Abstract
Poly(A) status is the major determinant of mRNA stability, even in endosymbiotic organelles. Poly(A) specific ribonuclease (PARN) is distributed widely among eukaryotes and has been shown to regulate the poly(A) status of cytoplasmic mRNA in various organisms. Surprisingly, our recent study revealed that PARN also directly regulates poly(A) status of mitochondrial mRNA in Arabidopsis. In this addendum, we discuss whether this mitochondrial function of PARN is common in plants and why PARN has been assigned such a unique function.
Introduction
Eukaryotes have endosymbiotic organelles, namely mitochondria and plastids. The idea that these organelles originated from symbiont bacteria has been widely accepted. During the establishment of endosymbiotic organelles, most genes from the symbiont genome were lost or transferred to the nuclear genome, presumably allowing the host to control the behavior of the endosymbiotic organelle. Despite this, the endosymbiotic organelles retained some genes, and the large protein complexes required for respiration in mitochondria are composed of both nucleus- and organelle-encoded components in most eukaryotes.Citation1 This necessitates tightly coordinated gene expression between the nuclear and organellar genomes. It has been postulated that signaling from organelles to the nucleus/cytoplasm (retrograde signaling) and that from the nucleus/cytoplasm to organelles (anterograde signaling) is involved in achieving such coordinated gene expression.Citation2-4 Although neither signaling system has been fully elucidated, they are thought to involve regulation at the levels of transcription, post-transcription, and post-translation, in both the nucleus and organelles. Such regulatory events have been extensively studied for nuclear genome-encoded genes. By contrast, those for plant mitochondrial genes are poorly understood, although there has been progress from recent studies especially on regulation at the post-transcriptional level.Citation5 The current knowledge strongly implies that plants have evolved a mitochondrial post-transcriptional regulation system that is unique among eukaryotes in spite of the single origin of mitochondria in the Eukaryota, which is supported by phylogenetic and phylogenomic analyses.Citation6 For example, plants possess a large number of pentatricopeptide repeat (PPR) proteins that are involved in various steps of RNA metabolism such as intron splicing and RNA editing in mitochondria and plastids.Citation7 Recently, we obtained data offering more evidence for unique mitochondrial RNA regulation in plants. We found that the poly(A) specific ribonuclease (PARN) of Arabidopsis thaliana directly regulates the poly(A) status of mitochondrial mRNA.Citation8 This finding is surprising given that PARN is distributed widely among eukaryotes (other than budding yeast and DrosophilaCitation9) and has been shown to regulate the poly(A) status of cytoplasmic RNA in various organisms, but never in organelles.Citation10,11
Distinct functions of multiple deadenylases of eukaryotes
Poly(A) specific ribonuclease (PARN) is a eukaryotic deadenylase. Eukaryotes usually possess 3 types of deadenylases: the CCR4-NOT complex, the PAN2-PAN3 complex and PARN.Citation12,13 Recent studies have shown that the PAN2-PAN3 complex in mammalian cells initiates poly(A) tract shortening, which in turn facilitates further poly(A) removal by CCR4-NOT before mRNA degradation takes place.Citation11,14 The physiological role of PARN in animals seems slightly different from those of the other deadenylases. PARN removes the poly(A) tract of the target mRNA with unique cis elements but does not induce mRNA degradation. Instead, the resulting target mRNA lacking poly(A) is sequestered as stable, translationally silent mRNA in protein-mRNA complexes.Citation11
Plants also have multiple deadenylases. The Arabidopsis genome is predicted to contain 9 CCR4 genes, 11 CAF/NOT/POP2 genes, and 2 PARN genes. Although it is not clear whether these different types of deadenylases have any redundant functions, the results of genetic analyses suggest that they do have distinct functions. For example, the AtCAF1a-disrupted mutant shows reduced expression of biotic and abiotic stress-related genes, suggesting that this deadenylase has a pivotal (and non-redundant) role in stress responses in Arabidopsis.Citation15,16
PARN in plants
There are 2 genes annotated as PARN-like in Arabidopsis, but one of them, At3g25430, seems unlikely to encode a functional PARN enzyme because it would lack several conserved amino acid residues;Citation17 this remains to be tested experimentally, however. The other PARN gene, At5g55879, has been characterized in detail. This gene was reported to be indispensable for early development of Arabidopsis in 2 independent studies using T-DNA insertion mutants.Citation17,18 Both claimed that this Arabidopsis PARN was localized in the cytoplasm, consistent with observations in other eukaryotes. We genetically identified this gene as ABA HYPERSENSITIVE GERMINATION2 (AHG2) in a screen for ABA-hypersensitive mutants.Citation19,20 Interestingly, the ahg2–1 mutation also causes salicylic acid hypersensitivity.Citation21 The discrepancy between the phenotypes caused by the null mutations and those caused by ahg2–1 can be most simply explained by the weakness of the ahg2–1 mutation, which presumably only reduces the gene expression level.Citation20 Given the clear phenotype of the mutants, it has been postulated that Arabidopsis PARN has an important and specific role that cannot be compensated for by other deadenylases.
Taking advantage of the viability of the ahg2–1 mutant, we analyzed Arabidopsis PARN in detail. We found that (1) all of the known ahg2–1 phenotypes are suppressed by loss-of-function mutations in the AGS1 gene (At2g17580) encoding a bacterial-type poly(A) polymerase although the ags1 mutant allele alone does not cause any visible phenotypes, (2) mitochondrial mRNA is hyper-accumulated and abnormally polyadenylated in ahg2–1, (3) both AHG2/PARN- and AGS1-GFP fusion proteins localize to the mitochondria in transgenic plants, (4) removal of the putative N-terminal transit peptides of AHG2 or AGS1 reduces their activity in vivo.Citation8 We thus concluded that Arabidopsis PARN, along with AGS1, regulates mitochondrial mRNA poly(A) status.
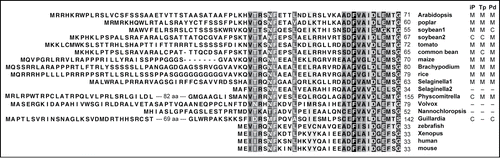
When we compared the amino acid sequence of Arabidopsis PARN with the predicted sequences of putative homologs from various species in the plant lineage, it became apparent that a non-conserved N-terminal extension sequence was common to PARNs of plants compared to those from animals (). Most of the higher plant PARNs were predicted to localize to mitochondria by 3 independent prediction programs, iPSORT, TargetP, and Predotar.Citation22-24 Selaginella moellendorffii (a lycophyte) has several PARN-like proteins. Interestingly, one of them (Selaginella1 in ) has an N-terminal extension while another (Selaginella2) does not. It is possible that the Selaginella1 PARN can function in mitochondria as well. In addition, the putative PARN from Physcomitrella (a bryophyte) has a 122-aa N-terminal extension that is predicted by TargetP and Predotar to direct the protein to mitochondria. Volvox also has a PARN with an N-terminal extension, and the genome database for the cryptophyte Guillardia theta contains a predicted PARN-like protein possessing a long N-terminal extension. Although none of the prediction programs supported their mitochondrial localization, there remains a possibility that the PARNs with long N-terminal extensions in these organisms localize to and function in mitochondria. In sum, most plant lineages appear to contain PARN genes encoding N-terminal extension sequences, strongly suggesting that PARN functions in mitochondria in these organisms, as we observed in Arabidopsis.
Figure 1. Comparison of the amino acid sequence of the N-terminal portion of poly(A) specific ribonucleases from various organisms. Highly conserved or relatively conserved amino acid residues are shaded in black and gray, respectively. The subcellular localization prediction results (M, mitochondria; C, plastids; –, other) from 3 prediction programs (iP, iPSORT; Tp, TargetP; Pd, Predotar) are summarized to the right of the species names. Accession numbers of PARN proteins: Arabidopsis, NP_175983.51; poplar, XP_002300380.2; soybean1, XP_003532241.1; soybean2, XP_003552994.1; tomato, XP_004244040.1; maize, NP_001169499.1; Brachypodium, XP_003581331.1; rice CAH66773.1; Selaginella1, XP_002969601.1; Selaginella2, XP_002987014.1; Physcomitrella, XP_001765586.1; Volvox, XP_002949475.1; Nannochloropsis, EWM30500.1; Guillardia, XP_005838739.1; zebrafish, AAQ97826.1; Xenopus, AAH73682.1; human, NP_002573.1; mouse, NP_083037.1.
Why is PARN used for mitochondrial post-transcriptional regulation in plants?
These observations raise the question of why plants use PARN for poly(A) regulation of mitochondrial mRNA. Although there is no clear answer at this moment, it could be related to the unique features of plant mitochondria, which have larger genomes and contain more protein-coding genes than do mitochondria from other eukaryotes. Presumably, the existence of another genome-possessing organelle, i.e., the plastid, plays a role in these plant-specific characteristics. In order to harbor 2 different types of symbiont (ancestral mitochondria and plastids), plants had to develop systems for tight and separate regulation of symbiont activities. For this purpose, it is possible that plants adopted PARN as a mitochondrion-specific post-transcriptional regulator controlling mRNA stability. Alternatively, PARN might be required to remove harmful non-coding RNAs that are transcribed from the mitochondrial genome. Plant mitochondrial genome is rearranged frequently by its high DNA recombination activity that is presumably required for maintenance of the genome.Citation25 But it in turn allows plant mitochondrial genomes to encode many non-functional genes and some intergenic regions of the mitochondrial genome to be transcribed.Citation26 Some of these transcripts or products could be deleterious to mitochondrial or cellular functions, eg. a case known as cytoplasmic male sterility. The idea that PARN has been recruited to remove these transcripts receives support from the fact that plants have a tremendous number of pentatricopeptide repeat proteins that are thought to suppress the expression of harmful mitochondrial genes or compensate for mutations occurring in mitochondrial genes at post-transcriptional levels, emphasizing the need for such protective activity.Citation7
Another question that arises is why PARN, from among the several types of deadenylases, has been selected for this function. One answer could be that PARN originally was not essential and/or had minor roles in the regulation of cytoplasmic mRNA. This seems plausible as PARN is involved in the poly(A) tail regulation of only a limited number of target mRNAs, and not in general mRNA decay. To date, the physiological importance of PARN has been described only for Xenopus oocytes and neuronal cells, in addition to Arabidopsis embryos. Such dispensability would allow plants to adapt PARN for use in different physiological roles, a kind of neofunctionalization.Citation27 It should be noted that PARN has been reported to function as a homo-dimer while other deadenylases act as large protein complexes. Therefore, it is possible that a mitochondrial post-translational regulation system involving PARN would be easier for plant cells to regulate than those with other deadenylases.
Conclusions and future perspectives
The AHG2-AGS1 regulatory system is novel and unique to plants. Therefore, the discovery of this system for mitochondrial mRNA poly(A) regulation has opened a new frontier for mitochondrial gene expression research. Mitochondria now attract more attention than ever, with studies suggesting mitochondrial involvement in cellular functions such as stress responses, in addition to respiration and metabolism. So far, the physiological relevance of the AHG2-AGS1 system is unclear. Interestingly, however, ahg2–1 causes abnormal responses to the stress-related plant hormones ABA and SA, implying that regulation by AHG2 or AGS1 is linked to stress responses. Further analysis of AHG2 and AGS1 should therefore shed light on the molecular nature of the link between mitochondria and stress responses in plants.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Grants-in-Aid for Scientific Research (B) (24370023) from MEXT Japan.
References
- Burger G, Gray MW, Franz Lang B. Mitochondrial genomes: anything goes. Trends Genet 2003; 19:709-16; PMID:14642752; http://dx.doi.org/10.1016/j.tig.2003.10.012
- Rhoads DM, Subbaiah CC. Mitochondrial retrograde regulation in plants. Mitochondrion 2007; 7:177-94; PMID:17320492; http://dx.doi.org/10.1016/j.mito.2007.01.002
- Nott A, Jung HS, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 2006; 57:739-59; PMID:16669780; http://dx.doi.org/10.1146/annurev.arplant.57.032905.105310
- Millar AH, Whelan J, Soole KL, Day DA. Organization and regulation of mitochondrial respiration in plants. Annu Rev Plant Biol 2011; 62:79-104; PMID:21332361; http://dx.doi.org/10.1146/annurev-arplant-042110-103857
- Hammani K, Giegé P. RNA metabolism in plant mitochondria. Trends Plant Sci 2014; 19:380-9; PMID:24462302; http://dx.doi.org/10.1016/j.tplants.2013.12.008
- Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol 2012; 4. a011403; PMID:22952398; http://dx.doi.org/10.1101/cshperspect.a011403
- Barkan A, Small I. PentatricopeptiderRepeat proteins in plants. Annu Rev Plant Biol 2014; 65:415-42; PMID:24471833; http://dx.doi.org/10.1146/annurev-arplant-050213-040159
- Hirayama T, Matsuura T, Ushiyama S, Narusaka M, Kurihara Y, Yasuda M, Ohtani M, Seki M, Demura T, Nakashita H, et al. A poly(A)-specific ribonuclease directly regulates the poly(A) status of mitochondrial mRNA in Arabidopsis. Nat Commun 2013; 4:2247; PMID:23912222
- Pavlopoulou A, Vlachakis D, Balatsos NAA, Kossida S. A comprehensive phylogenetic analysis of deadenylases. Evol Bioinforma Online 2013; 9:491-7
- Godwin AR, Kojima S, Green CB, Wilusz J. Kiss your tail goodbye: the role of PARN, nocturnin, and angel deadenylases in mRNA biology. Biochim Biophys Acta BBA–Gene Regul Mech 2013; 1829:571-9; http://dx.doi.org/10.1016/j.bbagrm.2012.12.004
- Norbury CJ. Cytoplasmic RNA: a case of the tail wagging the dog. Nat Rev Mol Cell Biol 2013; 14:643-53; PMID:23989958; http://dx.doi.org/10.1038/nrm3645
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 2007; 8:113-26; PMID:17245413; http://dx.doi.org/10.1038/nrm2104
- Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol 2008; 9:337-44; PMID:18334997; http://dx.doi.org/10.1038/nrm2370
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 2005; 12:1054-63; PMID:16284618; http://dx.doi.org/10.1038/nsmb1016
- Liang W, Li C, Liu F, Jiang H, Li S, Sun J, Wu X, Li C. The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res 2009; 19:307-16; PMID:19065152; http://dx.doi.org/10.1038/cr.2008.317
- Walley JW, Kelley DR, Nestorova G, Hirschberg DL, Dehesh K. Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol 2010; 152:866-875; PMID:19955262; http://dx.doi.org/10.1104/pp.109.149005
- Reverdatto SV, Dutko JA, Chekanova JA, Hamilton DA, Belostotsky DA. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA 2004; 10:1200-14; PMID:15247430; http://dx.doi.org/10.1261/rna.7540204
- Chiba Y, Johnson MA, Lidder P, Vogel JT, van Erp H, Green PJ. AtPARN is an essential poly(A) ribonuclease in Arabidopsis. Gene 2004; 328:95-102; PMID:15019988; http://dx.doi.org/10.1016/j.gene.2003.11.028
- Nishimura N, Yoshida T, Murayama M, Asami T, Shinozaki K, Hirayama T. Isolation and characterization of novel mutants affecting the abscisic acid sensitivity of Arabidopsis germination and seedling growth. Plant Cell Physiol 2004; 45:1485-99; PMID:15564532; http://dx.doi.org/10.1093/pcp/pch171
- Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T. Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J 2005; 44:972-84; PMID:16359390; http://dx.doi.org/10.1111/j.1365-313X.2005.02589.x
- Nishimura N, Okamoto M, Narusaka M, Yasuda M, Nakashita H, Shinozaki K, Narusaka Y, Hirayama T. ABA hypersensitive Germination2-1 causes the activation of both abscisic acid and salicylic acid responses in arabidopsis. Plant Cell Physiol 2009; 50:2112-22; PMID:19892832; http://dx.doi.org/10.1093/pcp/pcp146
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 2002; 18:298-305; PMID:11847077; http://dx.doi.org/10.1093/bioinformatics/18.2.298
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal Amino acid sequence. J Mol Biol 2000; 300:1005-16; PMID:10891285; http://dx.doi.org/10.1006/jmbi.2000.3903
- Small I, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. PROTEOMICS 2004; 4:1581-90; PMID:15174128; http://dx.doi.org/10.1002/pmic.200300776
- Marechal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol 2010; 186: 299-317; PMID:20180912; http://dx.doi.org/10.1111/j.1469-8137.2010.03195.x
- Holec S, Lange H, Kuhn K, Alioua M, Borner T, Gagliardi D. Relaxed transcription in arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol Cell Biol 2006; 26:2869-76; PMID:16537927; http://dx.doi.org/10.1128/MCB.26.7.2869-2876.2006
- Force A, Lynch M, Pickett FB, Amores A, Yan Y, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999; 151:1531-45; PMID:10101175
