Abstract
CREB3L1 has been recently proposed as a novel metastasis suppressor gene in breast cancer. Our current study highlights CREB3L1 expression, regulation, and function in bladder cancer. We demonstrate a significant downregulation of CREB3L1 mRNA expression (n = 64) in primary bladder cancer tissues caused by tumor-specific CREB3L1 promoter hypermethylation (n = 51). Based on pyrosequencing CREB3L1 methylation was shown to be potentially associated with a more aggressive phenotype of bladder cancer. These findings were verified by an independent public data set containing data from 184 bladder tumors. In addition, immunohistochemical evaluation showed that CREB3L1 protein expression is decreased in bladder cancer tissues as well. Interestingly, protein loss is predominately observed in the nuclei of aggressive tumor cells. Based on in vitro models we clearly show that CREB3L1 re-expression mediates suppression of tumor cell migration and colony growth of high grade and invasive bladder cancer cells. The candidate tumor suppressor and TGF-β signaling inhibitor HTRA3 was furthermore identified as putative target gene of CREB3L1 in both invasive J82 bladder cells and primary bladder tumors. Hence, our data provide for the first time evidence that the transcription factor CREB3L1 may have an important role as a putative tumor suppressor in bladder cancer.
Abbreviations
| ATCC | = | American Type Culture Collection |
| BMP-2 | = | bone morphogenetic protein 2 |
| CA | = | California |
| cDNA | = | copy number desoxyribonucleic acid |
| CIS | = | Carcinoma in situ |
| CREB3L1 | = | element binding protein 3-like 1 |
| DAB | = | 3-3′ diaminobenzidine |
| DAC | = | 5-aza-2′-deoxycytidine |
| DNA | = | desoxyribonucleic acid |
| EK | = | ethics committee |
| ER | = | endoplasmic reticulum |
| FC | = | fold change |
| FFPE | = | formalin fixed paraffin embedded |
| G1 | = | well differentiated |
| G2 | = | moderately differentiated |
| G3 | = | poorly differentiated |
| GAPDH | = | glyceraldehyde 3-phosphate dehydrogenase |
| HCV | = | Hepatitis C virus |
| HPV | = | human papilloma virus |
| HTRA (1-4) | = | high-temperature requirement factor A (1-4) |
| IQR | = | interquartile range |
| IRS | = | immunoreactive score |
| LMU | = | Ludwig-Maximilians-University |
| M | = | methylated |
| MIBC | = | muscle invasive bladder cancer |
| mRNA | = | messenger ribo nucleic acid |
| MSP | = | methylation specific PCR |
| n | = | number |
| NMIBC | = | non-muscle invasive bladder cancer |
| ns | = | not significant |
| NU | = | normal urothelium |
| OASIS | = | old astrocyte specifically-induced substance |
| PCR | = | polymerase chain reaction |
| pTa | = | papillary non-invasive tumors |
| RIP | = | regulated intramembrane proteolysis |
| RWTH | = | Rheinisch Westfälisch Technische Hochschule |
| s.e.m. | = | standard error of the margin |
| SP1 | = | site 1 protease |
| SP2 | = | site 2 protease |
| TCGA | = | The Cancer Genome Atlas |
| TGF-β | = | transforming growth factor beta |
| TSA | = | trichostatin A |
| TSS | = | transcription start site |
| U | = | unmethylated |
| UC | = | urothelial cell cancer |
| UPR | = | unfold protein response |
| USA | = | United States of America |
| WHO | = | World Health Organization |
| WI | = | Wisconsin |
Introduction
cAMP responsive element binding protein 3-like 1 (CREB3L1), also referred to as old astrocyte specifically induced substance (OASIS),Citation1 belongs to the CREB/ATF family of bZIP transcription factors in mammals. This family comprises additional 4 members (CREB3 and CREB3L2–4)Citation2,3 playing an important role in unfolded protein response (UPR).Citation4 Beyond the bZIP domain mediating dimerization and DNA binding,Citation5 CREB3L1 proteins contain a transmembrane domain anchored in the endoplasmic reticulum (ER) membrane similarly to the ATF6 structure.Citation3 Owing to the ER-anchored N-terminus facing the cytoplasm, the transcriptional activity of the CREB3L1 protein remains repressed. CREB3L1 is thought to be proteolytically cleaved by site 1 protease (SP1) and SP2 in the Golgi apparatus upon regulated intramembrane proteolysis (RIP) in response to ER stress,Citation6 although knowledge about the mechanism responsible for sensing ER stress by CREB3L1 is still lacking.Citation4 However, as a consequence, the released transcriptional active fragment can be translocated into the nucleus to subsequently induce UPR-associated target genes recognizing cAMP-responsive elements (CRE-related: TGACGTCA).Citation2,7
Recent studies have shown that CREB3L1 is involved in astrocyte differentiationCitation8 and bone formation.Citation9,10 Murakami and colleagues showed that expression of CREB3L1 in osteoblasts is induced by BMP-2 (bone morphogenetic protein 2), whereas CREB3L1 deficient mice were characterized by decreased type I collagen.Citation9 Besides collagen genes (Col1a1 and Col1a2), other genes implicated in matrix protein production such as Papss2 or Matn1 have been proposed as putative CREB3L1 target genes so far.Citation11 Interestingly, previous studies revealed that CREB3L1 silencing caused by aberrant DNA hypermethylation is a necessary step in Hepatitis C (HCV) virus infected cells, such as Huh7 contributing to sufficient cell proliferation.Citation12 According to that, Denard et al.Citation13 demonstrated in human hepatoma Huh7 cells that doxorubicin stimulated proteolytic cleavage of CREB3L1 activates the transcription of cell cycle inhibitors including p21 leading to increased doxorubicin resistance. In turn, overexpression of CREB3L1 in MCF-7 breast cancer cells enhanced doxorubicin sensitivity indicating a putative role of CREB3L1 in therapeutic cancer treatment. However, accumulating evidence suggests involvement of CREB3L1 in various aspects of tumor cell biology. On the one hand CREB3L1 has been identified as a fusion partner of FUS in low grade fibromyxoid sarcomaCitation14 or EWSR1 in osteosarcoma,Citation15 proposing an oncogenic character. On the other hand, Mellor et al. have shown that CREB3L1 is a metastasis suppressor in breast carcinoma whose activity impairs metastatic mechanisms such as tumor invasion and angiogenesis.Citation16
So far, the role of CREB3L1 is still unrevealed in bladder cancer, which is a very common malignant disease with more than 380,000 estimated new cases worldwide each year.Citation17 Lacking knowledge of the genes being involved in the complex mechanisms of bladder cancer invasion and metastasis, still leaves current therapeutic strategies insufficient,Citation18 and leads to high mortality rates in aggressive subtypes.Citation19,20 Hence, the identification and evaluation of molecules involved in these processes is a major task and the intended subject of this study. Here, we present a comprehensive analysis of CREB3L1 expression and regulation in human bladder cancer providing CREB3L1 as a novel tumor suppressor.
Results
CREB3L1 mRNA expression is downregulated in human bladder cancer
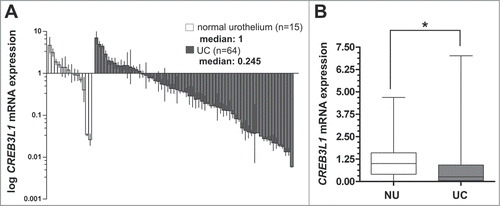
Previously, Wild and colleagues published expression profiles of 67 bladder tumors compared to 8 normal samples.Citation21 Based on that, we re-analyzed the DNA microarray data set by using in silico database mining procedures identifying novel candidates with differential expression in the course of bladder cancer progression. We frequently detected CREB3L1 expression loss in human bladder cancer specimens (data not shown). Therefore, we aimed to study, for the first time, CREB3L1 expression, regulation, and function in bladder cancer development. At first, real-time PCR analysis showed a clear downregulation of CREB3L1 mRNA expression in primary bladder tumors (UC) including carcinoma in situ (CIS) (overall n = 64) (median: 0.245; interquartile range (IQR): 0.04–0.89%; min-max range: 0.0–6.8) when compared to normal urothelium tissues (NU) (n = 15) (median: 1; IQR: 0.40–1.6; min-max range: 0.0–4.70) (). The significance (P = 0.0392, Mann Whitney Test) of the differential CREB3L1 mRNA expression is illustrated in . Overall, bladder cancer tissues showed a clear CREB3L1 mRNA expression loss by >4-fold.
Figure 1. Downregulation of CREB3L1 mRNA expression in human bladder cancer. (A) Real-time PCR based CREB3L1 mRNA expression analyses of 64 tumor samples (UC) compared with normal urothelium tissue (NU) samples (n = 15). Vertical lines: ± standard error of margin (s.e.m.). (B) Box plot demonstrating a significant association of CREB3L1 mRNA expression downregulation in UC. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak and minimum; ns: not significant, *P < 0.05.
CREB3L1 gene inactivation is associated with tumor-specific promoter hypermethylation and high-grade tumor stages
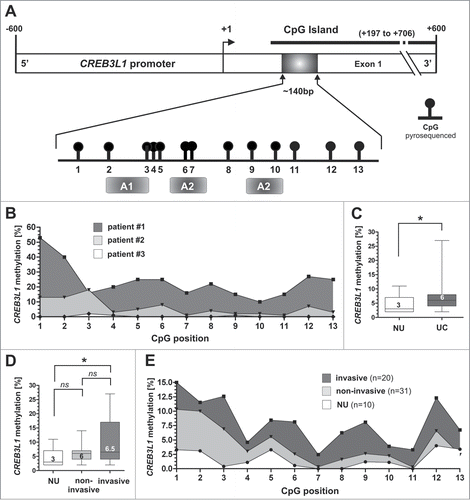
In recent studies, CREB3L1 promoter hypermethylation has been suggested as the molecular cause of CREB3L1 gene silencing linked with both, human papillomavirus (HPV)Citation22 and hepatitis C virus (HCV) induced cell transformations.Citation12 Therefore, we determined whether CREB3L1 promoter hypermethylation could be responsible for the observed mRNA loss in human bladder cancer. Analysis of the CREB3L1 gene promoter using the genomic DNA information (ENSEMBL contig ENSG00000157613) identified one CpG-rich island between genomic positions 46,299,409 and 46,299,918 (+197 bp to +706 bp relative to the expected transcription start site (TSS)) on chromosome 11p, which met the following criteria: DNA region: ≥ 200 bp; Obs/Exp: ≥ 0.6; %GC: ≥ 50. Using pyrosequencing technique, we subsequently analyzed the methylation status of 13 CpG sites within this CpG island that are closely associated to the transcription start site and encode potential regulatory sequences (). We determined the individual CpG sites in human bladder cancer samples (n = 51) and detected indeed altered DNA methylation levels in bladder cancer tissues when compared with NU (n = 10). The CREB3L1 methylation configurations of representative patients are shown in . The median CREB3L1 methylation level of normal tissues (n = 10) was 3% (IQR: 3–6%; min-max range: 2–11%), whereas the overall median methylation of UC samples was slightly but significantly (p = 0.0256, Mann Whitney Test) increased by twofold (median: 6%; IQR: 4–8%, min-max range: 2–27%) (). Classifying the UC samples by non-invasive and invasion subtypes, we clearly showed that increased CREB3L1 promoter methylation levels were significantly linked with invasive tumor stages (). Papillary non-invasive tumors showed a median methylation of only 6%. In contrast, invasive tumors exhibited a significantly increased (P < 0.05, Dunn's Multiple Comparison Test) median methylation of 6.5% when compared to NU. Clinico-pathological characteristics of the analyzed tumor samples were further correlated with CREB3L1 methylation for descriptive data analysis (). A close association between high CREB3L1 promoter hypermethylation and high grade UC was revealed by using a Fisher's exact test (P = 0.024). Besides that, a correlation of CREB3L1 promoter methylation and concomitant carcinoma in situ (CIS) was observed as well.
Table 1. Clinico-pathological parameters in relation to CREB3L1 promoter methylation
Figure 2. CREB3L1 promoter hypermethylation is associated with advanced bladder cancer stages. (A) Schematic map of the human CREB3L1 promoter region including the relative positions of pyrosequenced CpG dinucleotides (black boxes and circles). One predicted CpG island is located between base +197 and base +706. +1: CREB3L1 transcription start site. Gray boxes illustrating gene transcription-relevant regulatory elements statistically identified by using the Genomatix database (http://www.genomatix.de/): A1: activator protein site 2; A2: RNA polymerase II transcription factor IIB site. (B) Methylation status of representative bladder cancer patient specimens. (C) Box Plot demonstrating significant increase of CREB3L1 promoter methylation frequency in all analyzed bladder cancer samples. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak and minimum; *P < 0.05. (D) Box plot showing significant increase of CREB3L1 promoter methylation frequency in association to bladder cancer progression, i.e., in invasive tumors. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak and minimum; ns: not significant, *P < 0.05. (E) Mean methylation frequency for each pyrosequenced CpG site (1–13) of normal tissue (n = 10), non-invasive tumors (n = 31) and invasive tumors (n = 20) is shown.
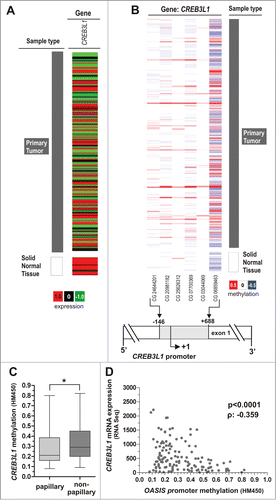
In order to assess the authenticity of these findings, we determined CREB3L1 promoter methylation and gene expression in a dataset of independent studies, in total representing 184 different bladder cancer samples. Based on data of The Cancer Genome Atlas (TCGA) we verified downregulation of CREB3L1 gene expression in bladder cancer in comparison to normal bladder tissues (). Furthermore, CREB3L1 promoter hypermethylation of CpG sites (located from −146 bp to +688 bp with respect to the TSS) was confirmed in bladder cancer samples () and prevalently found in the non-papillary, i.e. invasive subtype (). Of importance, a negative correlation (Spearman coefficient: −0.359) of CREB3L1 promoter methylation with CREB3L1 mRNA expression was demonstrated (P < 0.0001) ().
Figure 3. Tumor-specific CREB3L1 promoter hypermethylation is associated with CREB3L1 gene silencing and non-papillary tumor subtype in an independent TCGA data set. (A) CREB3L1 expression in bladder tumor samples from TCGA data portal. Red: high expression, black: mean expression and green: low expression. Left panel: sample type (dark gray: primary tumor; white: solid normal tissues). Right panel: CREB3L1 mRNA expression. (B) DNA methylation of the CREB3L1 promoter analyzed in bladder cancer samples from TCGA data portal. Red: high methylation, white: mean methylation; blue: low methylation. Right panel: sample type (dark gray: primary tumor; white: solid normal tissues). Left panels: values of CREB3L1 DNA methylation for each CG. The relative positions of 6 analyzed CpG duplets within the CREB3L1 promoter region are indicated. +1: CREB3L1 transcription start site. (C) Tumor samples stratified by subtypes. Box plot showed a significant association of increased CREB3L1 methylation with non-papillary UC. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak and minimum; *P < 0.05. (D) Negative correlation of CREB3L1 mRNA expression and its DNA methylation status in primary bladder cancer samples. ρ: Spearman correlation coefficient.
CREB3L1 promoter hypermethylation in human bladder cancer cell lines correlates with CREB3L1 mRNA expression
We next quantified the methylation frequency of 13 CpG sites within the CREB3L1 promoter in human bladder cell lines. The cell line HCV29, which is derived from tumor-associated NU,Citation23 showed a completely unmethylated CREB3L1 promoter status (). Compared to that, all analyzed malignant bladder cell lines (KK47, RT112, EJ28 and J82) exhibited a clear promoter hypermethylation. Quantitatively, the median methylation value of all 13 CpG dinucleotides was 1% (HCV29), 72% (RT112), 22% (KK47), 31% (EJ28) and 39% (J82), respectively.
Figure 4. Demethylation of the CREB3L1 promoter correlates with CREBL1 re-expression in vitro. (A) Graph illustrating quantitative CREB3L1 promoter methylation level of 13 CpG sites in the normal bladder cell line HCV29 and 4 human bladder cancer cell lines. (B) Pyrosequencing-based analysis of the quantitative methylation ratio measured for 13 CpG dinucleotides within the CREB3L1 promoter region determined prior (-DAC / -TSA; dark-gray-filled) and after in vitro demethylation agents treatment (+DAC / +TSA; gray-filled). A pyrogram is representatively shown for the bladder cancer cell line RT112. (C) Box plot analysis that shows effective reduction of the median methylation ratio within the CREB3L1 promoter region in human bladder cancer cell lines RT112 and J82 after DAC/TSA treatment (+) compared to both non-treated cell lines (−) and unmethylated normal cell line HCV29. Methylation frequencies were measured by pyrosequencing mirroring a methylation ratio of 13 analyzed CpG duplets. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak and minimum; **P < 0.01. (D) Real-time PCR analysis for CREB3L1 mRNA expression based on equal in vitro demethylation samples demonstrating a clear CREB3L1 re-expression after treatment with both DAC and TSA (+) for all analyzed bladder cell lines. Non-treated cells were set to 1. Error bars: + s.e.m.
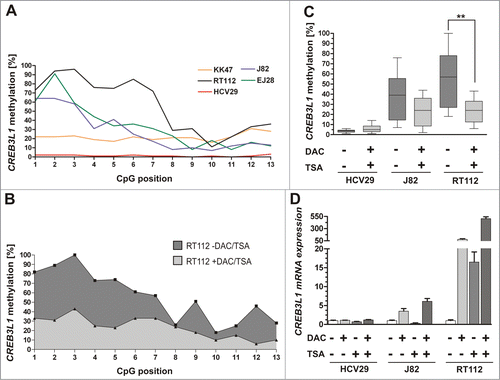
Having found that CREB3L1 loss is tightly associated with CREB3L1 promoter methylation in primary bladder tumors, we aimed to functionally confirm this epigenetic modification as the molecular cause for CREB3L1 gene silencing. Using pyrosequencing, we demonstrated a clear decrease in CREB3L1 promoter hypermethylation 72 h after the application of demethylation drugs to bladder tumor cell lines (J82 and RT112). The median methylation level of the analyzed CREB3L1 promoter region was significantly (p = 0.0021, Mann Whitney Test) reduced by 57.9% in RT112 tumor cells. A representative diagram showing the percentage of methylation of all 13 analyzed CpG sites of RT112 cells, before and after 5-aza-2′-deoxycytidine (DAC) and trichostatin A (TSA) treatment is shown in . As a consequence of the reduced methylation level (), we observed a clear upregulation of CREB3L1 mRNA in J82 (6-fold) and RT112 (503-fold) tumor cells after DAC and TSA treatment (). CREB3L1 mRNA in normal-like HVC29 bladder cells harboring an unmethylated CREB3L1 promoter served as control and were not further inducible by DAC/TSA. These findings support our notion that the epigenetic configuration of the CREB3L1 promoter has a major impact on the regulation of CREB3L1 expression.
CREB3L1 protein is decreased in nuclei of primary invasive bladder tumors
Given that CREB3L1 mRNA expression is abrogated in bladder cancer, we characterized CREB3L1 protein expression in NU as well as in bladder tumor tissues using immunohistochemistry. As expected for a transcription factor, we found CREB3L1 protein in the cytoplasm as well as frequently in the nuclei of normal urothelium (). According to that, most bladder tumor cells showed a similar cytoplasmatic protein level, but only sporadically nuclear protein localization was observed (). Interestingly, low-grade non-invasive bladder tumors retained high levels of CREB3L1 protein within the nucleus (see arrows in ), whereas higher pT stages mainly showed solely a cytoplasmatic staining (see arrow in ). This observation was confirmed by a quantification of stained CREB3L1 protein according to an adapted immunoreactive score (IRS) developed by Remmele and Stegner (1987) in both the cytoplasm and the nuclei. Overall, we compared samples of normal bladder tissues (n = 21) and bladder carcinomas (n = 96) evaluating the average CREB3L1 expression in a semi-quantitative manner. Compared to NU (median IRS: 6), cytoplasmatic CREB3L1 expression was not notably altered (median IRS: 6) in bladder tumors. In high-grade tumors, we observed an increased cytoplasmatic CREB3L1 protein level in comparison to low-grade tumors (Supplementary Table 1). In contrast, 59.4% (n = 57/96) of all analyzed bladder tumors exhibited a significant CREB3L1 protein downregulation within the nuclei of IRS <4 (median IRS: 2; p = 0.0104, Mann Whitney Test) compared to the nuclei of NU (median IRS: 4) ().
Figure 5. Loss of CREB3L1 protein expression in human UC. (A+B) Very strong CREB3L1 immunoreactivity in the cytoplasm and in nuclei of normal urothelial cells (arrow). (C) Strong nuclear CREB3L1 protein staining in papillary non-invasive low grade tumors. (D) Moderate CREB3L1 immunoreactivity in high grade, invasive tumor cells lacking nucleus staining. (E) Strong cytoplasmatic CREB3L1 staining in invasive tumor cells. (F-H) Low and very low CREB3L1 staining in the cytoplasm of muscle-invasive tumor cells that completely lack nuclear CREB3L1 protein expression. (I) Negative control of normal urothelium. The application of primary antibody was omitted. Scale bar: 50 μm. (J) Box plot demonstrating overall significant loss of CREB3L1 protein only in the nucleus of bladder tumors compared to NU. (K-L) Box plot graph illustrates that loss of CREB3L1 protein within nuclei is associated with high grade invasive bladder UC. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak and minimum; ns: not significant, *P < 0.05, ***P < 0.001.
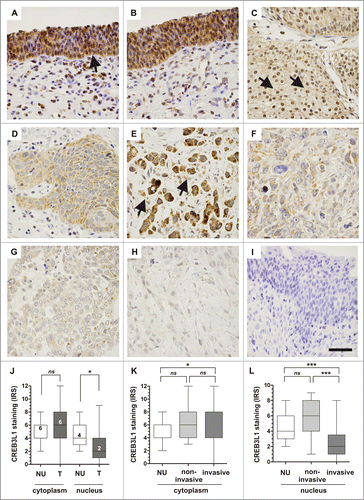
Stratifying the tumor samples by substages, we revealed that the observed loss of CREB3L1 protein in nuclei was assigned to invasive bladder tumors (P < 0.0001, Dunn's Multiple Comparison Test) (). The close association between loss of nuclear CREB3L1 protein and invasive tumor growth type, high grade tumors, and advanced stages of pT2 to pT4 tumors were significantly (P < 0.0001) confirmed by using a Fisher's exact test (Supplementary Table 2). Besides the protein loss regulated on transcription level, our data showed therewith an aberrant distribution between these compartments, potentially based on a failed translocation of CREB3L1 into the nucleus. Possibly, this translocation failure could be caused by gene mutations. To provide first insights into this hypothesis, we analyzed different public TCGA data sets, but revealed only a very low mutation frequency within the CREB3L1 gene sequence and no association with a mutated cleavage site (Supplementary Fig. 1).
CREB3L1 re-expression suppresses tumor cell migration and colony spread of high-grade and invasive bladder cancer cells
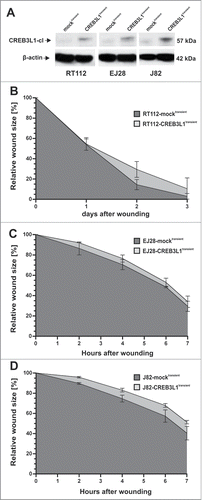
Since we found a clear downregulation of CREB3L1 expression in invasive bladder tumors, we aimed to assess the functional role of CREB3L1 in bladder cancer. We transiently re-expressed CREB3L1 in different malignant bladder cancer cell lines lacking endogenous CREB3L1 expression, using a full-length CREB3L1 cDNA pT-Rex-DEST30 expression vector to evaluate its function in a bulk of cells with a homogenously distributed genetic background. Ectopic CREB3L1 expression was verified by western blotting in comparison to control mock cells (empty vector) (). Based on these transiently transfected in vitro models (i.e., where still a large part of non-transfected cells is present), CREB3L1 re-expression led to a clear reduction of cell migration capacity in all analyzed bladder cancer cell lines, i.e., in the moderately differentiated cell line RT112 and the poorly differentiated cell lines EJ28 and J82, when compared to mock-transfected cells (). Already a few hours after scratching, CREB3L1–positive cells of both poorly differentiated bladder cancer cell lines (EJ28 and J82) repopulated the wounded area notably slower than mock-transfected cells. In RT112 cancer cells, a clear impairment of motile characteristics was observed from day 2.
Figure 6. Transient expression of CREB3L1 in bladder cancer cell lines suppresses cell migration. (A) Western blot analysis of the cleaved (cl) CREB3L1 protein re-expression in transiently CREB3L1-transfected RT112, EJ28 and J82 tumor cells compared to mock-transfected control cells. β-actin served as loading control. (B-D) Cell migration studies of transiently transfected tumor cell lines were performed by using a monolayer wound healing assay. Motile capacity of control cell sets (RT112-mocktransient, EJ28-mocktransient J82-mocktransient) and the CREB3L1 set (RT112-CREB3L1transient, EJ28-CREB3L1transient J82-CREB3L1transient) was assessed over 3 d (B) or 7 hours (C+D). Vertical lines: standard deviation (SD) of triplicates. Cell-free area on day 0 was set as 100% and used for standardization.
Given the limitations of these transient models, we performed further functional analysis using a stable in vitro model focusing on poorly differentiated invasive bladder cancer characteristics.Citation23 Upon transfection with the full-length CREB3L1 cDNA and clonal selection, its expression was stably restored in J82 cells (). Both the precursor full-length (84 kDa) and the cleaved (57 kDa) CREB3L1 protein were verified in J82-CREB3L1st cells, underlining the functionality of this in vitro system. Indeed, CREB3L1 expression caused a decreased proliferation rate in J82 cancer cells (). In agreement with the transient migration studies, we furthermore observed that CREB3L1 expression in J82 cells reduced cell migration compared to mock-transfected control clones (). After 24 h in culture, the CREB3L1-expressing cell clones repopulated solely on average 45.0% (mock clones 27.1%) of the wounded area. Representative images of the analyzed wound area 24 h after scratching are shown in .
Figure 7 (See previous page). Stable CREB3L1 re-expression mediates inhibition of tumor cell migration and colony growth of invasive single cell clones (A) CREB3L1 gain-of-function in vitro model: Stable CREB3L1 expression in J82 clones was confirmed by quantitative real-time PCR and protein gel blotting. Upper graph: Relative CREB3L1 mRNA expression of J82-mock clone #4 and #14 as well as J82-WT (wild type, set to 1) and CREB3L1-transfected J82 clones #2, #13, #17 is shown. Column: Mean of triplicate determinations. Error bars, ±SEM. Bottom: Corresponding CREB3L1 protein expression in J82 clones. A specific signal of the ectopic CREB3L1 protein (FL: full-length and cl: cleaved) is detectable only in CREB3L1st clones. β-actin served as loading control. (B) XTT proliferation assay was performed. J82-CREB3L1st clones (#2, #13, #17) showed reduced cell proliferation compared with mock control clones (#4, #14). Vertical lines: standard deviation (SD) of triplicate experiments. (C) Tumor cell migration based on stably independent J82-mockst and J82-CREB3L1st clones. Comparison of wound closure of a control cell set (n = 3) and J82-CREB3L1st clones (n = 3) over 3 d Vertical lines: standard deviation (SD) of triplicates. Cell-free area on day 0 was set as 100% and used for standardization. (D) Left graph: Comparison of wound area for each clone after 24 h. Right panel: Wound documentation of a magnified area by phase-contrast microscopy 24 h after scratching. Scale bar = 100 μm. Arrows: indicate scratch direction. White border: cell-free wound area. (E) Colony formation assay. Representative 6-well plates of J82-mockst (#4, #14) and J82-CREB3L1st clones (#13, #17) are shown 2 weeks after cell seeding. (F) Densitometrical analysis of grown colonies. Box plot demonstrates averages of colony growth of triplicate experiments for each clone. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak and minimum, ***P < 0.001.
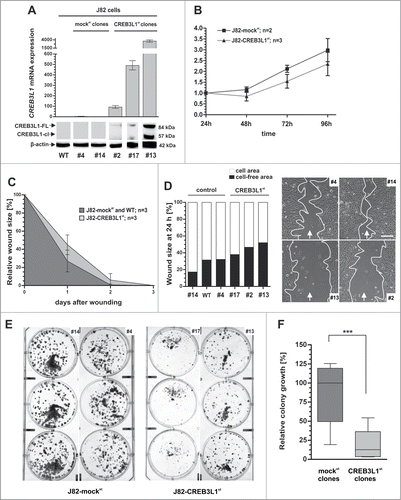
Subsequently, we studied the impact of CREB3L1 expression on colony formation that mirrors the clonogenic capacity of spreading cancer cells. Two weeks after cell seeding, J82 clones with CREB3L1 expression showed clearly reduced colony growth (). Densitometric analysis revealed a highly significant retardation in colony forming by 85.1% (p = 0.005, Mann Whitney Test) of CREB3L1-expressing cells compared to controls whose median colony growth was defined as 100% ().
HTRA3 is a putative target gene of CREB3L1 in human bladder cancer
As CREB3L1 is a well-known transcription factor,Citation2 we finally had a closer look on putative target genes whose CREB3L1-dependent induction may explain the tumor suppressive mechanisms mediated by CREB3L1 in bladder cancer. At first, we analyzed previously defined potential CREB3L1 targetsCitation11 that have further been associated with cancer progression. We screened the promoter regions of putative target genes for cAMP-responsive element binding sites (CREB). Indeed, we confirmed with highly statistical reliability 2 CRE-like sites in the promoter region of HTRA3, a putative TGF-β signaling modulator,Citation24 using Genomatix data base analysis (). Real-time PCR of HTRA3 and CREB3L1 mRNA further showed a strong expression of both genes in the wild type (WT) papillary, non-invasive bladder cancer cell line KK47 (). Invasive tumor cell lines EJ28, as well as J82, exhibited very weak expression of both genes at an approximately similar level. In turn, a transiently re-expression of ectopic CREB3L1 in these cell lines caused a clear upregulation in J82 tumor cells whereas KK47 cells exhibiting abundant endogenous CREB3L1 expression showed no further HTRA3 stimulation (). In accordance with these findings, we significantly confirmed increased HTRA3 mRNA (median FC: 62.5; p = 0.0347, Mann Whitney Test) expression in independent stable J82-CREB3L1st clones when compared to J82-CREB3L1-negative control cells ().
Figure 8. HTRA3 expression is tightly associated with CREB3L1 expression in bladder cancer. (A) Schematic map of the human HTRA3 promoter including the relative positions of used forward and reverse MSP primers (gray arrows). One predicted CpG island is located between base −192 and base +652. +1: HTRA3 transcription start site. A: potential CRE-like sites statistically identified by using the Genomatix database (http://www.genomatix.de/). (B) HTRA3 and CREB3L1 mRNA expression in WT bladder cancer cell lines. Error bars, +SEM (C) Mean HTRA3 mRNA induction in transiently CREB3L1-transfected bladder cancer cell lines when compared to mock controls (set to 1) based on 2 independent experiments. Error bars, +SEM (D) Left box plot: HTRA3 mRNA is significantly up regulated in stable J82-CREB3L1st clones compared to J82-mockst control clones (set to 1). Right box plot: CREB3L1 expression in J82-CREB3L1st clones compared to J82-mockst control clones. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak and minimum, *P < 0.05, **P < 0.01. (E) Representative MSP analyses show the HTRA3 promoter methylation status of human bladder cell lines KK47 and J82 and various primary bladder tumors. Bands labeled with U and M reflect unmethylated and methylated DNA, respectively. Bisulphite-converted unmethylated, genomic (U-co) and polymethylated, genomic (M-co) DNA were used as positive controls. NTC: non-template control. (F) Association of CREB3L1 mRNA expression and HTRA3 mRNA expression in 48 primary bladder cancer samples. ρ: Spearman correlation coefficient. (G) DNA methylation of the HTRA3 promoter analyzed in bladder cancer samples from TCGA data portal. Red: high methylation, white: mean methylation; blue: low methylation. Right panel: sample type (dark gray: primary tumor; white: solid normal tissues). Left panels: values of HTRA3 DNA methylation for each CpG. The relative positions of 6 analyzed CpG duplets within the HTRA3 promoter region are indicated. +1: CREB3L1 transcription start site. (H) Correlation of CREB3L1 mRNA expression and HTRA3 mRNA expression in the TCGA data set of primary bladder cancer samples. ρ: Spearman correlation coefficient.
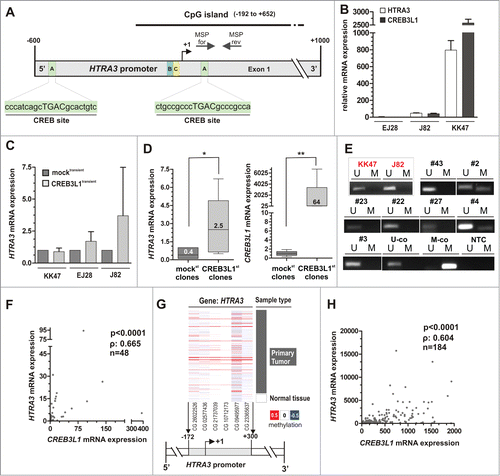
Next, we aimed to verify the expression axis of CREB3L1 and HTRA3 in primary bladder tumors. HTRA3 has been recently shown being silenced by epigenetic modifications, i.e., DNA methylation, in lung cancer.Citation25 To ensure an accessible promoter, we initially analyzed the DNA methylation status of a HTRA3 promoter locus close to the TSS and CREB site using MSP in human bladder cancer cell lines, as well as in primary bladder cancer tissues. As expected, both J82 and KK47 cells exhibited a completely unmethylated HTRA3 promoter (). In bladder tumor tissues, we found overall a weak HTRA3 methylation frequency of approximately 15% (5 out of 34). In turn, we revealed a strong positive correlation (Spearman coefficient: 0.665, P < 0.0001) between CREB3L1 and HTRA3 mRNA expression (). A minor methylation frequency of the HTRA3 promoter was confirmed using the independent TCGA bladder cancer tissue cohort (). Based on this data set, we also detected a clear association of CREB3L1 and HTRA3 mRNA (Spearman coefficient: 0.604, P < 0.0001) (), underscoring a physiological relevance of the CREB3L1-HTRA3 expression axis for bladder cancer development.
Discussion
So far, dysfunction of the highly conserved signaling pathway UPR in response to ER stress has been shown in various pathological processes, including metabolic as well as inflammatory disease, and cancer.Citation26-30 Even though a dual role of ER stress and UPR in cancer is controversially discussed,Citation31 processes such as hypoxia have clearly been identified to drive ER stress inducing UPR pathways during tumor cell growth.Citation32 It is well known that activation of BiP chaperones is increased in several tumor types that contributes to proliferation and survival of these tumor cellsCitation33. In light of that, CREB3L1, a member of CREB/ATF transcription factors, is also thought to regulate UPR to adapt and respond to ER stress conditions, thereby promoting tumor cell fitness.Citation34
However, a recent study revealed that CREB3L1 target genes are rather associated with extracellular matrix production than classical UPR genes.Citation11 Of interest, interaction of CREB3L1 with the chaperone BiP as described for ATF6 has not been detected so far.Citation4 This suggests a role of CREB3L1 beyond maintaining ER homeostasis. According to this notion, previous studies revealed that CREB3L1 activation has been associated with inhibition of cell growth of virus-infected cells.Citation35 Additionally, CREB3L1 was recently highlighted as a novel metastasis repressor in breast cancer by suppressing tumor invasion and angiogenesis.Citation16 Apart from that, too little is known about the putative role of the transcription factor CREB3L1 in human carcinogenesis. In the current study, we aimed to decipher for the first time CREB3L1 expression, regulation, and function in human bladder cancer development.
In primary bladder tumors, we demonstrated a significant CREB3L1 mRNA downregulation when compared with normal bladder urothelium. Loss of CREB3L1 expression has been further confirmed in an independent cohort of bladder cancer tissues. Focusing on the molecular mechanism that is causative for the observed gene silencing in bladder cancer we clearly identified aberrant CREB3L1 promoter hypermethylation. CREB3L1 hypermethylation was observed in a minor frequency in less aggressive bladder cancer subtypes, but the most abundant methylation level was notably associated with a more aggressive phenotype of bladder tumors, as demonstrated in 2 independent cohorts. Importantly, in both primary bladder tumors and in vitro cultured bladder cancer cell lines a strong correlation between CREB3L1 expression and its promoter hypermethylation has been revealed. So far, only 2 previous studies proposed aberrant methylation of the CREB3L1 promoter in virus-infected cells.Citation12,22 Promoter hypermethylation associated loss of CREB3L1 during carcinogenesis has not been described yet. Therewith, our data give first indications that this major epigenetic mechanism, known to promote tumor suppressor gene silencing,Citation36 might be important for CREB3L1 as well. Moreover, a suppressive role of CREB3L1 in the course of bladder cancer progression could be hypothesized as recently suggested for breast cancer.Citation16
In line with these findings, we subsequently verified decreased CREB3L1 expression in bladder tumors on protein level. Interestingly, the lack of protein was predominantly observed in the nuclei of high-grade tumor cells, whereas in the cytoplasm CREB3L1 was found expressed at an almost similar level. These data suggest that in addition to the inactivation of the CREB3L1 gene by aberrant promoter methylation during bladder cancer progression further mechanisms may exist to avoid CREB3L1 transcriptional activity, facilitating the aggressive character of tumor cells. In silico no mutations within the cleavage consensus sequences "RSLL" of CREB3L1 were detected that could serve as an explanation for a repressed release of the N-terminus. Bearing in mind that in case of activation the transcriptional active CREB3L1 fragments are thought to be unstable and rapidly degraded by the proteasomes.Citation3 Hence, a delay in translocation and stabilization of CREB3L1 as previously reportedCitation37 may indeed cause a significantly decreased amount of CREB3L1 protein within the nucleus of invasive bladder tumors.
However, these findings underline our hypothesis that CREB3L1 mediates tumor suppressive function, whereas loss of CREB3L1 may particularly drive invasive tumor characteristics. In fact, transient re-expression of CREB3L1 in different invasive bladder cancer cell lines (RT112, EJ28 and J82) causes an inhibition of cell migration. By stable CREB3L1 re-expression in poorly differentiated, invasive J82 bladder tumor cells we further restored CREB3L1 protein in single cell clones, allowing detailed investigation of CREB3L1 function in vitro. In this in vitro model, forced CREB3L1 expression reduces cell proliferation of J82 clones in line with the effects previously described for CREB3L1 in Huh-7 cells impaired by virus replication.Citation35 Additionally, a clear suppression of tumor cell motility was confirmed underscoring the potential role of CREB3L1 as repressor for the emergence of invasive tumor characteristics. This notion was strengthened by colony growth analysis of CREB3L1 expressing J82 clones in which CREB3L1 expression impaired the clonogenic capacity and led to highly significant suppression of colony formation in vitro. These data are in concordance with the proposed suppressive role of CREB3L1 in breast cancer,Citation16 therefore giving first insight into a putative similar function of CREB3L1 in bladder cancer progression.
As transcription factors play an eminent role in cancer metastasis by regulating a multitude of genes,Citation38 we identified HTRA3 as a putative target of CREB3L1 in bladder cancer cell lines as well as primary bladder tumors. HTRA3 belongs to a highly conserved High-Temperature Requirement Factor A (HTRA) family of serine proteases,Citation39 which modulates in a PDZ domain-dependent manner microtubule stability, mediating for example cell migration suppression as demonstrated for HTRA1.Citation40 Importantly, HTRA1–4 are known modulators of the metastasis-associated TGF-β signaling pathway.Citation24 Secreted HTRA3 proteins are thought to bind to various TGF-β receptor ligands inhibiting the activation of this signaling cascade. In line, HTRA3 has been shown to be downregulated in both cancer cell lines and primary tumors such as endometrial cancer.Citation41 Moreover, Yin et al. revealed a negative correlation of HTRA3 expression and lymph node metastasis in invasive mammary tumors.Citation42 Of interest, Singh and colleagues previously showed that inhibition of HTRA3 triggers HTR-8 cell invasion, i.e., loss of HTRA3 stimulates controlled invasion of invading interstitial trophoblasts.Citation43 Given these studies, HTRA3 could be proposed as a factor contributing to invasive cell characteristics, potentially acting as tumor suppressor in bladder cancer progression as well. In accordance to a previous study identifying HTRA3 promoter hypermethylation as the molecular cause for its downregulation in smoking-related lung cancer,Citation25 we also verified HTRA3 promoter methylation in bladder cancer even though the frequency of this epigenetic event was very low. Owing to that, the transcriptional HTRA3 repression could be mainly triggered by CREB3L1 inactivation. Moreover, HTRA3 induction fits to our observed suppressive function on tumor cell migration and colony growth in vitro, regulated by CREB3L1. However, it is likely that further mediators and target genes of CREB3L1 may also contribute to the observed phenotype and the detailed mechanisms behind have to be addressed in future studies.
In summary, we provide for the first time evidence that the CREB3L1 gene is inactivated by aberrant promoter hypermethylation in bladder cancer. CREB3L1 expression mediates inhibition of bladder cancer invasion, implying a critical role of this transcription factor as a putative metastasis suppressor. Irrespective of the underlying mechanism, our data highlight a linkage of CREB3L1 to the TGF-β signaling modulator and putative tumor suppressor HTRA3. Further pathway-based investigations might deliver novel molecular mechanisms and targets helping to improve treatment strategies for invasive bladder cancer.
Material and Methods
Patient samples
Formalin-fixed paraffin-embedded (FFPE) tissues of both primary bladder cancer and normal urothelium were obtained from the pathology archives of the Institute of Pathology RWTH Aachen University. The anonymized and retrospective study was approved by the local Ethics Committee (EK 122/04, 173/06 and 206/09). For cohort characteristics of analyzed samples see Supplementary Table 3. In order to enhance tumor cell content only manually micro-dissected samples were used for pyrosequencing and mRNA expression analysis. Evaluation of protein expression was performed on whole tissue sections or tissue microarrays with representative tissue cores of 1.5 mm diameter. For an independent correlation between CREB3L1 expression and DNA methylation public data from primary bladder cancer tissues and solid normal tissues from The Cancer Genome Atlas (TCGA) data portal were used. These comprise data of overall n = 184 patients from 2 independent platforms: Illumina Infinium DNA methylation chip (HumanMethylation450) and Illumina HiSeg gene expression. For cohort characteristics of analyzed TCGA samples in this current study see Supplementary Table 4.
Cell lines and reagents
Bladder cancer cell lines RT112 and J82 were originally obtained from the American Type Culture Collection (ATCC, Manassas, VA) and were resuscitated before using in experiments. The bladder cell lines HCV29, KK47 and EJ28 were a gift from Dr. Alexander Buchner (LMU München, Germany). All cell lines were cultured as previously described.Citation44 Using the PCR-based Venor® GeM Mycoplasma Detection Kit (Minerva Biolabs, Berlin, Germany) all cells were regularly tested for mycoplasma infection.
CREB3L1 immunohistochemistry
Immunohistochemical staining of CREB3L1 protein was performed according to the manufacturer's instructions using DAKO 5001 Kit (DAKO, Glostrup, Denmark). Subsequent to a microwave heating of FFPE sections in citrate buffer pH 6, these tissues were incubated at 4°C with a rabbit polyclonal anti-human CREB3L1 antibody (1:200) (ab33051, Abcam, Cambridge, UK) over night for 18 h. CREB3L1 protein staining was quantified by a pathologist using an adapted immuno-reactive scoring system (IRS) according to Remmele and Stegner.Citation45
Western blotting
Western blot analysis was performed as described previouslyCitation44 but slightly modified as follows: Blocked blots were probed with anti-β-actin (1:2,000, A5316, Sigma-Aldrich, Deisenheim, Germany) or anti-CREB3L1 (1:10,000, 11235–2-AP, Proteintech Europe, Manchester, UK) in blocking solution buffer, washed (0.01% (v/v) Tween20/PBS) and incubated with rabbit anti-mouse (DAKO, 1:8,000) or goat anti-rabbit (DAKO, 1:8,000) secondary peroxidase-conjugated antibody. Antibody detection was accomplished with Pierce ECL protein gel blotting substrate (Thermo Fisher Scientific, Bremen, Germany).
Nucleic acid extraction and reverse transcription PCR
DNA derived from FFPE bladder tissues was extracted using QIAmp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. Total RNA was isolated by using TRIzol reagent (Invitrogen, Carlsbad, CA). cDNA synthesis was performed by using the reverse transcription system (Promega, Madison, WI) as previously described.Citation46
Bisulphite-modification and methylation-specific PCR (MSP)
Bisulphite conversion and MSP reaction conditions of genomic and in vitro derived DNA was performed as specified previously.Citation47 For used HTRA3 MSP primers and cycle conditions see Supplementary Table 5.
Pyrosequencing
Pyrosequencing of 13 CpG sites within the CREB3L1 promoter region was performed by using the PyroMark PCR Kit (Qiagen) for initial fragment amplification. The PyroMark96 ID device and the PyroGoldSQA reagent Kit (Qiagen) were used as previously described.Citation44 The CREB3L1 assay was designed by using the Pyromark Assay Design Software (Qiagen) and all primers are listed in Supplementary Table 6.
Real-time PCR
Semiquantitative real-time PCR based on cDNAs was performed in an iCycler IQ5 (Bio-Rad Laboratories, Munich, Germany) using SYBR-Green PCR mix (Bio-Rad Laboratories) and quantified as previously shown.Citation46 Used CREB3L1 and GAPDH primers spanned at least one intron and are specified in Supplementary Table 7.
In vitro demethylation
Whole-genome demethylation of human bladder cell lines was carried out as recently shown.Citation46
Transfection of human bladder cancer cells
Human bladder cancer cell lines RT112, EJ28 and J82 were transiently transfected with either the pT-Rex-DEST30 (empty vector) or the CREB3L1-pT-Rex-DEST30 expression vector (Source BioScience, Berlin, Germany), containing the full-length human CREB3L1 cDNA, by using Fugene 6 according to the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany). In case of the transient in vitro model, 72 h after transfection, functional wound healing assays were performed. Furthermore, J82 cancer cells were also stably transfected with both, the empty vector or CRE3BL1 vector construct, respectively. In order to ensure independent clones with genomic CREB3L1 integration, 48 h after transfection, the cells were single cell cloned by limiting dilution, and grown in media containing 0.13 mg/ml G418 for 2 weeks. Resistant single cell clones were expanded to 75 cm2 flasks and characterized by both real-time PCR and western blot analyses for expression of CREB3L1.
XTT proliferation assay
Colony growth of CREB3L1-transfected human bladder cancer cells J82 was analyzed by using the XTT proliferation assay as performed previously.Citation47
Migration assay
Migration of in vitro cultured, CREB3L1-transfected human bladder cancer cells (RT112, EJ28 and J82) was assessed by a wound healing assay as previously specified.Citation48
Colony formation assay
The clonogenic capability of CREB3L1-transfected human bladder cancer cells J82 was tested by a colony formation assay as previously described.Citation49
Statistical analysis
All statistical analyses were performed by using SPSS 20.0 (SPSS, Chicago, IL) or GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA). Data were considered statistically significant when the 2 sided p-value was equal or below 5%. In order to compare 2 groups, the non-parametric Mann-Whitney U-test was implemented, whereas in case of more than 2 generated groups the Kruskal-Wallis test and Dunn's multiple comparison test was used. Descriptive Fisher's exact tests and 2-sided log-rank tests were performed in order to correlate clinico-pathological parameters with CREB3L1 promoter methylation or CREB3L1 protein expression, respectively. Correlation between CREB3L1 expression (TCGA Illumina HiSeq platform) and CREB3L1 methylation data (TCGA HM450 platform) or HTRA3 expression (TCGA Illumina HiSeq platform) was performed by calculating a Spearman correlation coefficient.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Table_S7.doc
Download MS Word (36 KB)Table_S6.doc
Download MS Word (28.5 KB)Table_S5.doc
Download MS Word (31.5 KB)Table_S4.doc
Download MS Word (49 KB)Table_S3.doc
Download MS Word (62 KB)Table_S2.doc
Download MS Word (63 KB)Table_S1.doc
Download MS Word (57.5 KB)Figure_S1.doc
Download MS Word (102.5 KB)Acknowledgments
The excellent technical assistance of Roswitha Davtalab and Sonja von Serényi is thankfully acknowledged.
Funding
This work was supported by the START-program of the Medical Faculty of the RWTH Aachen University (grant number: 149/08).
References
- Honma Y, Kanazawa K, Mori T, Tanno Y, Tojo M, Kiyosawa H, Takeda J, Nikaido T, Tsukamoto T, Yokoya S, et al. Identification of a novel gene, OASIS, which encodes for a putative CREB/ATF family transcription factor in the long-term cultured astrocytes and gliotic tissue. Brain Res Mol Brain Res 1999; 69:93-103; http://dx.doi.org/10.1016/S0169-328X(99)00102-3
- Omori Y, Imai J, Suzuki Y, Watanabe S, Tanigami A, Sugano S. OASIS is a transcriptional activator of CREB/ATF family with a transmembrane domain. Biochem Biophys Res Commun 2002; 293:470-7; http://dx.doi.org/10.1016/S0006-291X(02)00253-X
- Chan CP, Kok KH, Jin DY. CREB3 subfamily transcription factors are not created equal: recent insights from global analyses and animal models. Cell Biosci 2011; 1:6; PMID:21711675; http://dx.doi.org/10.1186/2045-3701-1-6
- Asada R, Kanemoto S, Kondo S, Saito A, Imaizumi K. The signalling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellular physiology. J Biochem 2011; 149:507-18; http://dx.doi.org/10.1093/jb/mvr041
- Kondo S, Saito A, Asada R, Kanemoto S, Imaizumi K. Physiological unfolded protein response regulated by OASIS family members, transmembrane bZIP transcription factors. IUBMB Life 2011; 63:233-9; http://dx.doi.org/10.1002/iub.433
- Murakami T, Kondo S, Ogata M, Kanemoto S, Saito A, Wanaka A, Imaizumi K. Cleavage of the membrane-bound transcription factor OASIS in response to endoplasmic reticulum stress. J Neurochem 2006; 96:1090-100; http://dx.doi.org/10.1111/j.1471-4159.2005.03596.x
- Schubert SW, Abendroth A, Kilian K, Vogler T, Mayr B, Knerr I, Hashemolhosseini S. bZIP-Type transcription factors CREB and OASIS bind and stimulate the promoter of the mammalian transcription factor GCMa/Gcm1 in trophoblast cells. Nucleic Acids Res 2008; 36:3834-46; PMID:18495750; http://dx.doi.org/10.1093/nar/gkn306
- Saito A, Kanemoto S, Kawasaki N, Asada R, Iwamoto H, Oki M, Miyagi H, Izumi S, Sanosaka T, Nakashima K, et al. Unfolded protein response, activated by OASIS family transcription factors, promotes astrocyte differentiation. Nat Commun 2012; 3:967
- Murakami T, Saito A, Hino S, Kondo S, Kanemoto S, Chihara K, Sekiya H, Tsumagari K, Ochiai K, Yoshinaga K, et al. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol 2009; 11:1205-11
- Murakami T, Hino S, Nishimura R, Yoneda T, Wanaka A, Imaizumi K. Distinct mechanisms are responsible for osteopenia and growth retardation in OASIS-deficient mice. Bone 2011; 48:514-23; PMID:21047569; http://dx.doi.org/10.1016/j.bone.2010.10.176
- Vellanki RN, Zhang L, Guney MA, Rocheleau JV, Gannon M, Volchuk A. OASIS/CREB3L1 induces expression of genes involved in extracellular matrix production but not classical endoplasmic reticulum stress response genes in pancreatic β-cells. Endocrinology 2010; 151:4146-57; PMID:20668028; http://dx.doi.org/10.1210/en.2010-0137
- Chen Q, Denard B, Huang H, Ye J. Epigenetic silencing of antiviral genes renders clones of Huh-7 cells permissive for hepatitis C virus replication. J Virol 2013; 87:659-65; http://dx.doi.org/10.1128/JVI.01984-12
- Denard B, Lee C, Ye J. Doxorubicin blocks proliferation of cancer cells through proteolytic activation of CREB3L1. Elife 2012; 1:e00090; PMID:23256041; http://dx.doi.org/10.7554/eLife.00090
- Mertens F, Fletcher CD, Antonescu CR, Coindre JM, Colecchia M, Domanski HA, Downs-Kelly E, Fisher C, Goldblum JR, Guillou L, et al. Clinicopathologic and molecular genetic characterization of low-grade fibromyxoid sarcoma, and cloning of a novel FUS/CREB3L1 fusion gene. Lab Invest 2005; 85:408-15; PMID:15640831; http://dx.doi.org/10.1038/labinvest.3700230
- Debelenko LV, McGregor LM, Shivakumar BR, Dorfman HD, Raimondi SC. A novel EWSR1-CREB3L1 fusion transcript in a case of small cell osteosarcoma. Genes Chromosomes Cancer 2011; 50:1054-62; http://dx.doi.org/10.1002/gcc.20923
- Mellor P, Deibert L, Calvert B, Bonham K, Carlsen SA, Anderson DH. CREB3L1 is a metastasis suppressor that represses expression of genes regulating metastasis, invasion, and angiogenesis. Mol Cell Biol 2013; 33:4985-95
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69-90
- Hurst CD, Knowles MA. Molecular subtyping of invasive bladder cancer: time to divide and rule? Cancer Cell 2014; 25:135-6; PMID:24525229; http://dx.doi.org/10.1016/j.ccr.2014.01.026
- Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer 2005; 5:713-25; PMID:16110317; http://dx.doi.org/10.1038/nrc1697
- Knowles MA. Molecular subtypes of bladder cancer: Jekyll and Hyde or chalk and cheese? Carcinogenesis 2006; 27:361-73; PMID:16352616; http://dx.doi.org/10.1093/carcin/bgi310
- Wild PJ, Herr A, Wissmann C, Stoehr R, Rosenthal A, Zaak D, Simon R, Knuechel R, Pilarsky C, Hartmann A. Gene expression profiling of progressive papillary noninvasive carcinomas of the urinary bladder. Clin Cancer Res 2005; 11:4415-29
- Steenbergen RD, Ongenaert M, Snellenberg S, Trooskens G, van der Meide WF, Pandey D, Bloushtain-Qimron N, Polyak K, Meijer CJ, Snijders PJ, et al. Methylation-specific digital karyotyping of HPV16E6E7-expressing human keratinocytes identifies novel methylation events in cervical carcinogenesis. J Pathol 2013; 231:53-62; http://dx.doi.org/10.1002/path.4210
- Masters JR, Hepburn PJ, Walker L, Highman WJ, Trejdosiewicz LK, Povey S, Parkar M, Hill BT, Riddle PR, Franks LM. Tissue culture model of transitional cell carcinoma: characterization of twenty-two human urothelial cell lines. Cancer Res 1986; 46:3630-6; PMID:3708594
- Tocharus J, Tsuchiya A, Kajikawa M, Ueta Y, Oka C, Kawaichi M. Developmentally regulated expression of mouse HtrA3 and its role as an inhibitor of TGF-β signaling. Dev Growth Differ 2004; 46:257-74; http://dx.doi.org/10.1111/j.1440-169X.2004.00743.x
- Beleford D, Liu Z, Rattan R, Quagliuolo L, Boccellino M, Baldi A, Maguire J, Staub J, Molina J, Shridhar V. Methylation induced gene silencing of HtrA3 in smoking-related lung cancer. Clin Cancer Res 2010; 16:398-409
- Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 2006; 124:587-99; PMID:16469704; http://dx.doi.org/10.1016/j.cell.2005.11.040
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 2008; 134:743-56; PMID:18775308; http://dx.doi.org/10.1016/j.cell.2008.07.021
- Zhang C, Wang G, Zheng Z, Maddipati KR, Zhang X, Dyson G, Williams P, Duncan SA, Kaufman RJ, Zhang K. Endoplasmic reticulum-tethered transcription factor cAMP responsive element-binding protein, hepatocyte specific, regulates hepatic lipogenesis, fatty acid oxidation, and lipolysis upon metabolic stress in mice. Hepatology 2012; 55:1070-82; PMID:22095841; http://dx.doi.org/10.1002/hep.24783
- Tsutsumi S, Namba T, Tanaka KI, Arai Y, Ishihara T, Aburaya M, Mima S, Hoshino T, Mizushima T. Celecoxib upregulates endoplasmic reticulum chaperones that inhibit celecoxib-induced apoptosis in human gastric cells. Oncogene 2006; 25:1018-29; PMID:16205636; http://dx.doi.org/10.1038/sj.onc.1209139
- Bobrovnikova-Marjon E, Grigoriadou C, Pytel D, Zhang F, Ye J, Koumenis C, Cavener D, Diehl JA. PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene 2010; 29:3881-95; PMID:20453876; http://dx.doi.org/10.1038/onc.2010.153
- Wang WA, Groenendyk J, Michalak M. Endoplasmic reticulum stress associated responses in cancer. Biochim Biophys Acta 2014.
- Fels DR, Koumenis C. The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther 2006; 5:723-8.
- Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res 2007; 67:3496-9; PMID:17440054; http://dx.doi.org/10.1158/0008-5472.CAN-07-0325
- Kadowaki H, Nishitoh H. Signaling pathways from the endoplasmic reticulum and their roles in disease. Genes (Basel) 2013; 4:306-33; PMID:24705207
- Denard B, Seemann J, Chen Q, Gay A, Huang H, Chen Y, Ye J. The membrane-bound transcription factor CREB3L1 is activated in response to virus infection to inhibit proliferation of virus-infected cells. Cell Host Microbe 2011; 10:65-74; http://dx.doi.org/10.1016/j.chom.2011.06.006
- Esteller M. Epigenetics in cancer. N Engl J Med 2008; 358:1148-59; http://dx.doi.org/10.1056/NEJMra072067
- Kondo S, Hino SI, Saito A, Kanemoto S, Kawasaki N, Asada R, Izumi S, Iwamoto H, Oki M, Miyagi H, et al. Activation of OASIS family, ER stress transducers, is dependent on its stabilization. Cell Death Differ 2012; 19:1939-49.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/10.1016/j.cell.2011.02.013
- Nie GY, Hampton A, Li Y, Findlay JK, Salamonsen LA. Identification and cloning of two isoforms of human high-temperature requirement factor A3 (HtrA3), characterization of its genomic structure and comparison of its tissue distribution with HtrA1 and HtrA2. Biochem J 2003; 371:39-48
- Chien J, Ota T, Aletti G, Shridhar R, Boccellino M, Quagliuolo L, Baldi A, Shridhar V. Serine protease HtrA1 associates with microtubules and inhibits cell migration. Mol Cell Biol 2009; 29:4177-87
- Singh H, Li Y, Fuller PJ, Harrison C, Rao J, Stephens AN, Nie G. HtrA3 is downregulated in cancer cell lines and significantly reduced in primary serous and granulosa cell ovarian tumors. J Cancer 2013; 4:152-64
- Yin Y, Wu M, Nie G, Wang K, Wei J, Zhao M, Chen Q. HtrA3 is negatively correlated with lymph node metastasis in invasive ductal breast cancer. Tumour Biol 2013; 34:3611-7.
- Singh H, Makino SI, Endo Y, Nie G. Inhibition of HTRA3 stimulates trophoblast invasion during human placental development. Placenta 2010; 31:1085-92; PMID:21035848; http://dx.doi.org/10.1016/j.placenta.2010.10.003
- Rose M, Gaisa NT, Antony P, Fiedler D, Heidenreich A, Otto W, Denzinger S, Bertz S, Hartmann A, Karl A, et al. Epigenetic inactivation of ITIH5 promotes bladder cancer progression and predicts early relapse of pT1 high-grade urothelial tumours. Carcinogenesis 2014; 35:727-36; PMID:24265292; http://dx.doi.org/10.1093/carcin/bgt375
- Remmele W, Stegner HE. [Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue]. Pathologe 1987; 8:138-40.
- Noetzel E, Rose M, Sevinc E, Hilgers RD, Hartmann A, Naami A, Knuchel R, Dahl E. Intermediate filament dynamics and breast cancer: aberrant promoter methylation of the Synemin gene is associated with early tumor relapse. Oncogene 2010; 29:4814-25; PMID:20543860; http://dx.doi.org/10.1038/onc.2010.229
- Veeck J, Chorovicer M, Naami A, Breuer E, Zafrakas M, Bektas N, Durst M, Kristiansen G, Wild PJ, Hartmann A, et al. The extracellular matrix protein ITIH5 is a novel prognostic marker in invasive node-negative breast cancer and its aberrant expression is caused by promoter hypermethylation. Oncogene 2008; 27:865-76; PMID:17653090; http://dx.doi.org/10.1038/sj.onc.1210669
- Kristiansen G, Hu J, Wichmann D, Stiehl DP, Rose M, Gerhardt J, Bohnert A, ten HA, Moch H, Raleigh J, et al. Endogenous myoglobin in breast cancer is hypoxia-inducible by alternative transcription and functions to impair mitochondrial activity: a role in tumor suppression? J Biol Chem 2011; 286:43417-28; http://dx.doi.org/10.1074/jbc.M111.227553
- Noetzel E, Rose M, Bornemann J, Gajewski M, Knuchel R, Dahl E. Nuclear transport receptor karyopherin-alpha2 promotes malignant breast cancer phenotypes in vitro. Oncogene 2012; 31:2101-14; PMID:21909132; http://dx.doi.org/10.1038/onc.2011.403
