Abstract
The JmjC-domain-containing 2-oxoglutarate-dependent oxygenases catalyze protein hydroxylation and Nϵ-methyllysine demethylation via hydroxylation. A subgroup of this family, the JmjC lysine demethylases (JmjC KDMs) are involved in histone modifications at multiple sites. There are conflicting reports as to the substrate selectivity of some JmjC oxygenases with respect to KDM activities. In this study, a panel of modified histone H3 peptides was tested for demethylation against 15 human JmjC-domain-containing proteins. The results largely confirmed known Nϵ-methyllysine substrates. However, the purified KDM4 catalytic domains showed greater substrate promiscuity than previously reported (i.e., KDM4A was observed to catalyze demethylation at H3K27 as well as H3K9/K36). Crystallographic analyses revealed that the Nϵ-methyllysine of an H3K27me3 peptide binds similarly to Nϵ-methyllysines of H3K9me3/H3K36me3 with KDM4A. A subgroup of JmjC proteins known to catalyze hydroxylation did not display demethylation activity. Overall, the results reveal that the catalytic domains of the KDM4 enzymes may be less selective than previously identified. They also draw a distinction between the Nϵ-methyllysine demethylation and hydroxylation activities within the JmjC subfamily. These results will be of use to those working on functional studies of the JmjC enzymes.
Abbreviations and acronyms
| 2OG | = | 2-oxoglutarate |
| FIH | = | Factor Inhibiting HIF |
| H3 | = | histone 3 |
| HIF | = | Hypoxia Inducible Factor |
| JmjC | = | Jumonji C-terminal |
| JmjN | = | Jumonji N-terminal |
| KDM | = | Lysine Demethylase |
| LSD | = | Lysine Specific Demethylase |
| MALDI-TOF MS | = | Matrix Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry |
| MINA53 | = | Myc-Induced Nuclear Antigen with a molecular mass of 53 kDa |
| NO66 | = | Nucleolar protein 66 |
| PHD | = | Plant Homeodomain |
| Rp | = | Ribosomal protein |
| TPR | = | Tetratricopeptide repeat |
Introduction
Post-oligomerization modifications to the nucleic acid and protein components of chromatin play central roles in transcriptional regulation. While DNA methylation is associated with gene silencing, methylation of histone proteins can be activating or repressive depending on the context of the modification.Citation1 The introduced methyl groups can undergo further modification, often by oxidation; the largest identified class of such enzymes are the Fe(II)- and 2-oxoglutarate- (2OG) dependent oxygenases, which catalyze oxidation of 5-methylcytosine bases, and of N-methyl groups in DNA (and RNA) and histone proteins.Citation2
Two families of Nϵ-methyllysine demethylases (KDMs) have been identified: the Fe(II)- and 2OG-dependent JmjC KDMs () and the flavin-dependent lysine specific demethylases (KDM1s or LSDs).Citation3 JmjC KDMs act on all 3 Nϵ-methyllysine methylation states (), whereas the KDM1s can only act on di- and mono-methylation states. The JmjC KDMs are part of the wider JmjC subfamily of 2OG oxygenases, which also catalyze protein hydroxylation not involving demethylation.Citation4 For several JmjC enzymes, (JMJD6, MINA53 and NO66), there are reports of both demethylation and hydroxylation activities.Citation4-9 Substrate promiscuity is well-established in 2OG oxygenase biochemistry, including for the JmjC hydroxylase factor inhibiting hypoxia inducible factor (FIH), which catalyzes hydroxylation of an asparaginyl residue in Hypoxia Inducible Factor α (HIF-α) as well as multiple asparagine-, aspartate- and histidine-residues in ankyrin repeat-domain proteins.Citation10-13 Given the different reports of the reactions catalyzed by some JmjC oxygenases, we considered that it would be useful to carry out a systematic study on the biochemical selectivity of the catalytic domains of human JmjC KDMs. We appreciate that other factors, for example, ancillary non-catalytic domains, targeting proteins, protein conformation, co-substrate and inhibitor availability, substrate length (peptide/protein), and cellular compartmentalization, may alter hydroxylation and demethylation catalysis and substrate selectivities in a cellular context. However, these results provide an in vitro comparative substrate profiling of JmjC oxygenases that will be useful to other researchers in the field.
Figure 1. JmjC Oxygenases share sequence homology and catalytic mechanisms. (A) Phylogenetic analysis of the catalytic domains of human JmjC oxygenases. Reported catalytic functions are indicated by colored circles. MINA53, NO66 and JMJD6 have been reported to be demethylases but have subsequently been shown to have hydroxylase activities.Citation4-6,8,9 KDM6C (UTY) was recently identified as a histone demethylase in vitro, acting on H3 peptide fragments methylated at H3K27.Citation43 HR is Hairless Protein, a recently identified H3K9 demethylase.Citation44 Enzymes used in this work are underlined. (B) Outline mechanism of JmjC oxygenase catalysis. Oxidative decarboxylation of 2-oxoglutarate (2OG) in the active site forms a highly reactive iron(IV)-oxo intermediate, which hydroxylates the substrate. In the case of demethylation (X = N), the hydroxylated product is unstable and fragments to produce the demethylated species and formaldehyde. The exact protonation states of water molecules complexed to the iron(II) species are unknown.
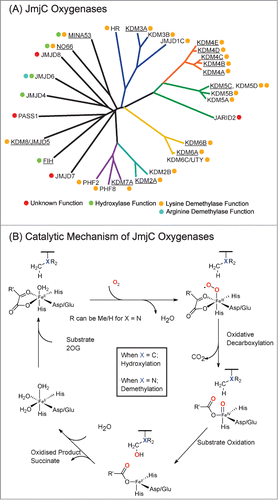
In general, the results with JmjC KDMs support the reported selectivities,Citation14-20 as well as current structural studies,Citation21 which show that JmjC KDMs and hydroxylases are different subfamilies within the 2OG oxygenase family, with little overlap in hydroxylation and demethylation activities. However, we observed that the KDM4 subfamily catalytic domains could not only catalyze demethylation of H3 peptides methylated at K9 and K36, but also at K27, implying that the selectivities of the JmjC KDMs may extend beyond the already characterized substrates.
Results
To compare the selectivities of the catalytic domains of the JmjC oxygenases we screened a set of H3 peptides (either from Alta Bioscience, or synthesized in house) with the isolated catalytic domains of 15 JmjC oxygenases using MALDI-TOF Mass Spectrometry (MS, ). The enzymes were chosen to represent different subfamilies of JmjC oxygenases () and based on the availability of active recombinant proteins.Citation22 The H3 fragment peptides screened were 21 residues in length and included mono-, di-, and tri-methyllysines at K4, K9, K27, and K36 sites (Table S1). Standard conditions of 1 μM enzyme, 200 μM 2OG, 10 μM Fe(II) and 100 μM ascorbate were used throughout (i.e., large excesses of cofactors and substrates),Citation23 but the assay buffer conditions were optimized to enhance activity and MS signal, as well as to reduce protein precipitation (Table S2).
Table 1. Summary of initial activity screening of JmjC oxygenases and methylated histone H3 fragment peptides. KDM activity (i.e., greater than 10% substrate demethylation as detected by MALDI mass spectrometry) was observed for all tested members of the KDM enzymes (ticks), including at reported methylation sites (green boxes) and, in the case of the KDM4 enzymes, at novel methylation sites (yellow boxes). No demethylation activity within limits of detection was observed for the putative demethylases MINA53, NO66 or JMJD5 (crosses, red boxes; note for JMJD5 construct-dependent demethylation activity is reported by Hsia et al (dashed red box) 7), while hydroxylation of reported non-histone substrates were observed for MINA53 and NO66 For JMJD5, prime substrate uncoupled 2OG turnover was observed indicating active construct (ticks, green boxes). In some cases, demethylation of the me2/1 state (to the me1/0 state respectively) was only observed when the me2/1 peptide was produced as an intermediate during demethylation of the me3/2 state (dashed green/yellow boxes)
The results with the catalytic domains of KDM2A, KDM3A, KDM5C, KDM6A, and KDM6B support literature assignments.Citation15,16,18,19,24-28 Thus, under standard conditions, and as summarized in , (i) KDM2A only accepted H3K36me2 and H3K36me1 (Figure S1), (ii) KDM3A accepted H3K9me2 and H3K9me1 (Figure S2), (iii) KDM5C accepted H3K4me3, H3K4me2, and H3K4me1 (Figure S3), and (iv) KDM6B acted on H3K27me3, H3K27me2 and H3K27me1, while KDM6A acted (under our conditions) on H3K27me3 and H3K27me2, but not H3K27me1 (Figure S4). The latter result is consistent with previous reports which demonstrate that the tetratricopeptide repeat (TPR) domain of KDM6A is required for effective H3K27me1 demethylation, possibly by promoting enhanced substrate binding.Citation16 KDM7A is reported to catalyze the demethylation of di- and mono-methylated forms of both H3K9 and H3K27 in the absence of substrate methylation at H3K4 (H3K4me3), but only at H3K27 when H3K4 is trimethylated (i.e., H3K4 methylation promotes demethylation at H3K27).Citation29,30 We observed the same substrate selectivity for peptides not methylated at H3K4 with our construct of KDM7A, which contains both the JmjC and PHD domains (Figure S5, S6).
In the case of the tested JmjC oxygenases assigned as hydroxylases (FIH and the recently reassigned NO66 and MINA53),Citation4,12,13 no KDM activity was observed with any of the tested H3 peptides under our assay conditions (, D, E, S7, S8). This is of note as NO66 and MINA53 were initially characterized as histone demethylases.Citation8,9 Recent work, however, has shown MINA53 and NO66 to be histidyl-hydroxylases acting on the ribosomal proteins Rpl27a and Rpl8, respectively.Citation31 In positive controls, we found these reported non-histone peptide substrates to be hydroxylated by MINA53 and NO66 (). JMJD5 has also been reported to act as an H3K36me2 demethylase;Citation7 however, under our assay conditions, JMJD5-catalyzed demethylation of H3K36me2 peptide was not observed (, S10).
Figure 2. JmjC Oxygenases MINA53, NO66 and JMJD5 do not catalyze demethylation of histone peptides. In addition to putative demethylation activities, MINA53 and NO66 have been characterized as hydroxylases acting on ribosomal proteins Rpl27a and Rpl8 respectively. Hydroxylation activities were observed for MINA53 and NO66, acting on Rpl27a and Rpl8 peptide fragments respectively (A and C); no demethylation was observed with methylated histone peptides (B, D and E). Prime-substrate uncoupled turnover of 2OG by JMJD5 (residues 1–416) was observed in a [14C]-labeled 2OG assay, which was dependent on the presence of iron(II) and inhibited by the broad-spectrum 2OG oxygenase inhibitor 2,4-pyridinedicarboxylic acid (2,4 PDA) (F). However, demethylation of an H3K36me2 histone peptide was not observed (G). Control reactions without added protein are in red.
![Figure 2. JmjC Oxygenases MINA53, NO66 and JMJD5 do not catalyze demethylation of histone peptides. In addition to putative demethylation activities, MINA53 and NO66 have been characterized as hydroxylases acting on ribosomal proteins Rpl27a and Rpl8 respectively. Hydroxylation activities were observed for MINA53 and NO66, acting on Rpl27a and Rpl8 peptide fragments respectively (A and C); no demethylation was observed with methylated histone peptides (B, D and E). Prime-substrate uncoupled turnover of 2OG by JMJD5 (residues 1–416) was observed in a [14C]-labeled 2OG assay, which was dependent on the presence of iron(II) and inhibited by the broad-spectrum 2OG oxygenase inhibitor 2,4-pyridinedicarboxylic acid (2,4 PDA) (F). However, demethylation of an H3K36me2 histone peptide was not observed (G). Control reactions without added protein are in red.](/cms/asset/48731be3-2c8b-4c06-a001-aa50e7c6781b/kepi_a_983381_f0002_c.gif)
In the case of the KDM4 subfamily, initial screening results with KDM4A suggested that they may be more promiscuous than reported (, , Figure S11);Citation14 hence, we analyzed all 4 human KDM4 subfamily members (KDM4A-D) and the human pseudogene KDM4E (, Figures S11–15). As reported, all the KDM4 members acted on H3K9me3 and H3K9me2, although we only observed clear demethylation of H3K9me1 to the H3K9me0 state by KDM4E (although not apparent under the standard conditions, prolonged incubations with KDM4 enzymes leads to demethylation of H3K9me1) (Figure S15).Citation14 Also, as reported, KDM4A-C catalyzed demethylation of H3K36me3/H3K36me2 but KDM4D/E did not (Figure S14–S15).Citation14 Surprisingly, most KDM4 catalytic domains also acted on H3K27me3 and in one cases H3K27me2 (Figures S11–15).
Figure 3. KDM4A catalyzes demethylation of histone fragment peptides methylated at H3K27. MALDI-TOF spectra of KDM4A (1–359) catalyzed demethylation of (A) H3K27me3 peptide (Biotin-Ahx(aminohexanoic acid)-KAPRKQLATKAARKme3SAPATGG), (B) H3K9me3 peptide (Biotin-Ahx-ARTKQTARKme3STGGKAPRKQLA). MALDI-TOF spectra of full-length KDM4A (1–1064) catalyzed demethylation of (C) H3K27me3 peptide (KAPRKQLATKAARKme3SAPATGG) and (D) H3K9me3 peptide (Biotin-Ahx-ARTKQTARKme3STGGKAPRKQLA). FLAG-tagged full-length KDM4A (1–1064) was produced in HEK293T cells and purified from cell lysates using anti-FLAG beads prior to reaction with the histone peptide. MALDI-TOF spectra of reactions with (standard conditions, black) and without added enzyme (red) for (A)–(D) are shown. (E) Kinetic parameters determined for KDM4A (1–359) catalyzed demethylation of Nϵ-trimethylated H3K9, H3K27 and H3K36 fragment peptides using FDH assay (Figure S16).
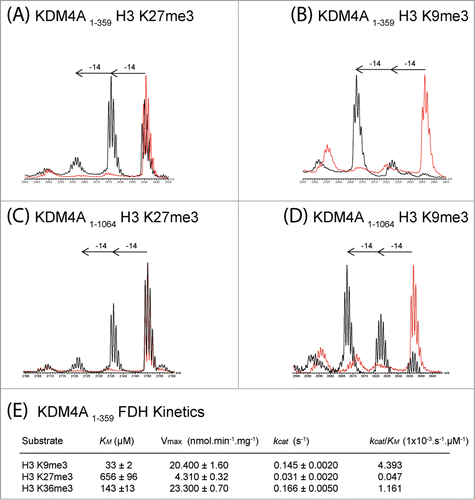
To investigate the unexpected results with respect to KDM4 selectivity, kinetic analyses were then undertaken using the KDM4A catalytic domain (, S16). The H3K9me3 and H3K36me3 modified peptides had similar kcat values (kcat = 0.145 s−1 and 0.166 s−1, respectively), but H3K9me3 manifested the highest kcat/Km (0.004393 s−1.μM−1 for H3K9me3 and 0.001161 s−1.μM−1 for H3K36me3), consistent with previous reports.Citation32,33 The H3K27me3 peptide was a poorer substrate than both the H3K9me3 and H3K36me3 peptides but still displayed substantial activity (kcat = 0.031 s−1). As anticipated, the Km value was lowest for H3K9me3 (Km = 33 μM) followed by H3K36me3 (Km = 143 μM). Competition experiments, monitoring demethylation of 2 equimolar peptides simultaneously, support the reduced demethylation efficiency of KDM4A toward H3K27me3 relative to H3K9me3/H3K36me3 (Figure S17). The majority of H3K9me3 and H3K36me3 peptides were demethylated to the monomethyllysine state before demethylation of H3K27me3 was observed.
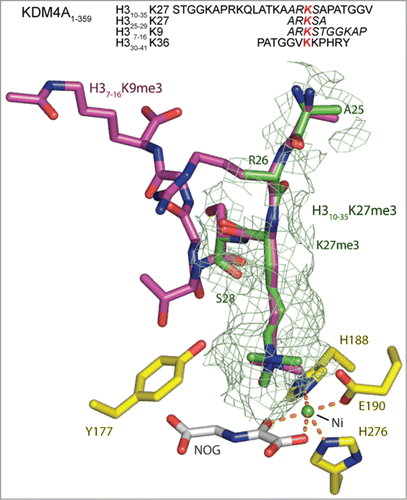
To investigate the mode of KDM4A binding to the H3K27me3 substrate, we solved crystal structures of the KDM4A catalytic domain complexed with both 25 residue and 5 residue H3K27me3 peptides (, S18, S19). In both cases, the H3K27me3 residue was observed to bind in a near identical manner to both H3K9me3 and H3K36me3 residues in H3 peptides, supporting the proposed activity at H3K27me3 (, S18). Notably, in the longer 25 residue H3K27me3 peptide complex, density was observed for the trimethyllysine and the 2 adjacent peptide residues (H3R26 and H3S28 respectively), but not for the other residues, while density was observed for all residues of the shorter 5 residue H3K27me3 peptide (Figure S19). The lack of observed density for the longer peptide may reflect relatively weaker binding relative of the H3K27me3 compared to the H3K9me3 and H3K36me3 peptides, consistent with the kinetic results (, S16).
Figure 4. View from an X-ray crystal structure of the catalytic domain of KDM4A in complex with an H3K27me3 fragment peptide (PDB ID: 4V2W). The active site residues are highlighted in yellow. The visible residues of the 25mer H310–35K27me3 (green) peptide is shown overlaid with H3K9me3 (pink), as complexed with KDM4A (PDB ID: 2OQ6, nickel substituted for iron, and N-oxalylglycine substituted for 2OG). The position of the H3K27me3 residue of the fragment peptide correlates closely with that of H3K9me3 (and H3K36me3, Figure S18). However, in the H310–25K27me3 derived structure only the electron density for the tri-methyl lysine and the residues either side (H3R26 and H3S28) are clearly defined, suggesting the other residues are bound less tightly than the comparable H3K9 and H3K36 substrates. A second structure of a shorter 5 residue H325–29K27me3 peptide in complex with KDM4A (PDB ID: 4V2V) overlays well with that of the 25 residue peptide (Figure S19). The sequences for the H3K9, K27 and K36 peptides present in these crystal structures are included with residues for which electron density is observed in italics. The red lysine residue marks the position of the Nϵ-trimethylated lysine.
To investigate whether KDM4A promiscuity is a result of using truncated protein (i.e., constructs containing only the catalytic domain), we used immunoprecipitated FLAG-tagged full-length recombinant KDM4A (which contains JmjN, JmjC, double PHD and tandem tudor domains, Figure S5) exogenously over-expressed in HEK 293T cells and tested for demethylation activity. The full-length KDM4A catalyzed demethylation of H3K9me3, K36me3 and K27me3 ().
Discussion
Overall, the results support most of the reported substrate selectivities for the catalytic domains of the JmjC KDMs.Citation15,16,18,19,24-29 It is important to note that in cases of apparent discrepancies with the literature, these may reflect the use of different constructs and/or conditions and, in some cases, may arise from the omission of non-catalytic domains that would otherwise alter substrate selectivity (e.g., a TPR domain in KDM6A increases activity toward H3K27me1).Citation29 Nonetheless, given that in some cases conflicting reports already exist in the literature, we hope that our results will provoke further substrate profiling work for KDMs in vitro and in cells. Within the subset of 6 ‘well-characterized’ subfamilies of JmjC KDMs, our results were supportive of the literature, except for the KDM4 subfamily. In the case of this subfamily, where we studied the catalytic domains of 5 KDM4s, we observed clear evidence for demethylation of H3K27me3/2 peptides. The full-length KDM4A was also able to catalyze demethylation at H3K27me3/me2. Interestingly, weak demethylase activity toward H3K27me3 was previously reported for recombinant mouse KDM4s.Citation34 We emphasize that, based on our studies alone, we cannot propose that the observed KDM activity against H3K27me3/2 is of ‘biological relevance’. Nonetheless, given the promiscuity of some other 2OG oxygenases acting on proteins (e.g., FIH),Citation10,11,35 there is clear potential for KDM4 subfamily members to act on H3K27me3/2 or indeed other presently unidentified substrates.
We did not observe any KDM activity with the JmjC oxygenases for which we have observed hydroxylase activity (i.e., the JmjC enzymes FIH, MINA53 and NO66) or JMJD5. From a physiological/evolutionary perspective, this is unsurprising because they belong to a different subfamily from the canonical JmjC KDMs ().Citation21 However, we do not rule out the possibility that the JmjC hydroxylases can act as KDMs; as indicated above, some 2OG oxygenases can be very promiscuous with regard to both their in vitro and in vivo substrates. As we have found with FIH, results with isolated proteins do not always predict cellular substrate selectivities and vice versa.
Materials and methods
Protein production Catalytic domains of KDM2A,Citation36 KDM4A-E,Citation33,37,38 KDM6B,Citation39 KDM7A,Citation39 NO66,Citation31 MINA53Citation31, and FIHCitation40 were produced in, and purified from E. coli as previously described. KDM3A and KDM5C were expressed and purified from Sf9 cells as described.Citation39 JMJD5(1–416) in pET28a(+) (Novagen) was transformed into Rosetta DE3 E. coli and grown in 2 × TY media. Cells were induced at mid log phase (OD600 = 0.6) with 0.5 mM IPTG and grown for a further 18 hours at 25°C and harvested by centrifugation. The pellet was resuspended in 50 mM HEPES pH 7.5, 500 mM NaCl, 5 mM imidazole and 5% glycerol with an EDTA-free protease inhibitor tablet (Roche). Cells were lysed by sonication and the lysate clarified by high speed centrifugation. Lysate was purified by application to a HisTrap™ HP 5 mL column (GEHealthcare). The Ni-Sepharose column was washed with 20 column volumes of 50 mM HEPES pH 7.5, 500 mM NaCl, 20 mM imidazole and 5% glycerol, eluting with a gradient up to 250 mM imidazole over 10 column volumes. Fractions containing JMJD5 were concentrated and applied to a 300 mL Superdex 75 prep grade column pre-equilibrated in 50 mM HEPES pH 7.5, 500 mM NaCl, 1 mM DTT and 5% glycerol. Enzyme activity was assessed by 2OG turnover assay (Fig. S13).
2OG turnover radioactivity assay Recombinant JMJD5 was tested for ability to decarboxylate 1-[14C]-labeled 2OG, as described for other 2OG oxygenases.Citation13 Reactions (100 μL) contained 4 mM sodium ascorbate, 0.288 mM 2OG, 0.0036 mM [14C]-2OG, 0.005 mM Fe(II), 1mM DTT, 2% catalase, 40 μM JMJD5, (10 μM inhibitor) in 50 mM HEPES pH 7.5. [14C]-CO2 radioactivity was recorded using a Beckman LS4500 liquid scintillation counter. Data shown () are mean values from one assay performed in triplicate. Error bars give standard error of mean.
Peptide Synthesis 21-mer H3 fragment peptides trimethylated at positions K4, K9, K27 and K36 were synthesized via fluorenylmethyloxycarbonyl (Fmoc)-mediated solid phase peptide synthesis (on MBHA resin) using a CS Bio 336X peptide synthesizer. Peptides were cleaved from the resin using 97.5% trifluoroacetic acid / 2.5% triisopropylsilane solution (3 hr incubation) before precipitation with cold diethyl ether. Peptides were purified by reverse-phase high-performance liquid chromatography using a Vydac C18 column (Solvent A = 0.1% trifluoroacetic acid in H2O, Solvent B = 0.1% trifluoroacetic acid in acetonitrile) to >95% purity as determined by MS.
MALDI-TOF activity assays Demethylation activities on peptides were measured by Matrix-assisted laser-desorption/ionization (MALDI) time-of-flight (TOF) mass spectrometry (MS), using a Waters Micromass MALDI micro MXTM mass spectrometer in negative ion, reflectron mode. Reaction conditions varied between enzymes as optimized for each reaction (Table S2). Concentrations of 2OG used were maintained above the Km values for each enzyme. All reactions used 1 μM enzyme, 10 μM peptide, 2-oxoglutarate (2OG), FeSO4.7H2O and sodium ascorbate. Reactions were incubated for 60–120 min at 37°C, and were quenched with 1:2 MeOH by volume. An aliquot of the quenched reaction (1 μL) was mixed with 1 μL α-cyano-4-hydroxycinnamic acid (CHCA) matrix (Sigma) and spotted on a MALDI-TOF plate for analysis.
MALDI competition assays Reaction conditions for competition assays were as for MALDI-TOF activity assays, but included 2 histone peptides (10 μM each). Reactions were initiated by mixing the samples before incubation at room temperature. At each time point, 5 μL of the reaction mixture was removed, quenched with a 1:2 ratio of sample to MeOH and snap frozen in liquid nitrogen. The zero time point was quenched prior to addition of enzyme.
Formaldehyde Dehydrogenase (FDH) Assays Production of formaldehyde as a by-product of demethylation was observed using the FDH/NAD+ coupled assay.Citation41 Assays contained buffer (50 mM HEPES pH 7.5, 0.01% Tween20), Fe(II) ammonium sulfate (10 μM), sodium ascorbate (100 μM), 2OG (200 μM), NAD+ (500 μM), enzyme (0.2–2 μM) with FDH enzyme (0.025 U, Sigma) per assay (total volume 25 μL) and were conducted at room temperature in clear-bottom black 384-well plate (Greiner). Reactions were monitored over 30 min using a PHERAstar FS (BMG Labtech) plate reader with 355 nm excitation and 460 nm emission. The Km and Vmax values were calculated from the reaction rate during the linear phase of formaldehyde production, as determined by the slope of the fluorescence signal recorded.
Cell culture HEK 293T cells were cultured at 37°C with 5% CO2 in DMEM (Sigma) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% Glutamax (Invitrogen). Full length KDM4A with FLAGx3 tag in pCDNA3 (kindly provided by Yi ZhangCitation42) was transiently transfected into HEK293T using Fugene (50 μg DNA, 90 uL PEI per 15 cm diameter plate, 80% confluency, PAA Laboratories) and harvested after 24 hr. Cells were washed in PBS, lysed in 50 mM HEPES, 150 mM NaCl, 0.01% NP40, 1% protease inhibitor cocktail (SIGMA), DNAse (10 μg/ml) and incubated with 60 μL FLAG-Magnetic Beads (SIGMA) for 4 hr at 4°C. Beads were washed 4 times with lysis buffer (600 μL), 2 times with 50 mM HEPES (300 μL) to remove NP40 and NaCl before MS analysis. Beads were re-suspended in an initial volume of 60 μL for use in further assays. Demethylation reactions were conducted with 1 μL of the immobilized anti-FLAG beads with peptide (10 μM), 2OG (200 μM), FeSO4.7H2O (10 μM), sodium ascorbate (100 μM). Reactions were incubated for 120 min at 37°C and quenched with 1:2 MeOH by volume. The demethylation products were observed by MALDI.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
SI.pdf
Download PDF (3.5 MB)Acknowledgments
We thank the Structural Genomics Consortium for providing KDM3A and KDM5C proteins, Dr Adam Hardy for FIH protein, Dr Wei Ge for MINA53 and NO66 proteins, and Tristan Smart for KDM2A protein. We thank Dr. Domen Zafred for assisting with the data collection for PDB ID: 4V2W.
Funding
This work was supported by the Wellcome Trust (091857/Z/10/Z), the Engineering and Physical Sciences Research Council (EP/L003376/1), the BHF Center of Research Excellence Oxford (RE/13/1/30181) and Cancer Research UK (C8717/A18245). RJH acknowledges a William R. Miller Junior Research Fellowship, St. Edmund Hall, Oxford. AK is supported by a Royal Society Dorothy Hodgkin Fellowship (DH120028).
References
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13:343-57; PMID:22473383; http://dx.doi.org/10.1038/nrg3173
- Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol 2008; 4:152-6; PMID:18277970; http://dx.doi.org/10.1038/nchembio0308-152
- Walport LJ, Hopkinson RJ, Schofield CJ. Mechanisms of human histone and nucleic acid demethylases. Curr Opin Chem Biol 2012; 16:525-34; PMID:23063108; http://dx.doi.org/10.1016/j.cbpa.2012.09.015
- Ge W, Wolf A, Feng T, Ho CH, Sekirnik R, Zayer A, Granatino N, Cockman ME, Loenarz C, Loik ND, et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat Chem Biol 2012; 8:960-2; PMID:23103944; http://dx.doi.org/10.1038/nchembio.1093
- Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science 2007; 318:444-7; PMID:17947579; http://dx.doi.org/10.1126/science.1145801
- Webby CJ, Wolf A, Gromak N, Dreger M, Kramer H, Kessler B, Nielsen ML, Schmitz C, Butler DS, Yates JR, 3rd, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 2009; 325:90-3; PMID:19574390; http://dx.doi.org/10.1126/science.1175865
- Hsia DA, Tepper CG, Pochampalli MR, Hsia EY, Izumiya C, Huerta SB, Wright ME, Chen HW, Kung HJ, Izumiya Y. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc Natl Acad Sci US A 2010; 107:9671-6; PMID:20457893; http://dx.doi.org/10.1073/pnas.1000401107
- Sinha KM, Yasuda H, Coombes MM, Dent SY, de Crombrugghe B. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J 2010; 29:68-79; PMID:19927124; http://dx.doi.org/10.1038/emboj.2009.332
- Lu Y, Chang Q, Zhang Y, Beezhold K, Rojanasakul Y, Zhao H, Castranova V, Shi X, Chen F. Lung cancer-associated JmjC domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3. Cell cycle 2009; 8:2101-9; PMID:19502796; http://dx.doi.org/10.4161/cc.8.13.8927
- Yang M, Chowdhury R, Ge W, Hamed RB, McDonough MA, Claridge TD, Kessler BM, Cockman ME, Ratcliffe PJ, Schofield CJ. Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the post-translational hydroxylation of histidinyl residues within ankyrin repeat domains. FEBS J 2011; 278:1086-97; PMID:21251231; http://dx.doi.org/10.1111/j.1742-4658.2011.08022.x
- Yang M, Ge W, Chowdhury R, Claridge TD, Kramer HB, Schmierer B, McDonough MA, Gong L, Kessler BM, Ratcliffe PJ, et al. Asparagine and aspartate hydroxylation of the cytoskeletal ankyrin family is catalyzed by factor-inhibiting hypoxia-inducible factor. J Biol Chem 2011; 286:7648-60; PMID:21177872; http://dx.doi.org/10.1074/jbc.M110.193540
- Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev 2002; 16:1466-71; PMID:12080085; http://dx.doi.org/10.1101/gad.991402
- Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, Welford RW, Elkins JM, Oldham NJ, Bhattacharya S, Gleadle JM, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J Biol Chem 2002; 277:26351-5; PMID:12042299; http://dx.doi.org/10.1074/jbc.C200273200
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 2006; 125:467-81; PMID:16603238; http://dx.doi.org/10.1016/j.cell.2006.03.028
- Tsukada Y-i, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006; 439:811-6; PMID:16362057; http://dx.doi.org/10.1038/nature04433
- Yamane K, Toumazou C, Tsukada Y-i, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-Containing H3K9 Demethylase, Facilitates Transcription Activation by Androgen Receptor. Cell 2006; 125:483-95; PMID:16603237; http://dx.doi.org/10.1016/j.cell.2006.03.027
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin Jr WG. The retinoblastoma binding protein RBP2 Is an H3K4 demethylase. Cell 2007; 128:889-900; PMID:17320163; http://dx.doi.org/10.1016/j.cell.2007.02.013
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine Johnathan R, Bonni A, Roberts TM, Shi Y. The X-Linked Mental Retardation Gene SMCX/JARID1C Defines a Family of Histone H3 Lysine 4 Demethylases. Cell 2007; 128:1077-88; PMID:17320160; http://dx.doi.org/10.1016/j.cell.2007.02.017
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 2007; 449:689-94; PMID:17851529; http://dx.doi.org/10.1038/nature06192
- Tsukada Y-i, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev 2010; 24:432-7; PMID:20194436; http://dx.doi.org/10.1101/gad.1864410
- McDonough MA, Loenarz C, Chowdhury R, Clifton IJ, Schofield CJ. Structural studies on human 2-oxoglutarate dependent oxygenases. Curr Opin Struct Biol 2010; 20:659-72; PMID:20888218; http://dx.doi.org/10.1016/j.sbi.2010.08.006
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 2006; 7:715-27; PMID:16983801; http://dx.doi.org/10.1038/nrg1945
- Hopkinson RJ, Tumber A, Yapp C, Chowdhury R, Aik W, Che KH, Li XS, Kristensen JBL, King ONF, Chan MC, et al. Five-Carboxy-8-hydroxyquinoline is a broad spectrum 2-oxoglutarate oxygenase inhibitor which causes iron translocation. Chem Sci 2013; 4:3110-7; http://dx.doi.org/10.1039/c3sc51122g
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A 2007; 104:18439-44; PMID:18003914; http://dx.doi.org/10.1073/pnas.0707292104
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 2007; 447:601-5; PMID:17468742; http://dx.doi.org/10.1038/nature05823
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007; 449:731-4; PMID:17713478; http://dx.doi.org/10.1038/nature06145
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007; 130:1083-94; PMID:17825402; http://dx.doi.org/10.1016/j.cell.2007.08.019
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 2007; 318:447-50; PMID:17761849; http://dx.doi.org/10.1126/science.1149042
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol 2010; 17:38-43; PMID:20023638; http://dx.doi.org/10.1038/nsmb.1753
- Loenarz C, Ge W, Coleman ML, Rose NR, Cooper CDO, Klose RJ, Ratcliffe PJ, Schofield CJ. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nϵ-dimethyl lysine demethylase. Human Mol Genet 2010; 19:217-22; PMID:19843542; http://dx.doi.org/10.1093/hmg/ddp480
- Ge W, Wolf A, Feng T, Ho CH, Sekirnik R, Zayer A, Granatino N, Cockman ME, Loenarz C, Loik ND, et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat Chem Biol 2012; 8:960-2; PMID:23103944; http://dx.doi.org/10.1038/nchembio.1093
- Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol 2007; 14:689-95; PMID:17589523; http://dx.doi.org/10.1038/nsmb1273
- Hillringhaus L, Yue WW, Rose NR, Ng SS, Gileadi C, Loenarz C, Bello SH, Bray JE, Schofield CJ, Oppermann U. Structural and Evolutionary Basis for the Dual Substrate Selectivity of Human KDM4 Histone Demethylase Family. J Biol Chem 2011; 286:41616-25; PMID:21914792; http://dx.doi.org/10.1074/jbc.M111.283689
- Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T, Reinberg D. Dynamic Histone H1 Isotype 4 Methylation and Demethylation by Histone Lysine Methyltransferase G9a/KMT1C and the Jumonji Domain-containing JMJD2/KDM4 Proteins. J Biol Chem 2009; 284:8395-405; PMID:19144645; http://dx.doi.org/10.1074/jbc.M807818200
- Cockman ME, Webb JD, Ratcliffe PJ. FIH-dependent asparaginyl hydroxylation of ankyrin repeat domain-containing proteins. Ann New York Acad Sci 2009; 1177:9-18; PMID:19845602; http://dx.doi.org/10.1111/j.1749-6632.2009.05042.x
- Rotili D, Altun M, Kawamura A, Wolf A, Fischer R, Leung IK, Mackeen MM, Tian YM, Ratcliffe PJ, Mai A, et al. A photoreactive small-molecule probe for 2-oxoglutarate oxygenases. Chem Biol 2011; 18:642-54; PMID:21609845; http://dx.doi.org/10.1016/j.chembiol.2011.03.007
- Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, Bray JE, Savitsky P, Gileadi O, von Delft F, et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 2007; 448:87-91; PMID:17589501; http://dx.doi.org/10.1038/nature05971
- Rose NR, Ng SS, Mecinovic J, Lienard BM, Bello SH, Sun Z, McDonough MA, Oppermann U, Schofield CJ. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases. J Med Chem 2008; 51:7053-6; PMID:18942826; http://dx.doi.org/10.1021/jm800936s
- Rose NR, Woon EC, Tumber A, Walport LJ, Chowdhury R, Li XS, King ON, Lejeune C, Ng SS, Krojer T, et al. Plant growth regulator daminozide is a selective inhibitor of human KDM2/7 histone demethylases. J Med Chem 2012; 55:6639-43; PMID:22724510; http://dx.doi.org/10.1021/jm300677j
- Hewitson KS, Holmes SL, Ehrismann D, Hardy AP, Chowdhury R, Schofield CJ, McDonough MA. Evidence that two enzyme-derived histidine ligands are sufficient for iron binding and catalysis by factor inhibiting HIF (FIH). J Biol Chem 2008; 283:25971-8; PMID:18611856; http://dx.doi.org/10.1074/jbc.M804999200
- Sakurai M, Rose NR, Schultz L, Quinn AM, Jadhav A, Ng SS, Oppermann U, Schofield CJ, Simeonov A. A miniaturized screen for inhibitors of Jumonji histone demethylases. Mol BioSystems 2010; 6:357-64; PMID:20094655; http://dx.doi.org/10.1039/b912993f
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine[thinsp]9 and lysine[thinsp]36. Nature 2006; 442:312-6; PMID:16732292; http://dx.doi.org/10.1038/nature04853
- Walport LJ, Hopkinson RJ, Vollmar M, Madden SK, Gileadi C, Oppermann U, Schofield CJ, Johansson C. Human UTY(KDM6C) Is a Male-specific N-Methyl Lysyl Demethylase. J Biol Chem 2014; 289:18302-13; PMID:24798337; http://dx.doi.org/10.1074/jbc.M114.555052
- Liu L, Kim H, Casta A, Kobayashi Y, Shapiro LS, Christiano AM. Hairless is a histone H3K9 demethylase. FASEB J 2014; 28:1534-42; PMID:24334705; http://dx.doi.org/10.1096/fj.13-237677
