Abstract
The SALK_135513 line of Arabidopsis thaliana is annotated by GenBank to have the T-DNA insertion in the fourth exon of NBR1 (At4g24690). Careful molecular analyses of the homozygous plants of SALK_135513 line indicated the place of T-DNA insertion in the fourth intron. Unexpectedly, 2 kinds of NBR1 transcripts, the wild-type and the mutated, resulting from alternative splicing events, were detected in those plants. Our findings explain the problems encountered by us with phenotypic evaluation of this line and emphasize the necessity for independent verification of the exact insertion site followed by careful expression studies when working with Arabidopsis T-DNA insertional mutants.
Abbreviations
| NBR1 | = | Neighbor of BRCA1 |
| WT | = | wild-type |
| T-DNA | = | transferred DNA |
| KO | = | knock-out |
| MV | = | methyl viologen |
| sqRT-PCR | = | semiquantitative RT-PCR |
| qRT-PCR | = | quantitative RT-PCR |
Introduction
Transferred DNA (T-DNA) from Agrobacterium tumefaciens Ti plasmids is nowadays popular tool for plant genetic manipulation.Citation1,2 T-DNA insertional mutagenesis helps in identification of the gene responsible for an observed phenotype and allows to establish its function. Currently, there are several collections of the Arabidopsis T-DNA mutants freely distributed to all researchers.Citation3
The protein product of the NBR1 (At4g24690) gene was recently identified as a hybrid ortholog of the mammalian NBR1 (Neighbor of BRCA1) and p62 proteins which are selective autophagy cargo receptors participating in recycling of cellular components.Citation4 The selective autophagy process involving NBR1-like proteins in plants is now very intensively studied.Citation4–6 The phenotypic and genetic studies of the effective knockout (KO) mutants could be very helpful in understanding the in vivo functions of NBR1. Although there are several insertion mutant lines available for NBR1, there are reports that they might be ineffective in knocking out the transcription.Citation4 Only recently, a mutant line SALK_135513 was characterized as showing clear phenotypic differences in several stress-generating conditions.Citation5 The aim of our study was to further characterize the NBR1 function based on the genetic and phenotypic observations of the SALK_135513 mutant. However, the precise mapping of the insertion site led us to conclude that T-DNA locates in the fourth intron instead of previously annotated fourth exon of NBR1 (GenBank Acc. No BZ384415.1). Additionally, we proved that the wild-type (WT) mRNA of NBR1 is present in SALK_135513 making the mutant ineffective in aborting the transcription. This explains why in our laboratory the mutant plants exhibited no obvious phenotypic and developmental defects under normal and unfavorable growth conditions.
Materials and Methods
The Arabidopsis thaliana SALK_135513 T-DNA insertion line was obtained from NASC. The plants (accession Columbia (WT) and SALK_135513 (homozygous)) were grown in soil. Total DNA and RNA were isolated from rosette leaves of 4- and 8-week-old plants, respectively. Genomic DNA was extracted as described by Edwards et al.Citation7 Total RNA (5 μg) isolated independently from 2 leaves per plant with TRI Reagent (Molecular Research Center, Inc.) was used for cDNA synthesis as described earlier.Citation8,9 The list of oligonucleotides used as primers is shown in Table S1. The PCR conditions were adjusted depending on the primers and are shown in Table S2.
For methyl viologen (MV) treatment, heat, drought and salt stresses the plants were 5 weeks old grown in soil in short-day conditions (8 h light-22°C/16 h dark-19°C; in greenhouse or growth chamber with a light intensity of 157.67 ± 19.78 μE m−2 s−1). For the oxidative stress the plants were sprayed with 20 μM MV and kept well watered for 2 d under constant light in a growth chamber (181.33 ± 13.32 μE m−2 s−1 light intensity). The heat stress was conducted by placing the well watered plants for 24 h under 4 Dual 2700 K/6500 K Red/Blue spectrum lamps generating a temperature of 45°C; the pictures were taken after 3 d of recovery. The dark stress was performed by keeping the plants in a complete darkness for 6 d followed by 4 d of recovery. For drought stress plants received no water for 4 d and for salt stress they were kept on 0.6 M NaCl for 4 d and next the pictures were taken.
Results and Discussion
Four PCR primers applied in 3 PCR combinations were used for initial genotyping of the SALK_135513 mutant. The details of PCR1, 2 and 3, each performed with genomic DNA of the WT and 3 individual SALK_135513 plants, are listed in Table S2, while the overall scheme and results of the analysis is shown in . The products of the expected size with the primers flanking the T-DNA insertion (PCR1 and PCR3) were obtained only in the case of the WT genomic DNA indicating homozygosity of the SALK_135513 plants. The small amounts of the shorter than expected PCR product in PCR1 in the case of SALK_135513 result from an unspecific reaction as proved by sequencing (data not shown). Furthermore, presence of the PCR2 product in SALK_135513 confirms that the analyzed plants indeed contain T-DNA insert within NBR1.
Figure 1. Mapping of the T-DNA insert in SALK_135513. (A) Scheme of the expected PCR products and their localization in the At4g24690 chromosomal region. (B) PCR results with the pairs of S13LP/S13RP (lanes 1; PCR1), LBb1.3/S13RP (lanes 2; PCR2) and S13LP/S05LP (lanes 3; PCR3) oligonucleotides as primers and genomic DNA isolated from the WT or 3 independent SALK_135513 plants (S13.2, S13.3 and S13.7) as templates. Bands marked with a black arrowhead (S13.7 plant, PCR1) and a white arrowhead (3 SALK_135513 plants, PCR2), were sequenced. Notice, that a non-specific band of 0.6 kb (PCR1) was amplified also in WT; L denotes 1kb DNA Ladder GeneRuler (Fermentas, cat No SMO311), with sizes of the selected bands indicated.
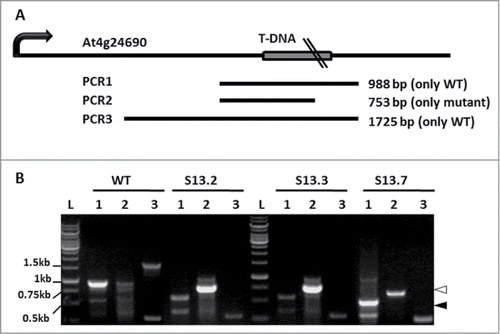
All PCR fragments were sequenced and aligned to the corresponding region of the WT genomic DNA allowing the precise insertion site of the T-DNA (GenBank Acc. No KC820135) (Fig. S1 and Fig. S2). Surprisingly, the insertion mapped to the fourth intron but not to the fourth exon of NBR1 as annotated (GenBank Acc. No BZ384415.1). This incorrect annotation might be a result of the rapid sequencing method used for indexing the SALK mutant collection.
The next step was to investigate if the T-DNA insert affects NBR1 transcription level. It has been reported that only 0.7% of the T-DNA intronic insertions do not have any effect on transcript level.Citation10 The semi-quantitative RT-PCR (sqRT-PCR) was carried out using 3 combinations of the NBR1-specific oligonucleotides with cDNA prepared from WT and SALK_135513 plants. The expected NBR1-specific PCR products for 2 kinds of the transcripts (WT and truncated) are schematically marked in . Expression of the PetC (photosynthetic electron transfer c; At4g03280) served as a quantitative reference.Citation11 Reactions were performed separately and pooled before loading on to a gel to facilitate analysis. The PCR products of the size expected for the cDNA corresponding to the WT transcript (1522, 1155 and 145 bp) were obtained for all plants indicating the presence of the WT transcript in the homozygous SALK_135513 line (). Sequencing of those PCR fragments (GenBank Acc No KC848420) confirmed their 100% identity with relevant fragments of WT NBR1 mRNA. The subsequent quantitative RT-PCR (qRT-PCR) using slightly different primers combinations () than the combination used for sqRT-PCR indicated the similar amounts of transcripts corresponding to the region upstream to the mapped insertion and much reduced amount of transcripts corresponding to the region downstream the T-DNA insert in the SALK_135513 line ().
Figure 2. Two variants of NBR1 transcripts in SALK_135513. (A) Scheme of the expected PCR fragments and their localization on the expected mRNAs. The exons of NBR1 (represented as boxes) and the T-DNA position is marked. The size of the expected PCR products is indicated. (B) Results of semiquantitative RT-PCR indicating the presence of the WT transcript in the homozygous SALK_135513 plants. The PCR was performed in duplicate using 2 pools of independently prepared cDNA for the indicated plants (WT or the mutant). The expected sizes of the PCR products in WT are (1522, 1155 and 145 bp). (C) Results of semiquantitative RT-PCR showing the presence of mutated mRNA in SALK_135513, spliced within the T-DNA insert. The oligonucleotide pairs LBb/S05LP (lanes labeled as (1) and TUBF/TUBR (lanes labeled as (2) were used as primers. The PCR products marked with black arrowheads were cut off from the gel and sequenced. L, 1kb DNA Ladder GeneRuler (Fermentas, cat No SMO311).
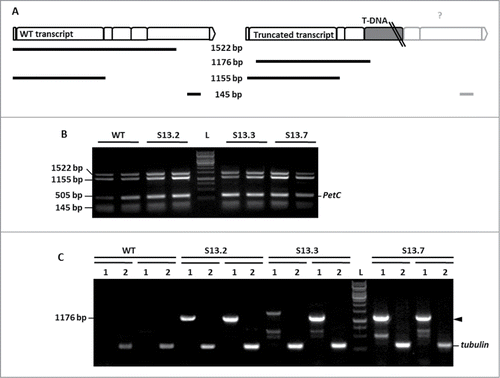
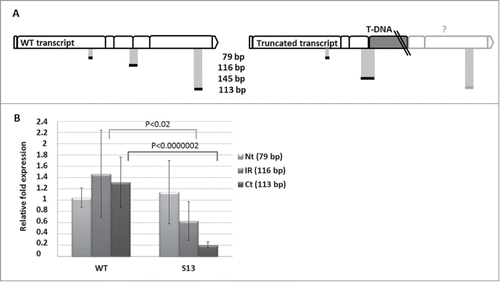
Figure 3. Quantitative RT-PCR for detection of 3 different parts of NBR1 transcript (Nt, N-terminal region of the cDNA, 79 bp; IR, intermediate region of the cDNA, 116 bp, Ct, C-terminal region of the cDNA, 113 bp) in non-stressed WT (Col-0) and SALK_135513 plants. (A) Schematic location of the expected PCR fragments on the WT and mutated mRNA of NBR1 (boxes represent individual exons); (B) Results of qRT-PCR in WT (Col-0) and SALK_135513 (S13) plants indicating the similar level of Nt but reduced amounts of IR and Ct in SALK_135513 as compared to WT. Data bars represent the mean (±SD) of 2 biological repeats performed in technical triplicate for WT and 4 biological repeats performed in technical triplicate for SALK_135513.
Further work aimed to check if the chimeric transcripts containing truncated NBR1 and fragments of T-DNA insert can be detected in SALK_135513 plants. The results of this analysis are shown in and the expected product of sqRT-PCR (1176 bp) is schematically marked in . The expression of tubulin α3 served as a quantitative reference. The results of the sqRT-PCR confirmed the presence of the chimeric transcripts in SALK_135513. Sequencing of the 1176 bp product indicated that splicing occurred within the T-DNA (removing its 97-nt fragment). The remaining T-DNA stayed inserted into the fourth intron of the NBR1 transcript (GenBank Acc. No KC848419), however the exact length of such truncated mRNA is not known because its sequence downstream of the T-DNA insertion point has not been verified.
We have demonstrated that mixture of the WT and the truncated transcripts exists in homozygous SALK_135513. Such cases have been reported before in T-DNA insertion mutants.Citation10 The mechanisms of splicing in plants are not completely understood but from preliminary studiesCitation12 and comparison to other organisms it is possible to distinguish some common features which may serve as recognition sites for the plant splicing machinery. There are several features required for splicing within the intron: a donor site (5′ end of the intron), a branch site (near the 3′ end of the intron) and an acceptor site (3′ end of the intron) composed of specific nucleotides. The alignment of the acceptor sites of the introns present in At4g24690 indicated a limited similarity and allowed the distinguishing of 2 groups. The putative site of the splicing in the T-DNA insert resembles the acceptor sites in introns 2, 3 and 6 of NBR1 (). These sequences coincide with the less restrictive definition criteria of the acceptor site which identified AG as acceptor sites for plants.Citation13
Table 1. Alignment of acceptor sites of the introns in NBR1 gene and a sequence of T-DNA insertion functioning as an acceptor site of an intron
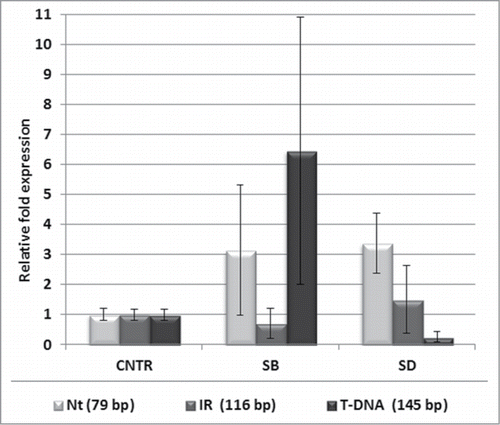
Zhou et al.Citation5 have reported that SALK_135513 mutant has increased sensitivity to various abiotic stresses. However, in our laboratory, no visible differences between the WT and SALK_135513 plants were noticed in any of the conditions of applied oxidative, heat, drought, salt or dark stresses (Fig. S3). We followed the expression of both types of NBR1 transcripts (WT and mutated) in SALK_135513 plants challenged by salt and drought stresses in comparison to the control conditions. Interestingly, the ratio between both types of NBR1 mRNA appeared to be differentially affected by stresses (). In plants challenged with the salt stress there was more of the mutated NBR1 produced while in drought stress the splicing machinery favored the WT NBR1 assembly.
Figure 4. Stress-dependent changes of the relative abundance of 2 variants of NBR1 transcript in SALK_135513 plants. Localization of the PCR product specific for SALK_135513 WT (Nt, 145 bp; IR, 116 and T-DNA, 145 bp) transcripts is as in Fig. 3A. The amounts of the indicated PCR products in SALK_135513 plants treated by salt stress (SB) and drought stress (SD) is shown in relation to the amount of the same products in SALK_135513 plants grown in the control conditions (CNTR). Data bars represent the mean (±SD) of 2 biological repeats performed in technical duplicate.
The observations of the phenotypes of SALK_135513 described in this work disagree with phenotypes reported by Zhou et al.Citation5 These discrepancies might be attributed to differences in growing conditions that might affect the splicing machinery. Similarly to long intronic non-coding RNA causing epigenetic changes in the level of gene expression, T-DNA insertion may affect the proper splicing.Citation14
Recently, it was demonstrated for the intronic T-DNA insertion mutant opr3 that it shows clear KO phenotype after wounding while upon fungal infection there is no effect on the normal transcription of OPR3 probably due to the splicing of the T-DNA-containing intron.Citation15 Ulker et al. 2008Citation16 reported that introns containing T-DNA inserts are less efficiently spliced than WT introns in the various mutant lines. These findings corroborates our data showing the influence of different conditions on the WT copy of the transcript in the SALK_135512 ().
Conclusions
Our results emphasize the need for detailed molecular analyses of Arabidopsis T-DNA insertion lines. This characterization should include the precise mapping of the T-DNA insert to confirm annotated position within the gene of interest as well as expression analysis with gene-specific primers to ensure the absence of WT mRNA copies in T-DNA mutant line. An extra care has to be taken when working with intronic T-DNA insertions because in some of them the expression may undergo environmental changes complicating interpretations of the mutant phenotype.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Material.pdf
Download PDF (519 KB)Supplemental Material
Supplemental data for this article can be accessed on thepublisher's website.
Additional information
Funding
References
- Galbiati M, Moreno MA, Nadzan G, Zourelidou M, Dellaporta SL. Large-scale T-DNA mutagenesis in Arabidopsis for functional genomic analysis. Funct Integr Genomics 2000; 1:25-34; PMID:11793219
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 1999; 11:2283-90; PMID:10590158; http://dx.doi.org/10.1105/tpc.11.12.2283
- Alonso JM, Ecker JR. Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Genet 2006; 7:524-36; PMID:16755288; http://dx.doi.org/10.1038/nrg1893
- Svenning S, Lamark T, Krause K, Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62SQSTM1. Autophagy 2011; 7:993-1010; PMID:21606687; http://dx.doi.org/10.4161/auto.7.9.16389
- Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, Chen Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet 2013; 9:e1003196; PMID:23341779; http://dx.doi.org/10.1371/journal.pgen.1003196
- Zientara-Rytter K, Lukomska J, Moniuszko G, Gwozdecki R, Surowiecki P, Lewandowska M, Liszewska F, Wawrzynska A, Sirko A. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 2011; 7:1145-58; PMID:21670587; http://dx.doi.org/10.4161/auto.7.10.16617
- Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 1991; 19:1349; PMID:2030957; http://dx.doi.org/10.1093/nar/19.6.1349
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162:156-9; PMID:2440339; http://dx.doi.org/10.1016/0003-2697(87)90021-2
- Lewandowska M, Wawrzynska A, Moniuszko G, Lukomska J, Zientara K, Piecho M, Hodurek P, Zhukov I, Liszewska F, Nikiforova V, et al. A Contribution to Identification of Novel Regulators of Plant Response to Sulfur Deficiency: Characteristics of a Tobacco Gene UP9C, Its Protein Product and the Effects of UP9C Silencing. Mol Plant 2010; 3:347-60; PMID:20147370; http://dx.doi.org/10.1093/mp/ssq007
- Wang YH. How effective is T-DNA insertional mutagenesis in Arabidopsis? J Biochem Tech 2008; 1:11-20
- Kihara T, Zhao CR, Kobayashi Y, Takita E, Kawazu T, Koyama H. Simple identification of transgenic Arabidopsis plants carrying a single copy of the integrated gene. Biosci Biotechnol Biochem 2006; 70:1780-3; PMID:16861815; http://dx.doi.org/10.1271/bbb.50687
- Barbazuk WB, Fu Y, McGinnis KM. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res 2008; 18:1381-92; PMID:18669480; http://dx.doi.org/10.1101/gr.053678.106
- Reddy AS. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 2007; 58:267-94; PMID:17222076; http://dx.doi.org/10.1146/annurev.arplant.58.032806.103754
- Gao Y, Zhao Y. Epigenetic suppression of T-DNA insertion mutants in Arabidopsis. Mol Plant 2013; 6:539-45; PMID:22973063; http://dx.doi.org/10.1093/mp/sss093
- Chehab EW, Kim S, Savchenko T, Kliebenstein D, Dehesh K, Braam J. Intronic T-DNA insertion renders Arabidopsis opr3 a conditional jasmonic acid-producing mutant. Plant Physiol 2011; 156:770-8; PMID:21487047; http://dx.doi.org/10.1104/pp.111.174169
- Ulker B, Peiter E, Dixon DP, Moffat C, Capper R, Bouche N, Edwards R, Sanders D, Knight H, Knight MR. Getting the most out of publicly available T-DNA insertion lines. Plant J 2008; 56:665-77; PMID:18644000; http://dx.doi.org/10.1111/j.1365-313X.2008.03608.x
