Abstract
Abscisic acid (ABA) is an important regulator of guard cell ion channels and stomatal movements in response to drought stress. Pyruvate is the final product of glycolysis in the cytosol, and could be transported by mitochondrial pyruvate carriers (MPCs) into mitochondrion for consequent cellular substance and energy metabolism. We recently characterized the first putative mitochondrial pyruvate carrier, NRGA1, in planta, and found that this small protein is involved in the negative regulation of drought and ABA induced guard cell signaling in Arabidopsis thaliana. The findings revealed a probable link between mitochondrial pyruvate transport and guard cell signaling. It has also been shown that NRGA1 protein product was directed to the mitochondria, and co-expression of MPC1 and NRGA1 functionally complement the absence of a native pyruvate transport protein in yeast. Here, we further demonstrated that MPC1 showed similar sub-cellular localization pattern to NRGA1. Quantitative RT-PCR analysis showed that the transcription of both NRGA1 and MPC1 were induced by pyruvate or ABA, and pyruvate strengthened the ABA induced transcription of these 2 genes. The similarity in subcellular localization and gene expression to ABA strongly suggests that MPC1 may associate with NRGA1 for mitochondrial pyruvate transport and is involved in ABA mediated stomatal movements in Arabidopsis.
Pyruvate is the final product of cytosolic glycolysis and also a key node in the control of secondary metabolism of glucose, fatty acid and amino acid. Although this metabolite has been known to cross the inner mitochondrial membrane for decades, it is only recently that proteins (mitochondrial pyruvate carriers, MPCs) necessary for this activity have been identified.Citation1,2 In mammals, many studies have provided important information about the relationship with gluconeogenesis, MPC control by hormones and drugs and included relevance to pathological conditions including hyperthyroidism, aging, and diabetes.Citation3-7 However, very little is known about the actual biological functions and molecular mechanisms for MPCs in planta. We recently identified a putative MPC gene, Negative Regulator of Guard Cell ABA Signaling 1 (NRGA1), which mediates ABA regulation of guard cell ion channels and drought stress responses in Arabidopsis.Citation8 The Arabidopsis MPC family comprises 5 members (At4G05590, At4G14695, At5G20090, At4G22310 and At4G26780), and the phylogenetic analysis showed that these proteins are classified into 3 categories: MPC1 (At5G20090), MPC2-like proteins (At4G14695, At4G22310 and At4G05590) and At4G26780 (). A complementation analysis in yeast showed that AtMPC1 and NRGA1 together functionally complement the absence of a native pyruvate transport protein.Citation8 To assess whether MPC1 has direct interaction with NRGA1 or other putative MPC proteins, we performed yeast 2-hybrid assay. However, the result turned out that, no interaction was found between any of the different MPC protein combinations ().
Figure 1. Gene structure and protein interactions in yeast. (A) Phylogenetic analysis of putative MPC proteins in Arabidopsis. The amino acid sequences of these proteins were aligned by CLUSTALX, and the phylogenetic tree was constructed using the neighbor-joining method with the following parameters: bootstrap (replicates 1000), poisson model and complete deletion. The numbers at the nodes indicate the bootstrap values. (B) The interaction between the different MPC protein pairs. 32: pDEST32 for generation of the bait plasmid, 22: pDEST22 for construction of the prey plasmid. wt (pEXP22-RalGDS-wt), m1(pEXP22-RalGDS-m1) or m2 (pEXP22-RalGDS-m2) is control plasmid which shows strong, weak and undetectable interaction with pEXP32-Krev1, respectively. The transformed yeast cells grow on SC/-Leu-Trp (SC-L-T), SC/-Leu-Trp-Ura (SC-L-T-U), or SC/-Leu-Trp-His (SC-L-T-H) medium containing 50 mM 3-AT and aX-gal assay for β-galactosidase activity of the transformants grown on YPAD. (C) Test of protein interactions using a “swapped” 2-hybrid approach. The system is the same as shown in (B).
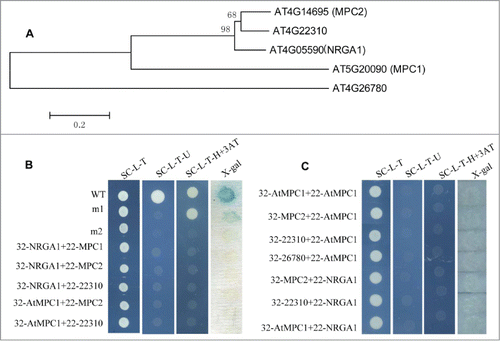
Immunoprecipitation and native gel electrophoresis conducted by Bricker et al.Citation1 suggest that the MPC is a large heterocomplex, potentially with unequal stoichiometry between the 2 proteins, thus the interaction between MPC1 and NRGA1 might needs the help of other proteins essential for mitochondrial pyruvate transport in Arabidopsis. Alternatively, the yeast 2-hybrid system based on transcriptional activation may not be suitable to detect the interaction of MPCs for that their interactions may not occur in the nucleus. Other experimental systems, such as co-immunoprecipitation or bimolecular fluorescence complementation (BiFC), will be needed further to identify the protein complex of MPCs in Arabidopsis.
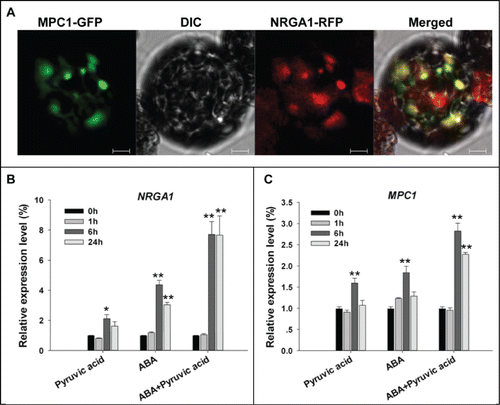
Functional analysis with the high-affinity MPC inhibitor UK5099Citation9 implicated MPC1 as a crucial mediator of pyruvate transport activity. Since the yeast mpc2Δmpc3Δ cells were rescued by coexpressing MPC1 and NRGA1,Citation8 the products of these genes presumably had a similar subcellular localization pattern. The subcellular localization of MPC1 was determined by transiently expressing in Arabidopsis mesophyll protoplasts a fusion of GFP to the MPC1 terminus driven by the CaMV 35S promoter. Previously 35S::NRGA1-GFP was shown to be targeted to mitochondria.Citation8 To confirm the location of MPC1, the 35S::MPC1-GFP was co-transformed into protoplasts along with 35S::NRGA1-RFP. As the GFP and RFP signals substantially overlapped one another (), the conclusion was that MPC1 is also deposited in the mitochondria and co-localized with NRGA1. Our previous work has demonstrated that ABA treatment induced NRGA1 transcription, we next asked if MPC1 had a similar expression pattern to NRGA1 in response to ABA. We also show that external application of pyruvate is able to regulate stomatal movements and enhance the ABA sensitivity.Citation8 To make clear the role of pyruvate itself or together with ABA in regulating the transcription of NRGA1 and MPC1, quantitative RT-PCR was performed by using the wild type seedlings over a time course exposed to ABA, pyruvate or ABA and pyruvate. The result revealed that, the transcription of NRGA1 was induced significantly by 200 μM pyruvate or 100 uM ABA at 6 h and 24 h, and pyruvate obviously enhanced the ABA induced NRGA1 transcription (). Although pyruvate- or ABA- induced transcription of MPC1 was not as obvious as that of NRGA1, it showed similar response pattern to NRGA1, and MPC1 transcripts also increased significantly when pyruvate and ABA were added simultaneously (). The combined data suggest that MPC1 may associate with NRGA1 and act in mitochondrial pyruvate transport and ABA mediated stomatal movements in Arabidopsis, and we are investigating this further.
Figure 2. Subcellular localization of MPC1 and its gene transcription behavior. (A) Colocalization of GFP-tagged MPC1 and RFP-tagged NRGA1. The yellow signal indicates areas of overlap between the green and red signals. Quantitative RT-PCR of NRGA1 (B) and MPC1 (C) transcript under 200 μM pyruvic acid, 100 μM ABA or both were analyzed. Transcript levels relative to that of ACTIN2 are presented for each treatment. Each value represents the mean±SE of 3 independent measurements. The *, ** represent significant difference determined by the Student's t-test at P < 0.05 and P < 0.01 respectively. Bars = 10 μm.
Sugar signaling pathways closely interact with phytohormones.Citation10 Genes involved in ABA biosynthesis and signaling have been successfully isolated in sugar-response screens.Citation11-15 In Arabidopsis, the early seedling development repression has been widely used as a convenient way of identifying mutants affected in sugar-sensing or signaling processes.Citation13,14 In our laboratory, evidence that external application of pyruvate is able to inhibit the seed germination and enhance the ABA sensitivity has been acquired (data not shown). Together with the observation that pyruvate regulates stomotal movements and gene expression, we hypothesize that pyruvate serves as a signal molecule and crosstalk with ABA to regulate the plant growth, development and responses to the changing environment. More physiological, biochemical and molecular approaches will be further needed to study pyruvate responses in plants.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
We would like to thank the National Natural Science Foundation of China (31170236 and 31271506 to W.Z.), the key special project “Breeding and Cultivation of Novel GM varieties” (2014ZX08009–022B to W.Z.) for funding support.
References
- Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 2012; 337:96-100; PMID:22628558; http://dx.doi.org/10.1126/science.1218099
- Herzig S, Raemy E, Montessuit S, Veuthey JL, Zamboni N, Westermann B, Kunji ER, Martinou JC. Identification and functional expression of the mitochondrial pyruvate carrier. Science 2012; 337:93-6; PMID:22628554; http://dx.doi.org/10.1126/science.1218530
- Mendes-Mourao J, Halestrap AP, Crisp DM, Pogson CI. The involvement of mitochondrial pyruvate transport in the pathways of gluconeogenesis from serine and alanine in isolated rat and mouse liver cells. FEBS Lett 1975; 53:29-32; PMID:1140393; http://dx.doi.org/10.1016/0014-5793(75)80674-0
- Paradies G, Petrosillo G, Gadaleta MN, Ruggiero FM. The effect of aging and acetyl-L-carnitine on the pyruvate transport and oxidation in rat heart mitochondria. FEBS Lett 1999; 454:207-9; PMID:10431808; http://dx.doi.org/10.1016/S0014-5793(99)00809-1
- Proudlove MO, Beechey RB, Moore AL. Pyruvate transport by thermogenic-tissue mitochondria. Biochem J 1987; 247:441-7; PMID:3426546
- Titheradge MA, Coore HG. Hormonal regulation of liver mitochondrial pyruvate carrier in relation to gluconeogenesis and lipogenesis. FEBS Lett 1976; 72:73-8; PMID:11133; http://dx.doi.org/10.1016/0014-5793(76)80901-5
- Kielducka A, Paradies G, Papa S. A comparative study of the transport of pyruvate in liver mitochondria from normal and diabetic rats. J Bioenerg Biomembr 1981; 13:123-32; PMID:7309723; http://dx.doi.org/10.1007/BF00763834
- Li CL, Wang M, Ma XY, Zhang W. NRGA1, a putative mitochondrial pyruvate carrier, mediates ABA regulation of guard cell ion channels and drought stress responses in Arabidopsis. Mol Plant 2014; 7:1508-21; PMID:24842572; http://dx.doi.org/10.1093/mp/ssu061
- Halestrap AP. The mitochondrial pyruvate carrier. Kinetics and specificity for substrates and inhibitors. Biochem J 1975; 148:85-96; PMID:1156402
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 2003; 300:332-6; PMID:12690200; http://dx.doi.org/10.1126/science.1080585
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, Leon P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 2000; 14:2085-96; PMID:10950871
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S. The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant J 2000; 23:577-85; PMID:10972884; http://dx.doi.org/10.1046/j.1365-313x.2000.00822.x
- Laby RJ, Kincaid MS, Kim D, Gibson SI. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 2000; 23:587-96; PMID:10972885; http://dx.doi.org/10.1046/j.1365-313x.2000.00833.x
- Zhou L, Jang JC, Jones TL, Sheen J. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA 1998; 95:10294-9; PMID:9707641; http://dx.doi.org/10.1073/pnas.95.17.10294
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J 2001; 26:421-33; PMID:11439129; http://dx.doi.org/10.1046/j.1365-313X.2001.2641043.x
