino S, Clark TG, Stanier P, Kwiatkowski D, et al. High-throughput analysis of candidate imprinted genes and allele-specific gene expression in the human term placenta. BMC Genet 2010; 11:25; PMID:20403199; http://dx.doi.org/10.1186/1471-2156-11-25
- Yuen RKC, Jiang R, Penaherrera MS, McFadden DE, Robinson WP. Genome-wide mapping of imprinted differentially methylated regions by DNA methylation profiling of human placentas from triploidies. Epigenetics Chromatin 2011; 4:10; PMID:21749726; http://dx.doi.org/10.1186/1756-8935-4-10
- Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res 2007; 17:1723-30; PMID:18055845; http://dx.doi.org/10.1101/gr.6584707
- Wang X, Soloway PD, Clark AG. A survey for novel imprinted genes in the mouse placenta by mRNA-seq. Genetics 2011; 189:109-22; PMID:21705755; http://dx.doi.org/10.1534/genetics.111.130088
- Wang X, Miller DC, Harman R, Antczak DF, Clark AG. Paternally expressed genes predominate in the placenta. Proc Natl Acad Sci U S A 2013; 110:10705-10; PMID:23754418; http://dx.doi.org/10.1073/pnas.1308998110
- Chess A. Mechanisms and consequences of widespread random monoallelic expression. Nat Rev Genet 2012; 13:421-8; PMID:22585065; http://dx.doi.org/10.1038/nrg3239
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science 2007; 318:1136-40; PMID:18006746; http://dx.doi.org/10.1126/science.1148910
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 2013; 41:D64-9; PMID:23155063; http://dx.doi.org/10.1093/nar/gks1048
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001; 29:308-11; PMID:11125122; http://dx.doi.org/10.1093/nar/29.1.308
- Das R, Lee YK, Strogantsev R, Jin SN, Lim YC, Ng PY, Lin XM, Chng K, Yeo GSH, Ferguson-Smith AC, et al. DNMT1 and AIM1 Imprinting in human placenta revealed through a genome-wide screen for allele-specific DNA methylation. Bmc Genomics 2013; 14:685; PMID:24094292; http://dx.doi.org/10.1186/1471-2164-14-685
- Smallwood A, Papageorghiou A, Nicolaides K, Alley MK, Jim A, Nargund G, Ojha K, Campbell S, Banerjee S. Temporal regulation of the expression of syncytin (HERV-W), maternally imprinted PEG10, and SGCE in human placenta. Biol Reprod 2003; 69:286-93; PMID:12620933; http://dx.doi.org/10.1095/biolreprod.102.013078
- Sarras H, Alizadeh Azami S, McPherson JP. In search of a function for BCLAF1. S World J 2010; 10:1450-61; PMID:20661537; http://dx.doi.org/10.1100/tsw.2010.132
- Lee YY, Yu YB, Gunawardena HP, Xie L, Chen X. BCLAF1 is a radiation-induced H2AX-interacting partner involved in gammaH2AX-mediated regulation of apoptosis and DNA repair. Cell Death Dis 2012; 3:e359; PMID:22833098; http://dx.doi.org/10.1038/cddis.2012.76
- Call KM, Glaser T, Ito CY, Buckler AJ, Pelletier J, Haber DA, Rose EA, Kral A, Yeger H, Lewis WH, et al. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms’ tumor locus. Cell 1990; 60:509-20; PMID:2154335; http://dx.doi.org/10.1016/0092-8674(90)90601-A
- Jinno Y, Yun K, Nishiwaki K, Kubota T, Ogawa O, Reeve AE, Niikawa N. Mosaic and polymorphic imprinting of the WT1 gene in humans. Nat Genet 1994; 6:305-9; PMID:8012395; http://dx.doi.org/10.1038/ng0394-305
- Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem 2013; 288:33292-302; PMID:24100041; http://dx.doi.org/10.1074/jbc.M113.500397
- Tornehave D, Chemnitz J, Teisner B, Folkersen J, Westergaard JG. Immunohistochemical demonstration of pregnancy-associated plasma protein A (PAPP-A) in the syncytiotrophoblast of the normal placenta at different gestational ages. Placenta 1984; 5:427-31; PMID:6084247; http://dx.doi.org/10.1016/S0143-4004(84)80023-5
- Wang J, Qiu Q, Haider M, Bell M, Gruslin A, Christians JK. Expression of pregnancy-associated plasma protein A2 during pregnancy in human and mouse. J Endocrinol 2009; 202:337-45; PMID:19474058; http://dx.doi.org/10.1677/JOE-09-0136
- Giudice LC, Conover CA, Bale L, Faessen GH, Ilg K, Sun I, Imani B, Suen LF, Irwin JC, Christiansen M, et al. Identification and regulation of the IGFBP-4 protease and its physiological inhibitor in human trophoblasts and endometrial stroma: evidence for paracrine regulation of IGF-II bioavailability in the placental bed during human implantation. J Clin Endocrinol Metab 2002; 87:2359-66; PMID:11994388; http://dx.doi.org/10.1210/jcem.87.5.8448
- Overgaard MT, Boldt HB, Laursen LS, Sottrup-Jensen L, Conover CA, Oxvig C. Pregnancy-associated plasma protein-A2 (PAPP-A2), a novel insulin-like growth factor-binding protein-5 proteinase. J Biol Chem 2001; 276:21849-53; PMID:11264294; http://dx.doi.org/10.1074/jbc.M102191200
- Brambati B, Macintosh MC, Teisner B, Maguiness S, Shrimanker K, Lanzani A, Bonacchi I, Tului L, Chard T, Grudzinskas JG. Low maternal serum levels of pregnancy associated plasma protein A (PAPP-A) in the first trimester in association with abnormal fetal karyotype. Br J Obstet Gynaecol 1993; 100:324-6; PMID:7684253; http://dx.doi.org/10.1111/j.1471-0528.1993.tb12973.x
- Christians JK, Gruslin A. Altered levels of insulin-like growth factor binding protein proteases in preeclampsia and intrauterine growth restriction. Prenatal Diagn 2010; 30:815-20; PMID:20658698; http://dx.doi.org/10.1002/pd.2583
- Frost JM, Moore GE. The importance of imprinting in the human placenta. PLoS Genet 2010; 6:e1001015; PMID:20617174; http://dx.doi.org/10.1371/journal.pgen.1001015
- Goldstein OG, Hajiaghamohseni LM, Amria S, Sundaram K, Reddy SV, Haque A. Gamma-IFN-inducible-lysosomal thiol reductase modulates acidic proteases and HLA class II antigen processing in melanoma. Cancer Immunol, Immunother: CII 2008; 57:1461-70; PMID:18343923; http://dx.doi.org/10.1007/s00262-008-0483-8
- Becker JC, Schrama D. Control of central and peripheral tolerance to melanocyte differentiation antigens by GILT. J Invest Dermatol 2012; 132:15-7; PMID:22158609; http://dx.doi.org/10.1038/jid.2011.361
- Rausch MP, Hastings KT. GILT modulates CD4 +T-cell tolerance to the melanocyte differentiation antigen tyrosinase-related protein 1. J Invest Dermatol 2012; 132:154-62; PMID:21833020; http://dx.doi.org/10.1038/jid.2011.236
- Yang QS, Ying K, Yuan HL, Chen JZ, Meng XF, Wang Z, Xie Y, Mao YM. Cloning and expression of a novel human galectin cDNA, predominantly expressed in placenta(1). Biochimica Et Biophysica Acta 2002; 1574:407-11; PMID:11997112; http://dx.doi.org/10.1016/S0167-4781(01)00319-0
- Oomizu S, Arikawa T, Niki T, Kadowaki T, Ueno M, Nishi N, Yamauchi A, Hattori T, Masaki T, Hirashima M. Cell surface galectin-9 expressing Th cells regulate Th17 and Foxp3+ Treg development by galectin-9 secretion. PloS One 2012; 7:e48574; PMID:23144904; http://dx.doi.org/10.1371/journal.pone.0048574
- Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 2012; 482:414-8; PMID:22246324; http://dx.doi.org/10.1038/nature10744
- Kolundzic N, Bojic-Trbojevic Z, Radojcic L, Petronijevic M, Vicovac L. Galectin-8 is expressed by villous and extravillous trophoblast of the human placenta. Placenta 2011; 32:909-11; PMID:21862124; http://dx.doi.org/10.1016/j.placenta.2011.07.087
- Ramos S, Khademi F, Somesh BP, Rivero F. Genomic organization and expression profile of the small GTPases of the RhoBTB family in human and mouse. Gene 2002; 298:147-57; PMID:12426103; http://dx.doi.org/10.1016/S0378-1119(02)00980-0
- Babak T, Deveale B, Armour C, Raymond C, Cleary MA, van der Kooy D, Johnson JM, Lim LP. Global survey of genomic imprinting by transcriptome sequencing. Curr Biol 2008; 18:1735-41; PMID:19026546; http://dx.doi.org/10.1016/j.cub.2008.09.044
- Degner JF, Marioni JC, Pai AA, Pickrell JK, Nkadori E, Gilad Y, Pritchard JK. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 2009; 25:3207-12; PMID:19808877; http://dx.doi.org/10.1093/bioinformatics/btp579
- Rozowsky J, Abyzov A, Wang J, Alves P, Raha D, Harmanci A, Leng J, Bjornson R, Kong Y, Kitabayashi N, et al. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol Syst Biol 2011; 7:522; PMID:21811232; http://dx.doi.org/10.1038/msb.2011.54
- Karro H, Rahu M, Gornoi K, Baburin A. Sünnikaalu jaotumine raseduse kestuse järgi Eestis aastail 1992–1994. Eesti Arst 1997; 4:299-303
- Lahiri DK, Nurnberger JI, Jr. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res 1991; 19:5444; PMID:1681511; http://dx.doi.org/10.1093/nar/19.19.5444
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559-75; PMID:17701901; http://dx.doi.org/10.1086/519795
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11:R106; PMID:20979621; http://dx.doi.org/10.1186/gb-2010-11-10-r106
- David M, Dzamba M, Lister D, Ilie L, Brudno M. SHRiMP2: sensitive yet practical SHort Read Mapping. Bioinformatics 2011; 27:1011-2; PMID:21278192; http://dx.doi.org/10.1093/bioinformatics/btr046
- Kin T, Ono Y. Idiographica: a general-purpose web application to build idiograms on-demand for human, mouse and rat. Bioinformatics 2007; 23:2945-6; PMID:17893084; http://dx.doi.org/10.1093/bioinformatics/btm455
- Lambertini L, Diplas AI, Lee MJ, Sperling R, Chen J, Wetmur J. A sensitive functional assay reveals frequent loss of genomic imprinting in human placenta. Epigenetics 2008; 3:261-9; PMID:18769151; http://dx.doi.org/10.4161/epi.3.5.6755
- Pozharny Y, Lambertini L, Ma Y, Ferrara L, Litton CG, Diplas A, Jacobs AR, Chen J, Stone JL, Wetmur J, et al. Genomic loss of imprinting in first-trimester human placenta. Am J Obstet Gynecol 2010; 202:391 e1-8; PMID:20350649; http://dx.doi.org/10.1016/j.ajog.2010.01.039
- Lim AL, Ng S, Leow SC, Choo R, Ito M, Chan YH, Goh SK, Tng E, Kwek K, Chong YS, et al. Epigenetic state and expression of imprinted genes in umbilical cord correlates with growth parameters in human pregnancy. J Med Genet 2012; 49:689-97; PMID:23042810; http://dx.doi.org/10.1136/jmedgenet-2012-100858
- Liu Y, Hoyo C, Murphy S, Huang Z, Overcash F, Thompson J, Brown H, Murtha AP. DNA methylation at imprint regulatory regions in preterm birth and infection. Am J Obstet Gynecol 2013; 208:395 e1-7; http://dx.doi.org/10.1016/j.ajog.2013.02.006
- Okabe H, Satoh S, Furukawa Y, Kato T, Hasegawa S, Nakajima Y, Yamaoka Y, Nakamura Y. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res 2003; 63:3043-8; PMID:12810624
- Graf J, Hodgson R, van Daal A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutat 2005; 25:278-84; PMID:15714523; http://dx.doi.org/10.1002/humu.20143
- Newton JM, Cohen-Barak O, Hagiwara N, Gardner JM, Davisson MT, King RA, Brilliant MH. Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am J Hum Genet 2001; 69:981-8; PMID:11574907; http://dx.doi.org/10.1086/324340
- Hoshimoto S, Kuo CT, Chong KK, Takeshima TL, Takei Y, Li MW, Huang SK, Sim MS, Morton DL, Hoon DS. AIM1 and LINE-1 epigenetic aberrations in tumor and serum relate to melanoma progression and disease outcome. J Invest Dermatol 2012; 132:1689-97; PMID:22402438; http://dx.doi.org/10.1038/jid.2012.36
- Vainio P, Mpindi JP, Kohonen P, Fey V, Mirtti T, Alanen KA, Perala M, Kallioniemi O, Iljin K. High-throughput transcriptomic and RNAi analysis identifies AIM1, ERGIC1, TMED3 and TPX2 as potential drug targets in prostate cancer. PloS One 2012; 7:e39801; PMID:22761906; http://dx.doi.org/10.1371/journal.pone.0039801
- Saida T. Recent advances in melanoma research. J Dermatol Sci 2001; 26:1-13; PMID:11323215; http://dx.doi.org/10.1016/S0923-1811(01)00085-8
- Lamy L, Ngo VN, Emre NC, Shaffer AL 3rd, Yang Y, Tian E, Nair V, Kruhlak MJ, Zingone A, Landgren O, et al. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell 2013; 23:435-49; PMID:23541952; http://dx.doi.org/10.1016/j.ccr.2013.02.017
- Dyer MJ. The pathogenetic role of oncogenes deregulated by chromosomal translocation in B-cell malignancies. Int J Hematol 2003; 77:315-20; PMID:12774917; http://dx.doi.org/10.1007/BF02982637
- Kelly JL, Novak AJ, Fredericksen ZS, Liebow M, Ansell SM, Dogan A, Wang AH, Witzig TE, Call TG, Kay NE, et al. Germline variation in apoptosis pathway genes and risk of non-Hodgkin's lymphoma. Cancer Epidemiol, Biomar Prevent: A Pub Am Assoc Cancer Res, Cosponsored Am Soc Prev Oncol 2010; 19:2847-58; http://dx.doi.org/10.1158/1055-9965.EPI-10-0581
- McPherson JP, Sarras H, Lemmers B, Tamblyn L, Migon E, Matysiak-Zablocki E, Hakem A, Azami SA, Cardoso R, Fish J, et al. Essential role for Bclaf1 in lung development and immune system function. Cell Death Differ 2009; 16:331-9; PMID:19008920; http://dx.doi.org/10.1038/cdd.2008.167
- Liang X, Zhou D, Wei C, Luo H, Liu J, Fu R, Cui S. MicroRNA-34c enhances murine male germ cell apoptosis through targeting ATF1. PloS One 2012; 7:e33861; PMID:22479460; http://dx.doi.org/10.1371/journal.pone.0033861
- Faucher G, Guenard F, Bouchard L, Garneau V, Turcot V, Houde A, Tchernof A, Bergeron J, Deshaies Y, Hould FS, et al. Genetic contribution to C-reactive protein levels in severe obesity. Mol Genet Metab 2012; 105:494-501; PMID:22178353; http://dx.doi.org/10.1016/j.ymgme.2011.11.198
- Turcot V, Bouchard L, Faucher G, Tchernof A, Deshaies Y, Perusse L, Marceau P, Hould FS, Lebel S, Vohl MC. A polymorphism of the interferon-gamma-inducible protein 30 gene is associated with hyperglycemia in severely obese individuals. Hum Genet 2012; 131:57-66; PMID:21701784; http://dx.doi.org/10.1007/s00439-011-1043-4
- Lingueglia E, Renard S, Voilley N, Waldmann R, Chassande O, Lazdunski M, Barbry P. Molecular cloning and functional expression of different molecular forms of rat amiloride-binding proteins. Eur J Biochem FEBS 1993; 216:679-87; http://dx.doi.org/10.1111/j.1432-1033.1993.tb18188.x
- Song WB, Lv YH, Zhang ZS, Li YN, Xiao LP, Yu XP, Wang YY, Ji HL, Ma L. Soluble intercellular adhesion molecule-1, D-lactate and diamine oxidase in patients with inflammatory bowel disease. World J Gastroenterol: WJG 2009; 15:3916-9; PMID:19701972; http://dx.doi.org/10.3748/wjg.15.3916
- Hornemann T, Richard S, Rutti MF, Wei Y, von Eckardstein A. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J Biol Chem 2006; 281:37275-81; PMID:17023427; http://dx.doi.org/10.1074/jbc.M608066200
- Hicks AA, Pramstaller PP, Johansson A, Vitart V, Rudan I, Ugocsai P, Aulchenko Y, Franklin CS, Liebisch G, Erdmann J, et al. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet 2009; 5:e1000672; PMID:19798445; http://dx.doi.org/10.1371/journal.pgen.1000672
- Berthold J, Schenkova K, Rivero F. Rho GTPases of the RhoBTB subfamily and tumorigenesis. Acta Pharmacologica Sinica 2008; 29:285-95; PMID:18298893; http://dx.doi.org/10.1111/j.1745-7254.2008.00773.x
- Roberts AJ, Hedlund PB. The 5-HT(7) receptor in learning and memory. Hippocampus 2012; 22:762-71; PMID:21484935; http://dx.doi.org/10.1002/hipo.20938
- Salas-Vidal E, Meijer AH, Cheng X, Spaink HP. Genomic annotation and expression analysis of the zebrafish Rho small GTPase family during development and bacterial infection. Genomics 2005; 86:25-37; PMID:15894457; http://dx.doi.org/10.1016/j.ygeno.2005.03.010
