Abstract
The unconventional, lysine-63-linked ubiquitination has been shown to play a central role in regulating human and animal innate and adaptive immunity. By contrast, the role and mechanism of K63-linked ubiquitination in plant biology remain largely unexplored. The tomato (Solanum lycopersicum) Fni3 ubiquitin-conjugating enzyme and its co-factor, Suv ubiquitin E2 variant (Uev) were shown recently to catalyze K63-linked ubiquitination and are essential for protein Fen and other resistance protein-mediated plant immunity. In this study we detected the subcellular localization of Fen, Fni3 and Suv and confirmed the interaction of Fni3 with Suv in tomato protoplasts. Additionally, we identified 2 tomato Uev1 homologs, SlUev1C and SlUev1D, respectively and showed they are not required for Fen-mediated programmed cell death in Nicotiana benthamiana, suggesting Uev homologs play differential role in the cell.
Plants have evolved a sophisticated innate immune system to ward off infection by many pathogens.Citation1 Ubiquitination, a major post-translational protein modification process in eukaryotic cells, has emerged in recent years as a key component of plant immune system and the importance of ubiquitination in the regulation of plant immunity has been increasingly appreciated.Citation2-4
Conventionally, ubiquitination is known as lysine (K)-48-linked polyubiquitination that serves as the principal signal for 26S proteasome-mediated protein degradation.Citation5 Nevertheless, various types of unconventional ubiquitination including mono-ubiquitination and poly-ubiquitination linking through other lysine residues of the ubiquitin molecule have also been discovered to exist commonly and many serve as non-degradative, regulatory signals.Citation6 For example, the K63-linked polyubiquitination that is often catalyzed by the Ubc13 ubiquitin-conjugating enzyme (E2) and its co-factor, a ubiquitin E2 variant (Uev) has hitherto been shown to play non-proteolytic, regulative role in several physiological processes. In particular, K63-linked ubiquitination has been demonstrated to be a central player in the regulation of human and animal innate and adaptive immunity.Citation7
Tomato (Solanum lycopersicum) immunity against the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) is mainly conferred by the Ser/Thr kinase protein Pto, which is also dependent on a leucine-rich repeat-containing protein, Prf. In addition to Pto, another tomato kinase protein, Fen also confers immunity against certain strains of the Pst pathogen.Citation8 The Fen protein contains a putative N-myristoylation signal and a mutation in the signal renders the Fen protein functionally inactive.Citation9,10 The myristoylation signal usually targets a protein to membrane of the cell. To assess whether Fen is localized to the membrane region, we examined the localization of Fen-GFP fusion protein in tomato protoplasts derived from tomato pto11 plants.Citation8,11 Interestingly, Fen was found to localize to both nucleus and cytoplasm regardless the GFP protein is fused to the N- or C-terminal of Fen, which is similar to the localization of the GFP protein alone ().
Figure 1. Subcellular localization and bimolecular fluorescence complementation (BiFC) assay using tomato protoplasts. (A). The subcellular localization of Fen, Fni3 and Suv in tomato protoplast. Except for Fen-cGFP in which GFP was fused to the C-terminal of Fen, the GFP protein was in the N terminal in all other fusion proteins. (B) Examination of interaction of Fen and Fni3 with Suv using bimolecular fluorescence complementation (BiFC) assay. Presence of green fluorescence denotes the occurrence of interaction of the 2 proteins in the cell.
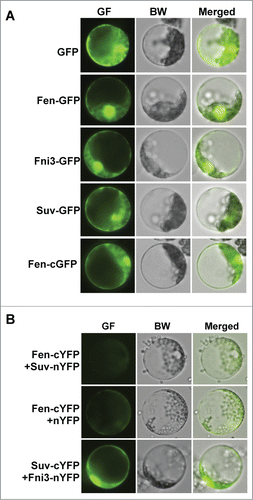
The tomato Fni3 protein is a homolog of the Ubc13 type ubiquitin E2 enzyme. The Suv Uev protein acted as the co-factor of Fni3 in catalyzing K63-specific ubiquitination.Citation12 Ubc13 and its homologs are unique among ubiquitin-conjugating enzymes in that they catalyze exclusively K63-linked ubiquitination and a Uev which serves as co-factor is essential for their catalytic activity. Fni3 was found to interact with Fen and Suv in yeast 2-hybrid (Y2H) and GST pull-down assay but Suv did not interact with Fen in the assays.Citation12 Similar to the subcellular localization of Fen, both Fni3 and Suv are targeted to nucleus and cytoplasm of tomato protoplast (). Using yellow fluorescence protein (YFP)-based bimolecular fluorescence complementation (BiFC) assay, we confirmed Fni3 interacts with Suv in tomato cell but no interaction between Fen and Suv was detected, which is in consistence with the findings in Y2H and pull-down assay ().
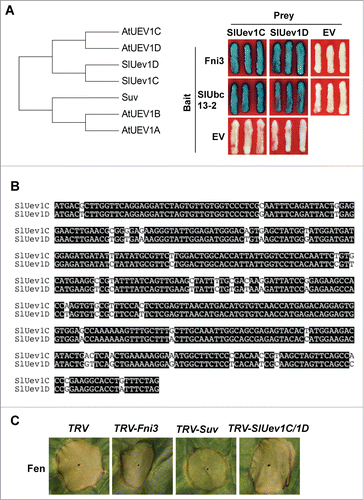
Previously four homologous Uev1 proteins, UEV1A-1D were identified from Arabidopsis and Uev1D was shown to be involved in DNA damage response.Citation13 Using Blast search of the Sol Genomics Network (SGN) database (http://solgenomics.net) we identified two homologs of Suv from tomato genome. Phylogenetic analysis indicated the two tomato Suv homologs show higher similarity to AtUEV1C and 1D than to AtUEV1A and 1B and they were named SlUev1C and 1D, respectively (, left panel). SlUev1C and 1D share 91.6% identity in nucleotide sequence () and 98.5% identity in amino acid sequence (data not shown). Compared to SlUev1C and SlUev1D, Suv is more homologous to AtUEV1B. We were, however, unable to identify the close tomato homolog of AtUEV1A from the SGN database. Since Uev proteins often interact with Ubc13 type ubiquitin E2 enzyme acting as a co-factor in catalyzing K63-linked ubiquitination, we tested whether SlUev1C and SlUev1D interact with Fni3 and its homolog, SlUbc13-2 using Y2H.Citation12 Indeed, both SlUev1C and SlUev1D interacted with Fni3 and SlUbc13-2, respectively (, right panel). The interaction of SlUev1C and SlUev1D with Fni3 and SlUbc13-2 prompted us to examine if, like Suv, the SlUev1C and SlUev1D genes are required for programmed cell death (PCD) induced by overexpression of Fen in Nicotiana benthamiana plants.Citation12 To this end we used the tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) system to silence SlUev1C and SlUev1D gene in N. benthamiana plants.Citation14 We then overexpressed the Fen protein on the leaves of these plants using Agrobacterium-mediated transient expression as described previously.Citation15 We also performed VIGS using the Fni3 and Suv gene and the TRV empty vector as control. As shown in , overexpression of Fen in TRV control plants resulted in strong cell death and silencing of the Fni3 and Suv gene significantly diminished PCD triggered by overexpression of Fen, which is in consistence with previous findings.Citation12 By contrast, silencing of SlUev1C and SlUev1D gene did not affect Fen-mediated PCD. This differential requirement of tomato Uev proteins for Fen-mediated PCD in N. benthamiana suggests that they may play different role in the cell.
Figure 2. SlUev1C and SlUev1D are not involved in Fen-mediated PCD in Nicotiana benthamiana. (A) Phylogenetic tree of AtUEV1A-1D and tomato Uev1 homologs. The Amino acid sequences of the proteins were aligned using Clustal W, which was followed by generation of phylogenetic tree using MEGA 6.Citation16,17 (B) Interaction of SlUev1C and SlUev1D with Fni3 and SlUbc13-2, respectively in yeast cells. (C) Alignment of the nucleotide sequence of SlUev1C and 1D using Clustal W. (D) Silencing SlUev1C and SlUev1D gene did not affect Fen-mediated cell death in N. benthamiana leaf.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Support for this work is in part from the National Science Foundation (grant #1052495), United States Department of Agriculture/National Institute of Food and Agriculture (grant # 2012-67014-19449) and University of Arkansas at Little Rock.
References
- Jones JDG, Dangl JL. The plant immune system. Nature 2006; 444:323-9; PMID:17108957; http://dx.doi.org/10.1038/nature05286
- Cheng YT, Li X. Ubiquitination in NB-LRR-mediated immunity. Curr Opin Plant Biol 2012; 15:392-9; PMID:22503756; http://dx.doi.org/10.1016/j.pbi.2012.03.014
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012; 486:228-32; PMID:22699612
- Zeng L-R, Vega-Sánchez ME, Zhu T, Wang G-L. Ubiquitination-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res 2006; 16:413-26; PMID:16699537; http://dx.doi.org/10.1038/sj.cr.7310053
- Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem 1992; 61:761-807; PMID:1323239; http://dx.doi.org/10.1146/annurev.bi.61.070192.003553
- Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans 2009; 37:937-53; PMID:19754430; http://dx.doi.org/10.1042/BST0370937
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature 2009; 458:430-7; PMID:19325622; http://dx.doi.org/10.1038/nature07959
- Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, Martin GB. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 2007; 448:370-4; PMID:17637671; http://dx.doi.org/10.1038/nature05966
- Loh YT, Zhou J, Martin GB. The myristylation motif of Pto is not required for disease resistance. Mol Plant Microbe Interact 1998; 11:572-6; PMID:9612955; http://dx.doi.org/10.1094/MPMI.1998.11.6.572
- Rommens CM, Salmeron JM, Baulcombe DC, Staskawicz BJ. Use of a gene expression system based on potato virus X to rapidly identify and characterize a tomato Pto homolog that controls fenthion sensitivity. Plant Cell 1995; 7:249-57; PMID:7734960; http://dx.doi.org/10.1105/tpc.7.3.249
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 2002; 31:375-83; PMID:12164816; http://dx.doi.org/10.1046/j.1365-313X.2002.01360.x
- Mural RV, Liu Y, Rosebrock TR, Brady JJ, Hamera S, Connor RA, Martin GB, Zeng L. The tomato fni3 lysine-63-specific ubiquitin-conjugating enzyme and suv ubiquitin e2 variant positively regulate plant immunity. Plant Cell 2013; 25:3615-31; PMID:24076975; http://dx.doi.org/10.1105/tpc.113.117093
- Wen R, Torres-Acosta JA, Pastushok L, Lai X, Pelzer L, Wang H, Xiao W. Arabidopsis UEV1D promotes Lysine-63-linked polyubiquitination and is involved in DNA damage response. Plant Cell 2008; 20:213-27; PMID:18178771; http://dx.doi.org/10.1105/tpc.107.051862
- Abramovitch RB, Kim Y-J, Chen S, Dickman MB, Martin GB. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J 2003; 22:60-9; PMID:12505984; http://dx.doi.org/10.1093/emboj/cdg006
- del Pozo O, Pedley KF, Martin GB. MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 2004; 23:3072-82; PMID:15272302; http://dx.doi.org/10.1038/sj.emboj.7600283
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947-8; PMID:17846036; http://dx.doi.org/10.1093/bioinformatics/btm404
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725-9; PMID:24132122; http://dx.doi.org/10.1093/molbev/mst197
