Abstract
Plant responses to abiotic stresses are controlled by a complex tier of epigenetic, transcriptional and post-transcriptional regulation. We have provided evidence that the DEAD-box RNA helicases, STRESS RESPONSE SUPPRESSOR (STRS) 1 and STRS2 are negative regulators of Arabidopsis thaliana stress-responsive transcription factors. Using GFP-STRS fusion proteins, we have demonstrated that the STRSs are localized to the nucleolus and chromocenters, and are rapidly removed to the nucleoplasm upon application of various abiotic stresses. The STRSs appear to act via RNA-directed DNA methylation to suppress Arabidopsis stress responses; this repressive epigenetic mechanism is abrogated by abiotic stress eventually leading to an open chromatin structure allowing expression of stress-responsive genes.
Abbreviations
| ABA | = | abscisic acid |
| STRS | = | STRESS RESPONSE SUPPRESSOR |
| RdDM | = | RNA-directed DNA methylation |
Plants responses to abiotic stresses involve a plethora of physiological and biochemical changes that are activated and integrated via global changes in gene expression. Reprogramming of gene expression in response to stress involves a complex network of regulatory proteins controlled at the epigenetic, transcriptional and post-transcriptional levels.Citation1-3
In a systems biology-based screen for novel abiotic stress regulatory proteins,Citation4,5 we identified genes encoding two DEAD-box RNA helicases that were rapidly down-regulated by multiple stresses and by abscisic acid (ABA).Citation6 Mutations in either gene caused increased tolerance to salt, osmotic and heat stresses, as well as enhancement of stress-induced gene expression, whereas overexpression yielded the opposite effect.Citation7 We therefore designated the genes, STRESS RESPONSE SUPPRESSOR1 (STRS1) and STRS2. We further demonstrated that STRS1 and STRS2 are part of the ABA-independent and ABA-dependent stress regulatory networks, and function to attenuate the stress-induced expression of transcriptional activator genes.
DEAD-box RNA helicases comprise the largest family of RNA helicases, with 58 members having been identified in the Arabidopsis thaliana genome.Citation8-10 Their name derives from one of 12 helicase core motifs that contains the characteristic Asp–Glu–Ala–Asp (DEAD) sequence. RNA helicases are major regulators of gene expression in prokaryotes and eukaryotes via their role in almost all aspects of RNA metabolism. They are often constituent parts of supramolecular complexes where they function in remodeling RNA and in assembly of ribonucleoprotein structures. RNA helicases possess RNA duplex unwinding activity and can promote duplex formation as well as displacement of proteins from RNA. They may also provide nucleation centers by acting as RNA clamps for assembly of large ribonucleoprotein complexes. In plants, DEAD-box RNA helicases have been associated with diverse molecular functions such as siRNA-mediated gene silencing, pre-mRNA splicing, chloroplast/mitochondrial intron splicing, nonsense-mediated mRNA decay, mRNA export, ribosome biogenesis and exon-junction complex functions.Citation11-22 Furthermore, it has been shown that these helicases play roles in various biological processes including abiotic stress responses.Citation6,12,15,19,23-29
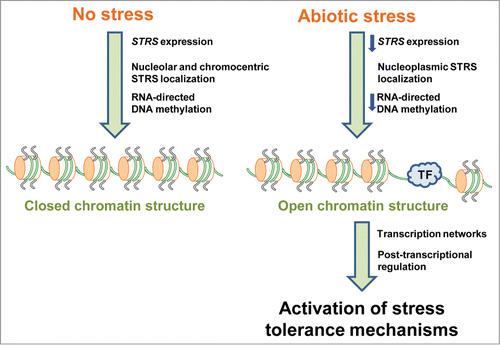
To further explore the function of the STRS DEAD-box RNA helicases in regulating abiotic stress-responsive gene expression, we examined STRS sub-cellular localization.Citation7 GFP-STRS fusion proteins localized to the nucleolus, nucleoplasm and chromocenters and exhibited relocalization in response to abscisic acid (ABA) treatment and various stresses. This relocalization was reversed when stress treatments were removed. Chromocenters are regions of heterochromatic DNA that are generally transcriptionally inactive,Citation30 and this observation led us to hypothesize that the STRSs may be involved in gene silencing consistent with their role as negative regulators of gene expression. Accordingly, the STRSs were mislocalized in specific gene silencing mutants; STRS2 displayed mislocalization in three mutants of the RNA-directed DNA methylation (RdDM) pathway, rdr2, dcl3 and drd1, while STRS1 was mislocalized in the hd2c mutant. HD2C, is a histone deacetylase that interacts with HDA6, a nuclear histone deacetylase that has been implicated in RdDM, is important for establishment of transcriptionally repressive CG methylation and regulates locus-directed heterochromatin silencing in cooperation with MET1.Citation31-35 Importantly, both HD2C and HDA6 are involved in ABA and salt stress responses.Citation35,36 Notably, heterochromatic RdDM target loci displayed reduced DNA methylation and increased expression in the strs mutants. Taken together, the evidence suggests that the STRSs are involved in RdDM-mediated epigenetic silencing of gene expression to bring about suppression of the Arabidopsis stress response ().
Figure 1. Model of STRS function in epigenetic suppression of Arabidopsis responses to abiotic stress. Blue arrows, decrease; TF, transcription factor.
A recent paper has also demonstrated a role for the RdDM pathway in plant abiotic stress responses.Citation37 Arabidopsis mutants defective in various genes encoding RdDM components exhibit reduced tolerance to heat stress. Transcriptome studies of mutants of NRPD2, encoding a subunit of both Pol IV and Pol V complexes, showed that these mutants respond normally to heat stress. However, during recovery from stress, heat stress-induced genes remain at elevated levels indicating an inability of the mutant to exit the heat stress transcriptional program. Intriguingly, the misexpression of several genes in nrpd2 is correlated with defective epigenetic regulation of adjacent transposons. For instance, the expression of At1g34220, which remains up-regulated during recovery in nrpd2, is correlated with reduced DNA methylation of an upstream transposon remnant and with heat-induced read-through transcription originating at the transposon.
It is clear from our findingsCitation7 as well as those from Popova et al.Citation37 and others that epigenetic silencing of specific genes is a crucial component in regulating plant abiotic stress responses.Citation2,38 A general theme appears to be promotion of stress-mediated chromatin modifications that are characteristic of open chromatin, linked with increased expression of stress-responsive genes. Thus, in Arabidopsis subjected to dehydration stress, the permissive histone marks, H3K9 acetylation (H3K9ac) and H3K4 trimethylation (H3K4me3), characteristic of open, active chromatin are enriched on the ABA- and drought-inducible RD20, RD29A and AtGOLS2 genes.Citation39 The presence of these marks is reduced during subsequent recovery from stress. The changes in histone marks are correlated with decreased and increased nucleosome density and gene transcription during stress induction and recovery, respectively. Other stresses such as salt stress or application of ABA lead to the enrichment of histone marks associated with open chromatin.Citation36,40 Further support for the idea of stress-mediated promotion of open, active chromatin comes from priming experiments whereby young Arabidopsis seedlings were exposed to a 24-hour mild salt priming treatment before being returned to control conditions for 10 dCitation41 Priming leads to a decrease in the repressive H3K27me3 mark that is associated with closed chromatin structure as well as to increased tolerance of primed plants to a subsequent drought treatment. Several genes that display changes in H3K27me3 due to priming also exhibit altered expression in response to the drought treatment.
It is interesting to note that chromatin decondensation as well as other features of stress responses such as disruption of nucleolar structure and a reduction in protein synthesis is characteristic of dedifferentiating cells such as protoplasts.Citation42 Indeed, plants exposed to a variety of stresses share a similar profile of altered transcription factor expression to cells undergoing dedifferentiation during protoplasting. We recently proposed therefore that acute stress directs plant cells to undergo dedifferentiation as an adaptive mechanism in preparation for the acquisition of a new cell fate upon perception of a subsequent stimulus.Citation38
How epigenetic silencing mechanisms such as the RdDM pathway remodel chromatin in response to abiotic stress in plants, and how specific components such as the STRS DEAD-box RNA helicases function to suppress the Arabidopsis stress response remains to be elucidated. The epigenetic control of abiotic stress transcriptional networks exemplifies the complex tiers of regulation required for plants to respond appropriately to adverse environmental conditions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Prof. Gideon Grafi for critical reading and insightful comments.
Additional information
Funding
References
- Covarrubias AA, Reyes JL. Post-transcriptional gene regulation of salinity and drought responses by plant microRNAs. Plant Cell Environ 2010; 33:481-9; PMID:19781008; http://dx.doi.org/10.1111/j.1365-3040.2009.02048.x
- Kim JM, To TK, Nishioka T, Seki M. Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ 2010; 33:604-11; PMID:19930132; http://dx.doi.org/10.1111/j.1365-3040.2009.02076.x
- Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci 2014; 5:170; PMID:24904597; http://dx.doi.org/10.3389/fpls.2014.00170
- Kant P, Gordon M, Kant S, Zolla G, Davydov O, Heimer YM, Khalifa-Caspi V, Shaked R, Barak S. Functional-genomics-based identification of genes that regulate Arabidopsis responses to multiple abiotic stresses. Plant Cell Environ 2008; 31:697-714; PMID:18182014; http://dx.doi.org/10.1111/j.1365-3040.2008.01779.x
- Ransbotyn V, Yeger-Lotem E, Basha O, Acuna T, Verduyn C, Gordon M, Chalifa-Caspi V, Hannah MA, Barak S A. combination of gene expression ranking and co-expression network analysis increases discovery rate in large scale mutant screens for novel Arabidopsis thaliana abiotic stress genes. Plant Biotech J 2014; In press; PMID:25370817; http://dx.doi.org/10.1111/pbi.12274
- Kant P, Kant S, Gordon M, Shaked R, Barak S. STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol 2007; 145:814-30; PMID:17556511; http://dx.doi.org/10.1104/pp.107.099895
- Khan A, Garbelli A, Grossi S, Florentin A, Batelli G, Acuna T, Zolla G, Kaye Y, Paul LK, Zhu JK, Maga G, Grafi G, Barak S. The Arabidopsis STRESS RESPONSE SUPPRESSOR DEAD-box RNA helicases are nucleolar- and chromocenter-localized proteins that undergo stress-mediated relocalization and are involved in epigenetic gene silencing. Plant J 2014; 79:28-43; PMID:24724701; http://dx.doi.org/10.1111/tpj.12533
- Linder P, Owttrim GW. Plant RNA helicases: linking aberrant and silencing RNA. Trends Plant Sci 2009; 14:344-52; PMID:19446493; http://dx.doi.org/10.1016/j.tplants.2009.03.007
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 2011; 12:505-16; PMID:21779027; http://dx.doi.org/10.1038/nrm3154
- Putnam AA, Jankowsky E. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim Biophys Acta 2013; 1829:884-93; PMID:23416748; http://dx.doi.org/10.1016/j.bbagrm.2013.02.002
- Gendra E, Moreno A, Mar Alba M, Pages M. Interaction of the plant glycine-rich RNA-binding protein MA16 with a novel nucleolar DEAD box RNA helicase protein from Zea mays. Plant J 2004; 38:875-86; PMID:15165181; http://dx.doi.org/10.1111/j.1365-313X.2004.02095.x
- Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu JK. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 2005; 17:256-67; PMID:15598798; http://dx.doi.org/10.1105/tpc.104.027557
- Yoine M, Ohto M, Onai K, Mita S, Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signaling in Arabidopsis. Plant J 2006; 47:49-62; PMID:16740149; http://dx.doi.org/10.1111/j.1365-313X.2006.02771.x
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 2007; 5:e57; PMID:17298187; http://dx.doi.org/10.1371/journal.pbio.0050057
- Koroleva OA, Calder G, Pendle AF, Kim SH, Lewandowska D, Simpson CG, Jones IM, Brown JWS, Shaw PJ. Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell 2009; 21:1592-606; PMID:19435936; http://dx.doi.org/10.1105/tpc.108.060434
- Kohler D, Schmidt-Gattung S, Binder S. The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana. Plant Mol Biol 2010; 72:459-67; PMID:19960362; http://dx.doi.org/10.1007/s11103-009-9584-9
- Asakura Y, Galarneau E, Watkins KP, Barkan A, van Wijk KJ. Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol 2012; 159:961-74; PMID:22576849; http://dx.doi.org/10.1104/pp.112.197525
- Chi W, He B, Mao J, Li Q, Ma J, Ji D, Zou M, Zhang L. The function of RH22, a DEAD RNA helicase, in the biogenesis of the 50S ribosomal subunits of Arabidopsis chloroplasts. Plant Physiol 2012; 158:693-707; PMID:22170977; http://dx.doi.org/10.1104/pp.111.186775
- Guan Q, Wu J, Zhang Y, Jiang C, Liu R, Chai C, Zhu J. A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell 2013; 25:342-56; PMID:23371945; http://dx.doi.org/10.1105/tpc.112.108340
- Kammel C, Thomaier M, Sorensen BB, Schubert T, Langst G, Grasser M, Grasser KD. Arabidopsis DEAD-box RNA helicase UAP56 interacts with both RNA and DNA as well as with mRNA export factors. PLoS ONE 2013; 8:e60644; PMID:23555998; http://dx.doi.org/10.1371/journal.pone.0060644
- Gu L, Xu T, Lee K, Lee KH, Kang H. A chloroplast-localized DEAD-box RNA helicaseAtRH3 is essential for intron splicing and plays an important role in the growth and stress response in Arabidopsis thaliana. Plant Physiol Biochem 2014; 82:309-18; PMID:25043599; http://dx.doi.org/10.1016/j.plaphy.2014.07.006
- Hsu YF, Chen YC, Hsiao YC, Wang BJ, Lin SY, Cheng WH, Jauh GY, Harada JJ, Wang CS. AtRH57, a DEAD-box RNA helicase, is involved in feedback inhibition of glucose-mediated abscisic acid accumulation during seedling development and additively affects pre-ribosomal RNA processing with high glucose. Plant J 2014; 77:119-35; PMID:24176057; http://dx.doi.org/10.1111/tpj.12371
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci USA 2002; 99:11507-12; PMID:12165572; http://dx.doi.org/10.1073/pnas.172399299
- Kim JS, Kim KA, Oh TR, Park CM, Kang H. Functional characterization of DEAD-box RNA helicases in Arabidopsis thaliana under abiotic stress conditions. Plant Cell Physiol 2008; 49:1563-71; PMID:18725370; http://dx.doi.org/10.1093/pcp/pcn125
- Li D, Liu H, Zhang H, Wang X, Song F. OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J Exp Bot 2008; 59:2133-46; PMID:18441339; http://dx.doi.org/10.1093/jxb/ern072
- Liu HH, Liu J, Fan SL, Song MZ, Han XL, Liu F, Shen FF. Molecular cloning and characterization of a salinity stress-induced gene encoding DEAD-box helicase from the halophyte Apocynum venetum. J Exp Bot 2008; 59:633-44; PMID:18272921; http://dx.doi.org/10.1093/jxb/erm355
- Chung E, Cho CW, Yun BH, Choi HK, So HA, Lee SW, Lee JH. Molecular cloning and characterization of the soybean DEAD-box RNA helicase gene induced by low temperature and high salinity stress. Gene 2009; 443:91-9; PMID:19463922; http://dx.doi.org/10.1016/j.gene.2009.05.005
- Macovei A, Vaid N, Tula S, Tuteja N. A new DEAD-box helicase ATP-binding protein (OsABP) from rice is responsive to abiotic stress. Plant Signal Behav 2012; 7:1138-43; PMID:22899052; http://dx.doi.org/10.4161/psb.21343
- Tuteja N, Sahoo RK, Garg B, Tuteja R. OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. IR64). Plant J 2013; 76:115-27; PMID:23808500.http://dx.doi.org/10.1111/tpj.12277
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci USA 2002; 99:14584-9; PMID:12384572; http://dx.doi.org/10.1073/pnas.212325299
- Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJ. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J 2002; 21: 832-41; http://dx.doi.org/10.1093/emboj/cdf663
- Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS. Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev 2010; 24:1119-32; PMID:20516197; http://dx.doi.org/10.1101/gad.1914110
- To TK, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, Tanaka M, Endo T, Kakutani T, Toyoda T, et al. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet 2011; 7:e1002055; PMID:21552333; http://dx.doi.org/10.1371/journal.pgen.1002055
- Liu X, Yu CW, Duan J, Luo M, Wang K, Tian G, Cui Y, Wu K. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol 2012; 158:119-29; PMID:21994348; http://dx.doi.org/10.1104/pp.111.184275
- Luo M, Wang YY, Liu X, Yang S, Lu Q, Cui Y, Wu K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot 2012; 63:3297-306; PMID:22368268; http://dx.doi.org/10.1093/jxb/ers059
- Chen LT, Luo M, Wang YY, Wu K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA salt stress response. J Exp Bot 2010; 61:3345-3353; PMID:20519338; http://dx.doi.org/10.1093/jxb/erq154
- Popova OV, Dinh HQ, Aufsatz W, Jonak C. The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol Plant 2013; 6:396-410; PMID:23376771; http://dx.doi.org/10.1093/mp/sst023
- Grafi G, Barak S. Stress induces cell dedifferentiation in plants. Biochim Biophys Acta 2014; S1874-9399; In press. PMID:25086338; http://dx.doi.org/10.1016/j.bbagrm.2014.07.015
- Kim JM, To TK, Ishida J, Matsui A, Kimura H, Seki M. Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol 2012; 53:847-56; PMID:22505693; http://dx.doi.org/10.1093/pcp/pcs053
- Song Y, Ji D, Li S, Wang P, Li Q, Xiang F. The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS ONE 2012; 7:e41274; PMID:22815985; http://dx.doi.org/10.1371/journal.pone.0041274
- Sani E, Herzyk P, Perrella G, Colot V, Amtmann A, Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol 2013; 14:R59; PMID:23767915; http://dx.doi.org/10.1186/gb-2013-14-6-r59
- Grafi G, Chalifa-Caspi V, Nagar T, Plaschkes I, Barak S, Ransbotyn V. Plant response to stress meets dedifferentiation. Planta 2011; 233:433-8; PMID:21312042; http://dx.doi.org/10.1007/s00425-011-1366-3
