Abstract
Powdery mildew fungi form feeding structures called haustoria inside epidermal cells of host plants to extract photosynthates for their epiphytic growth and reproduction. The haustorium is encased by an interfacial membrane termed the extrahaustorial membrane (EHM). The atypical resistance protein RPW8.2 from Arabidopsis is specifically targeted to the EHM where RPW8.2 activates haustorium-targeted (thus broad-spectrum) resistance against powdery mildew fungi. EHM-specific localization of RPW8.2 suggests the existence of an EHM-oriented protein/membrane trafficking pathway during EHM biogenesis. However, the importance of this specific trafficking pathway for host defense has not been evaluated via a genetic approach without affecting other trafficking pathways. Here, we report that expression of EHM-oriented, nonfunctional RPW8.2 chimeric proteins exerts dominant negative effect over functional RPW8.2 and potentially over other EHM-localized defense proteins, thereby compromising both RPW8.2-mediated and basal resistance to powdery mildew. Thus, our results highlight the importance of the EHM-oriented protein/membrane trafficking pathway for host resistance against haustorium-forming pathogens such as powdery mildew fungi.
Abbreviations
| EHM | = | extrahaustorial membrane |
| PM | = | plasma membrane |
| eds | = | enhanced disease susceptibility |
Introduction
Powdery mildew is one of the most common and important plant diseases caused by ascomycete fungi belonging to the order of Erysiphales.Citation1,2 Several distinct powdery mildew resistance mechanisms have been characterized in plants. For example, plants use intracellular immune receptors traditionally named resistance (R) proteins to detect the presence or virulence activity of a specific effector from powdery mildew and subsequently activate resistance to a strain of powdery mildew expressing the recognized effector.Citation3-6 In contrast, the disease resistance (R) gene locus RPW8 confers broad-spectrum resistance in Arabidopsis thaliana to powdery mildew fungi.Citation7 This locus contains 2 homologous atypical R genes, RPW8.1 and RPW8.2 (collectively referred to as RPW8 unless otherwise indicated). Protein expression and localization studies revealed that (i) both proteins are induced by powdery mildew infection, and (ii) RPW8.1 is only expressed in mesophyll cells while RPW8.2 is mainly expressed in the mildew-infected epidermal cells albeit detectable in the mesophyll cells underneath the infection site.Citation8,9 Our recent work showed that these 2 proteins seem to spatially collaborate to activate defense responses.Citation10 Most interestingly, RPW8.2 is specifically targeted to the extra-haustorial membrane (EHM), which is believed to be a host cell-derived interfacial membrane encasing the fungal haustorium.Citation9,11,12 The EHM-specific localization of RPW8.2 correlates with accumulation of hydrogen peroxide (H2O2) around the EHM and formation of a callose-enriched encasement of the haustorial complex, therefore providing a cell biological basis for RPW8-mediated broad-spectrum resistance.Citation9,13 More recently, we demonstrated that RPW8.2 (174 amino acids in total) possesses 2 basic residue–enriched EHM-targeting motifs, which together with the putative N-terminal signal peptide or transmembrane domain, constitute a minimum 60 amino acid sequence that is necessary and sufficient to target YFP to the EHM.Citation14 Because protein trafficking to other subcellular compartments such as the plasma membrane (PM) appears to be normal in the haustorium-invaded cells,Citation12,14 RPW8.2′s specific localization to the EHM suggests the existence of an EHM-oriented specific protein/membrane trafficking pathway. Our earlier work also showed that although defense activated by RPW8.2 requires the conserved salicylic acid (SA)-dependent signaling pathway,Citation15,16 EHM-targeting of RPW8.2 to the EHM is SA-independent.Citation9 For example, RPW8.2-YFP expressed in SA-signaling defective eds1-2 or pad4-1 mutant is still targeted to the EHM.Citation9 Conversely, when EHM-oriented trafficking is disrupted by application of a cytoskeleton inhibitor cytochalasin E or via overexpression of an actin depolymerizing factor (ADF6), RPW8.2 accumulates as punctate vesicles in the infected epidermal cells and these vesicles appear to correlate with cell death activation.Citation9 These observations suggest that defense function of RPW8.2 may not be contingent to its EHM-localization. However, since pharmacological or genetic inhibition of the actin cytoskeleton probably blocks multiple trafficking pathways, the cell death observed in affected plants expressing RPW8.2-YFP may not be (solely) attributable to accumulation of RPW8.2 vesicles along the EHM-oriented trafficking pathway. Thus, the role of the EHM-oriented trafficking in RPW8.2-mediated defense has yet to be unequivocally defined. Here, we show that several YFP-tagged fusion proteins in which a small PM protein Lti6bCitation17 is fused with N-terminally truncated RPW8.2 (i.e., YFP-Lti6b-RPW8.2ΔNt; Y6R82ΔNt for short) exert a dominant negative effect over the RPW8.2 wild-type protein. That is: expression of those fusion proteins that show EHM-docking not only compromises EHM-targeting of RPW8.2-YFP but also largely abrogates RPW8.2-mediated cell death and resistance. In addition, expressing such Y6R82ΔNt proteins in plants lacking RPW8 results in enhanced disease susceptibility compared with susceptible wild-type plants, implying the existence of other EHM-localized defense proteins whose trafficking is also affected. These observations highlight the importance of EHM-oriented protein trafficking for host resistance against haustorium-forming pathogens such as powdery mildew fungi.
Results
YFP-tagged LTI6b-RPW8 chimeric proteins show EHM-oriented docking
In the course of identifying EHM-targeting motifs in RPW8.2, we initially adopted a strategy deployed by Le Maout and colleagues to identify the targeting motif required for basolateral membrane localization of the inwardly rectifying potassium channel Kir 2.3 in certain renal epithelial cells.Citation18 That strategy involves fusion of the candidate C-terminal tail of Kir2.3 with CD4, a cell-surface protein without apparent polarity, followed by serial truncations to identify minimal size of the Kir2.3′s C-tail that renders basolateral membrane localization of CD4.Citation18 We chose Lti6b (At3g05890), a small (55 amino acids in total) PM-localized protein as a membrane cargo.Citation17 Lti6b was translationally fused with RPW8.2 that lacks its N-terminal transmembrane domain or signal peptide (amino acid 1 to 22), and YFP was added to the N-terminus of LTI6b to make a YFP-LTI6b-RPW8.2ΔNt22 fusion construct (designated Y6R82ΔNt22). The DNA construct was placed under control of the RPW8.2 promoter and stably expressed in Arabidopsis accession Col-0 lacking RPW8.Citation7 Confocal imaging showed that expression of Y6R82ΔNt22 was nicely induced by infection from powdery mildew isolate Golovinomyces cichoracearum (Gc) UCSC1. In ∼75% invaded epidermal cells, the fusion protein was detected in varied-sized puncta randomly scattered in invaded cells (). In ∼25% invaded cells however, the fusion protein was detected as puncta or small patches around the EHM (), suggesting that the vesicles carrying this fusion protein partially retains the ability to dock to the EHM. However, since limited diffuse YFP signal was detected in the EHM, it appears that the chimeric protein was largely unable to fuse with the EHM. Consistent with this observation, all 36 Col-0 T1 independent transgenic lines expressing this construct were fully susceptible to powdery mildew (not shown), suggesting that this chimeric protein is not functional in defense activation. Nevertheless, we speculated that the assumed EHM-targeting motif(s) in RPW8.2 is (partially) functional in Y6R82ΔNt22 to enable docking of this fusion protein to the EHM. We thus made 11 additional chimeric constructs in which different sized N-terminally truncated RPW8.2 variants were each translationally fused with YFP-Lti6b (), hoping to identify amino acid regions of RPW8.2 required for EHM-oriented targeting (docking).
Table 1. RPW8.2 DNA constructs and protein localization patterns
Figure 1. Localization of YFP-Lti6b–tagged RPW8.2 variants in haustorium-invaded cells. Six week-old transgenic plants were inoculated with Gc UCSC1 and infected leaves were subjected to confocal imaging. H, haustorium; P, penetration site. Bar represents 10 μm in A-D, 50 μm in E and F. (A) A representative confocal images (Z-stack) showing random localization of Y6R82ΔNt22 vesicles in ∼75% haustorium-invaded cells. (B) A representative confocal image (single optical section) showing EHM-docking of Y6R82ΔNt22 puncta in ∼25% haustorium-invaded cells. (C) A representative epifluorescent image showing EHM-oriented localization of Y6R82ΔNt102. Inset is a confocal image of one haustorium-invaded cell. (D) A representative confocal image (Z-stack) showing random localization of Y6R82ΔNt114 vesicles in all haustorium-invaded cells. (E) A representative confocal image (Z-stack) showing earliest (16-20 hpi) EHM-oriented vesicle docking of Y6R82ΔNt102. Note the large-sized vesicles (0.5–1.0 μm) in the inset. (F) A representative confocal image (single optical section) showing EHM-oriented vesicle strands of Y6R82ΔNt102 (see live targeting in Movie 1 in the online supplemental information).
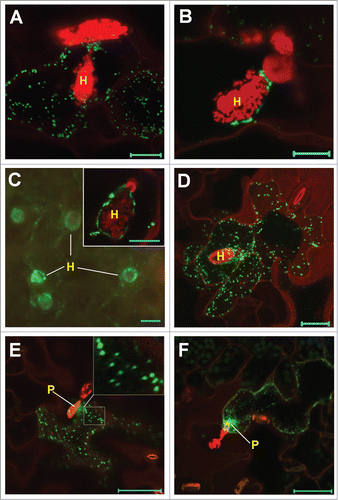
We generated at least 24 T1 transgenic lines for each construct and examined them for protein localization. Interestingly, construct #2 (Y6R82ΔNt31) to #7 (Y6R82ΔNt101) all produced fusion proteins that were mostly found in random puncta, with only occasional or rare (i.e., in ∼10% or fewer invaded cells) EHM-association as seen for Y6R82ΔNt22 (). Surprisingly, Y6R82ΔNt102 (which contains a shorter RPW8.2 C-tail) exhibited the most frequent (∼65%) EHM-association () with more apparent EHM-docking (inset in ). However, the fusion proteins from the remaining constructs only exhibited randomly distributed puncta unrelated to the EHM (see one example in ). We thus used transgenic line expressing Y6R82ΔNt102 for more detailed characterization of its EHM-docking. The earliest EHM-oriented vesicle aggregation of Y6R82ΔNt102 was detectable at ∼16 hrs post-inoculation (hpi) and more obvious at 20 hpi (), which roughly concurs with the earliest detection of RPW8.2-YFP as diffuse signal in the EHM.Citation9 Vesicles were seen rapidly moving to the periphery of the haustorium via EHM-oriented strands (which may represent actin cables) (; Supplemental movie 1). These vesicles (0.5-1.0 μm) were 2–5 times larger than those of RPW8.2-YFP (0.1-0.5 μm) that are rarely seen presumably because their trafficking is much quicker.Citation9
Our above observations seemed to suggest that amino acids 103 to 174 in RPW8.2 confers EHM-docking and that amino acids 103 to115 are most critical for this trafficking property. These results were different or even contradictory to what we later found using the functional (both in terms of defense and trafficking) RPW8.2-YFP fusion gene as template for a large-scale mutational analysis through which we identified amino acids 20–30 and 95–100 to be the core EHM-targeting signals in RPW8.2.Citation14 It is possible that the C-terminal portion (especially from amino acids103 to 115) of RPW8.2 may contain a cryptic EHM-targeting signal which when combined with the transmembrane domains from Lti6b may enable EHM-oriented trafficking. Alternatively, combining the RPW8.2 C-terminal 71 amino acids with YFP-Lti6b may generate a chimeric protein that somehow can more readily enter the EHM-oriented trafficking pathway.
None of the Col-0 T1 transgenic lines (>24 examined for each construct) expressing any of the YFP-Lti6b–tagged RPW8.2 variants showed resistance to powdery mildew. It is possible that the addition of Lti6b and/or the N-terminal tagging of YFP-Lti6b to RPW8.2 may change the conformation and/or topology of RPW8.2, resulting in nonfunctional fusion proteins dissimilar to the endogenous RPW8.2 protein or RPW8.2-YFP. We thus abandoned this approach for searching for physiologically relevant EHM-targeting signals.
Expression of YFP-Lti6B-RPW8 fusion proteins compromises basal resistance to powdery mildew
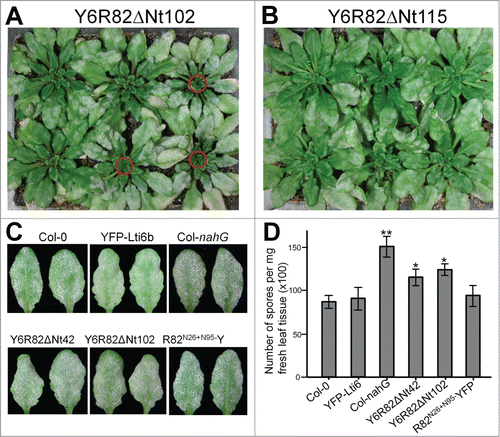
Unexpectedly, we observed by the naked eye that 20–35% of the T1 lines (>24 for each construct) expressing those YFP-Lti6b–tagged RPW8.2 variants with EHM-docking showed enhanced disease susceptibility (‘eds’) to powdery mildew isolate Gc UCSC1 (one example in ). However, no obvious ‘eds’ phenotype was observed in T1 lines expressing fusion proteins that lacked EHM-docking or expressing YFP-Lti6b alone (one example in ). We speculated that those YFP-Lti6b-R82 proteins capable of entering the EHM-oriented trafficking pathway might be able to produce a dominant negative effect over endogenous EHM-resident proteins, hence the ‘eds’. To test this idea, we did a comparative phenotypic analysis with Col-0 lines expressing Y6R82ΔNt42, Y6R82ΔNt102, R82N26+N95-YFP, or YFP-Lti6b, along with Col-0 wild-type and Col-nahG, a transgenic line known to have ‘eds’ to powdery mildew due to SA-deficiency.Citation16 The R82N26+N95-YFP is a DNA fusion construct in which amino acids 26–31 and 95–100 falling into the 2 EHM-targeting motifs of RPW8.2 were each replaced by the 6 amino acids “NARRIS” such that the fusion protein loses its EHM-targeting signal yet with minimal structural perturbation when compared to the functional RPW8.2-YFP protein.Citation14 Compared with Col-0, plants of selected representative homozygous T3 transgenic lines expressing Y6R82ΔNt42 or Y6R82ΔNt102 showed clear ‘eds’ (though not as strong as that of Col-nahG), whereas plants expressing YFP-Lti6b or R82N26+N95-YFP showed no or only marginally increased susceptibility (). These results suggest that Y6R82ΔNt42 and Y6R82ΔNt102 in particular may be more potent in interfering the normal EHM-oriented trafficking than R82N26+N95-YFP. Because Col-0 does not contain RPW8.1 and RPW8.2Citation7 but contains 4 homologs of RPW8 (HR1, HR2, HR3 and HR4), it is possible that these RPW8 homologs may be EHM-resident proteins and contribute to basal resistance in Col-0. Hence, expression of Y6R82ΔNt42 and Y6R82ΔNt102 might negatively affect EHM-targeting and function of these proteins.
Figure 2. Expression of YFP-Lti6b–tagged RPW8.2 variants results in enhanced disease susceptibility. Six week-old plants were inoculated with Gc UCSC1 and disease phenotypes were assessed at 10 dpi. (A) Representative T1 plants transgenic for Y6R82ΔNt102 (as an example). Plants with enhanced disease susceptibility (‘eds’) are indicated by red circles. (B) Representative T1 plants transgenic for Y6R82ΔNt115 (as an example). No plants with apparent ‘eds’ were seen. (C) Representative individual infected leaves of indicated genotypes. Col-nahG is SA-deficient and displays ‘eds’. (D) Quantification of disease susceptibility. Data represent means ± SE from one of 3 independent experiments. Student's t-test was used to compare value of other genotypes to that of Col-0. *P < 0.05; **P < 0.001.
Expression of YFP-Lti6B-RPW8 fusion proteins compromises RPW8-mediated resistance to powdery mildew
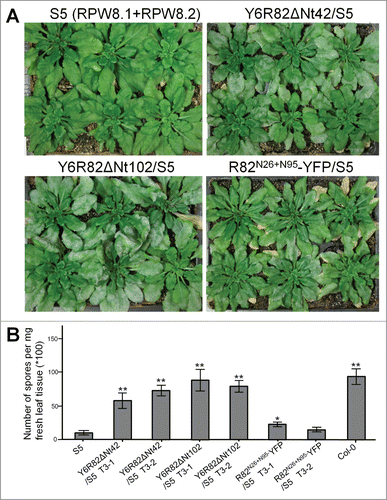
An extrapolation of the above results is that expression of Y6R82ΔNt42 and Y6R82ΔNt102 may exert a dominant negative effect over RPW8.2′s resistance function. To test this, we individually introduced Y6R82ΔNt42, Y6R82ΔNt102 and R82N26+N95-YFP into S5, which is a Col-0 line transgenic for a single copy of RPW8.1 and RPW8.2.Citation15 We examined the disease phenotypes of S5 plants and S5 transgenic T1 lines. As expected, we found that more than half of S5 T1 lines transgenic for Y6R82ΔNt42 or Y6R82ΔNt102 were susceptible or moderately susceptible to powdery mildew. In contrast, only 3 of the 24 S5 T1 lines transgenic for R82N26+N95-YFP were slightly more susceptible, the remaining lines were similar to S5 plants (). The disease phenotypes were confirmed with at least 2 independent homozygous T3 lines for each construct based on quantitative assays (). Based on these data, it is obvious that RPW8-mediated resistance was largely abolished due to ectopic expression of Y6R82ΔNt42 or Y6R82ΔNt102 in particular, whereas ectopic expression of R82N26+N95-YFP had little effect.
Figure 3. Expression of YFP-Lti6b–tagged RPW8.2 variants abrogates RPW8-mediated resistance. Six week-old plants were inoculated with Gc UCSC1 and disease phenotypes were assessed at 10 dpi. (A) Disease phenotypes of S5 (expressing both RPW8.1 and RPW8.2), and S5 lines transgenic for indicated DNA constructs. Note the differences in the amount of whitish powdery mildew in different trays. (B) Quantification of number of spores per mg fresh leaves for the indicated genotypes. Data represent means ± SE from one of 3 independent experiments. Student's t-test was used to compare value of other genotypes to that of S5. *P < 0.05; **P < 0.001.
Expression of YFP-Lti6b-RPW8.2 fusion proteins reduces EHM-targeting of RPW8.2
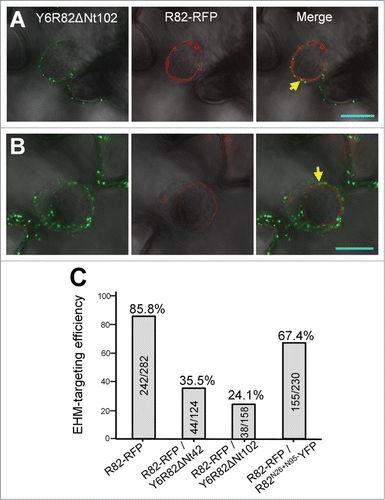
One likely mechanistic explanation for the dominant negative effect of Y6R82ΔNt42 or Y6R82ΔNt102 over RPW8.2 is that these fusion proteins could enter the EHM-oriented trafficking pathway and sequester relevant trafficking components engaged for RPW8.2′s EHM-targeting, thereby resulting in (partial) loss of RPW8.2′s EHM-localization and defense function. To obtain additional evidence for this speculation, we generated Col-0 transgenic lines co-expressing Y6R82ΔNt42 or Y6R82ΔNt102 and RPW8.2-RFP (R82-RFP) through agrobacterium-mediated stable transformation. While 30% (20 of 66) T1 transgenic lines expressing R82-RFP exhibited resistance to powdery mildew, none of the 6 T1 lines co-expressing Y6R82ΔNt42 with R82-RFP and the 10 T1 lines co-expressing Y6R82ΔNt102 with R82-RFP showed similar level of resistance, further validating the results from S5 transgenic lines (). Confocal imaging of leaves inoculated with Gc UCSC1 showed that Y6R82ΔNt102 was docked around the EHM labeled by R82-RFP but there was no diffuse YFP signal in the EHM (). When more Y6R82ΔNt102 puncta was detected in the haustorium-invaded cells, R82-RFP signal was in most cases very weak or undetectable (), suggesting likely interference of EHM-oriented trafficking of R82-RFP by Y6R82ΔNt102. By counting the number of R82-RFP–labeled (EHM) haustoria versus the total number of haustoria (stained by propidium iodide) in Gc UCSC1-inoculated leaves of 3 representative T1 lines for each construct, we found that EHM-targeting efficiency of R82-RFP was reduced from 85.8% to 35.5% due to co-expression of Y6R82ΔNt42 and to 24.1% due to co-expression of Y6R82ΔNt102. In contrast, co-expression of R82N26+N95-YFP caused only mild reduction (from 85% to 67.4%) in RPW8.2-RFP's EHM-localization ().
Figure 4. Expression of YFP-Lti6b–tagged RPW8.2 variants compromises EHM-targeting of RPW8.2-RFP. (A–B) Representative confocal image (single optical section) showing that strong RFP signal from RPW8.2-RFP at the EHM often coincided with no or weak YFP signal from Y6R82ΔNt102 around the EHM (A) or that strong YFP signal in puncta docked around the EHM often correlated with no or weak RFP signal from RPW8.2-RFP at the EHM (B). Bar represents 10 μm. (C) Quantification of EHM targeting efficiency in leaf epidermal cells of plants with indicated genotypes. Haustoria were visualized by propidium iodide staining. EHM targeting efficiency of RPW8.2-RFP was calculated as the percentage of haustoria with RFP–labeled EHM vs. total haustoria using the numbers in the columns.
Combined, these results indicate that disruption of EHM-localization of RPW8.2 by dominant negative Y6R82ΔNt42 or Y6R82ΔNt102 in particular inhibits RPW8.2′s EHM-targeting and its defense function, providing unequivocal evidence that EHM-localization of RPW8.2 is essential for activation of resistance to powdery mildew.
Discussion
In this study, we unexpectedly found that expression of several nonfunctional, YFP-Lti6b–tagged mutant RPW8.2 proteins resulted in ‘eds’ to powdery mildew in the absence of RPW8 and largely abrogated RPW8-mediated resistance. Since expression of those that show EHM-oriented docking produced a more obvious dominant-negative effect, our observations (i) suggest the existence of additional EHM-based defense proteins other than RPW8 in Arabidopsis and (ii) highlight the importance of a “healthy” smooth EHM-oriented trafficking pathway for mounting host defenses against powdery mildew pathogens.
What proteins might function at the EHM to confer basal resistance to powdery mildew in Arabidopsis accession Col-0 that lacks RPW8.1 and RPW8.2? Given that Col-0 contains 4 homologs of RPW8, namely HR1, HR2, HR3 and HR4 at the same ∼12Kb region where RPW8 is located,Citation7,19 it is reasonable to speculate that these RPW8 homologous proteins might also be EHM-localized defense proteins (albeit their defense activities are probably much lower compared to RPW8.2, accounting for Col-0′s susceptibility to Gc UCSC1). Hence, expression of “poisonous” Y6R82ΔNt102 might also interfere EHM-targeting of these RPW8 homologous proteins, resulting in loss of their function in basal resistance. If so, this might have circumvented the difficulty in assessing the likely redundant function of these genes through conventional genetic approaches (since these genes are tightly linked and have been under purifying selectionCitation19), thereby providing genetic evidence for their role in haustorium-targeted defense in Arabidopsis. Apparently, to substantiate this inference, we will need to examine the subcellular localization of these RPW8 homologs in Col-0 wild-type and their EHM-targeting efficiency in Col-0 lines expressing Y6R82ΔNt102 in our future experiments.
The dominant negative effect of Y6R82ΔNt102 might in part be attributable to its competition for the same set of trafficking components such as SNAREs or Rab proteins recruited for targeting EHM resident proteins. Consistent with this idea, a recent report showed that 2 homologous v-SNAREs Vamp721 and Vamp722 are also engaged in targeting RPW8.2-YFP to the EHMCitation20 in addition to their role in the default secretory pathway and the polarized trafficking to the fungal penetration site.Citation21 In most cases, genetic disruption of these conserved trafficking components results in severe deleterious effect in plant development,Citation22,23 which often hinders characterization of their role in a specific trafficking pathway. A dominant negative cargo protein like Y6R82ΔNt102 is a valuable tool in assessing the role of polar transport of the target cargo protein(s) and the engaged trafficking pathway in a specific biological process (i.e., haustorium-targeted resistance in the case of RPW8.2), as it would less likely affect other trafficking pathways. Consistent with this notion, we did not observe any developmental defect in the transgenic plants expressing Y6R82ΔNt102.
Since we did not detect obvious dominant negative effect from expression of YFP-RPW8.2 (which is defective in defense but retains proper EHM-localizationCitation13 (not shown), or YFP-Lti6b (), it is reasonable to speculate that inserting Lti6b between YFP and RPW8.2 may change the overall membrane topology of RPW8.2, making the fusion protein neither functional in defense nor capable of fusing with the EHM, yet “poisonous” to functional RPW8.2 and perhaps other RPW8 family members when incorporated into the EHM-oriented trafficking pathway. Given that expression of the non-EHM–localized R82N26+N95-YFP has no or little effect on RPW8-medated resistance, one might think that Y6R82ΔNt102 may reside in the same vesicles along with functional RPW8.2 thereby interfering with its normal vesicle traffic and fusion with the EHM. This agrees well with our observations that RPW8.2-RFP's EHM-targeting efficiency was more significantly reduced in Col-0 lines co-expressing Y6R82ΔNt102 than lines co-expressing R82N26+N95-YFP (). However, due to the fact that we rarely observed punctate vesicles for RPW8.2-RFP along the trafficking pathway (implying that the properties of RFP are different from those of YFP in manifestation of fluorescence signal), we were unable to examine if Y6R82ΔNt102 colocalizes with RPW8.2-RFP in punctate vesicles and interferes with its EHM-targeting. Hence, we cannot formally exclude other possibilities such as transgene-induced silencing of RPW8.2 for the loss of RPW8.2-mediated resistance in the mildew-susceptible S5 plants expressing Y6R82ΔNt102.
EHM docking of YFP-Lti6b–tagged mutant RPW8.2 proteins is difficult to explain, given that Y6R82ΔNt102, which contains neither of the 2 defined EHM-targeting motifs,Citation14 exhibited better EHM-docking than Y6R82ΔNt22, which contains both EHM-targeting motifs. It is possible that the C-terminal tail of RPW8.2 contains a cryptic EHM-targeting signal (and if so, amino acids from 103 to 115 of RPW8.2 may be critical since removal of these 13 amino acid in Y6R82ΔNt115 resulted in loss of EHM-docking; ) that may come to effect when the 2 proper EHM-targeting motifs are removed. Another possibility is that the EHM-oriented trafficking pathway is very active in haustorium–invaded cells during EHM biogenesis, and certain membrane proteins such as Y6R82ΔNt102 can more readily enter this pathway, and get passively docked to the EHM. The partial or complete exclusion of other YFP-Lti6b–tagged RPW8.2 fusion proteins from entering this pathway suggests that certain restrictions may apply with regard to what membrane protein cargos that can enter this pathway.
Our earlier studies indicated that the 2 functional arms of RPW8.2 (i.e., defense activation and EHM-localization) are under control of separate mechanisms. We speculated that integration of these 2 arms is key to activation of RPW8-mediated broad-spectrum resistance.Citation9,13 Results from this study fully support this inference, since expression of Y6R82ΔNt102 affects both the defense and EHM-localization of wild-type RPW8.2. How exactly Y6R82ΔNt102 affects RPW8.2′s defense function is not clear. One possibility is that Y6R82ΔNt102 may disrupt RPW8.2′s interaction with other defense-related proteins such as 14-3-3lambda or PAPP2C (a protein phosphatase type 2C)Citation24,25 at a specific stage of RPW8.2 vesicle transport or at the EHM. Notably, expression of Y6R82ΔNt102 almost completely abolished powdery mildew resistance of S5 plants expressing both RPW8.1 and RPW8.2 (). This result seems to suggest that RPW8.1′s defense function is also affected despite that RPW8.1 is normally expressed in mesophyll cells and functions either additively or co-operatively with RPW8.2 in the epidermal cells where haustoria invade.Citation10 One possible explanation is that expression of Y6R82ΔNt102 from the RPW8.2 promoter in mesophyll cells may also interfere with RPW8.1′s localization to the periphery of chloroplasts, Citation8 thereby also affecting its defense function. Alternatively, functional interference of HR1, HR2, HR3 and HR4 in the S5 background by Y6R82ΔNt102 may also in part account for the loss of resistance in S5 plants.
In summary, our results from this study provide genetic and cell biological evidence for the importance of a smooth EHM-oriented trafficking pathway for activating haustorium-targeted resistance against powdery mildew and imply that EHM resident proteins other than RPW8 may be engaged in basal resistance in susceptible Arabidopsis plants.
Materials and Methods
Plants growth and transformation
Seeds of Arabidopsis thaliana were sown in Metromix 360 (Maryland Plant and Suppliers, USA) and cold treated (4°C for 3 days), and seedlings were kept under 22°C, 75% relative humidity, short-day (8 h light at ∼125 μmol·m−2·s−1, 16 h dark) conditions for 5 to 6 weeks before pathogen inoculation or other treatments.
Arabidopsis thaliana accession Col-0 (or Col-gl carrying the glabrous mutation) was used for generation of all transgenic lines described in this study via conventional Agrobacterium-mediated stable transformation. Unless otherwise indicated, at least 24 independent T1 transgenic lines were generated and tested with powdery mildew.
DNA constructs
The methods used for cloning the DNA constructs under control of the RPW8.2 promoter (see ) were essentially the same as previously described.Citation9,14 Briefly, the RPW8.2 native promoter was amplified with primers EcoR82PF (5′-CAGAATTCACCGAAATTGTTAGTATTCA-3′) and BamR82PR (5′-ATGGATCCGAAATTAGTTTGTTAGCTCTCGAG-3′), and cloned into pPZP211 leading to the vector pPR82R5. YFP was amplified with primers BglYFPF1(5′-TCAGATCTATGGTGAGCAAGGGCGAG-3′) and BamYFPR1(5′- TGGGATCCGACTTGTACAGCTCGTCCATG-3′) and cloned into pPR82R5 resulting in the vector pP2BglYFP. Lti6b was amplified using LTI6btpF (5′-CACCAGATCTTAGTACAGCCACTTTCGTAGAGATTA-3′) and BamLTI6bR (5′-CAGGATCCCTTGGTGATGATATAAAGAGCGTAA-3′) primer pairs, and cloned into pP2BglYFP generating the vector pY6B. RPW8.2 fragments with desired N-terminal truncations were amplified by a high-fidelity thermostable DNA polymerase with appropriate forward primers (Supplemental Table 2 of Wang et al. 2013) and reverse primer BglR823′R (5′- TGAGATCTTTTGTTGTTTTTTACTCT-3′), and cloned into the BamHI site of pY6B containing the 3′-UTR of RPW8.2.
Pathogen infection and microscopy
Fresh mature spores of powdery mildew isolate Golovinomyces cichoracearum (Gc) UCSC1 were evenly inoculated on leaves of 6 week-old Arabidopsis plants and disease reaction phenotypes were assessed using methods as previously described.Citation16 Laser scanning confocal microscopy images were acquired as previously describedCitation9,14 by using the Zeiss LSM 710 microscope. The image data were processed using Zeiss ZEN microscope software (2012 edition) and Adobe Photoshop CS4.
Phenotypic evaluation and microscopic examination were done with all T1 lines for each DNA construct, and the results were confirmed with T2 or T3 generations for some selected T1 lines.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_movie_1.zip
Download Zip (1.1 MB)Acknowledgments
We thank R. Cooper for maintaining plant growth facility.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This project was supported by a National Science Foundation Grant (IOS-1146589) to S. X. and a grant from the National Natural Science Foundation of China (#31071670) to W. W.
References
- Huckelhoven R, Panstruga R. Cell biology of the plant-powdery mildew interaction. Curr Opin Plant Biol 2011; 14:738-46; PMID:21924669; http://dx.doi.org/10.1016/j.pbi.2011.08.002
- Spencer, M D. The Powdery Mildew. Academic Press Inc Ltd, London, 1978.
- Zhou F, Kurth J, Wei F, Elliott C, Vale G, Yahiaoui N, Keller B, Somerville S, Wise R, Schulze-Lefert P. Cell-autonomous expression of barley mla1 confers race-specific resistance to the powdery mildew fungus via a rar1-independent signaling pathway. Plant Cell 2001; 13:337-50; PMID:11226189; http://dx.doi.org/10.1105/tpc.13.2.337
- Jordan T, Seeholzer S, Schwizer S, Toller A, Somssich IE, Keller B. The wheat Mla homologue TmMla1 exhibits an evolutionarily conserved function against powdery mildew in both wheat and barley. Plant J 2011; 65:610-21; PMID:21208308; http://dx.doi.org/10.1111/j.1365-313X.2010.04445.x
- Halterman D, Zhou F, Wei F, Wise RP, Schulze-Lefert P. The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J 2001; 25:335-48; PMID:11208025; http://dx.doi.org/10.1046/j.1365-313x.2001.00982.x
- Yahiaoui N, Srichumpa P, Dudler R, Keller B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 2004; 37:528-38; PMID:14756761; http://dx.doi.org/10.1046/j.1365-313X.2003.01977.x
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 2001; 291:118-20; PMID:11141561; http://dx.doi.org/10.1126/science.291.5501.118
- Wang W, Devoto A, Turner JG, Xiao S. Expression of the membrane-associated resistance protein RPW8 enhances basal defense against biotrophic pathogens. Mol Plant Microbe Interact 2007; 20:966-76; PMID:17722700; http://dx.doi.org/10.1094/MPMI-20-8-0966
- Wang W, Wen Y, Berkey R, Xiao S. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 2009; 21:2898-913; PMID:19749153; http://dx.doi.org/10.1105/tpc.109.067587
- Ma X, Li Y, Sun J, Wang T, Fan J, Lei Y, Huang Y, Xu Y, Zhao J, Xiao S, et al. Ectopic expression of resistance to powdery mildew8.1 confers resistance to fungal and oomycete pathogens in arabidopsis. Plant Cell Physiol 2014; 55:1484-96; PMID:24899552; http://dx.doi.org/10.1093/pcp/pcu080
- Roberts AM, Mackie AJ, Hathaway V, Callow JA, Green JR. Molecular differentiation in the extrahaustorial membrane of pea powdery mildew haustoria at early and late stages of development. Physiol Mol Plant Pathol 1993; 43:147-60; http://dx.doi.org/10.1006/pmpp.1993.1047
- Koh S, Andre A, Edwards H, Ehrhardt D, Somerville S. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J 2005; 44:516-29; PMID:16236160; http://dx.doi.org/10.1111/j.1365-313X.2005.02545.x
- Wang W, Berkey R, Wen Y, Xiao S. Accurate and adequate spatiotemporal expression and localization of RPW8.2 is key to activation of resistance at the host-pathogen interface. Plant Signal Behav 2010; 5:1002-5; PMID:20864817; http://dx.doi.org/10.4161/psb.5.8.12230
- Wang W, Zhang Y, Wen Y, Berkey R, Ma X, Pan Z, Bendigeri D, King H, Zhang Q, Xiao S. A comprehensive mutational analysis of the arabidopsis resistance protein RPW8.2 reveals key amino acids for defense activation and protein targeting. Plant Cell 2013; 25:4242-61; PMID:24151293; http://dx.doi.org/10.1105/tpc.113.117226
- Xiao S, Brown S, Patrick E, Brearley C, Turner JG. Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 2003; 15:33-45; PMID:12509520; http://dx.doi.org/10.1105/tpc.006940
- Xiao S, Calis O, Patrick E, Zhang G, Charoenwattana P, Muskett P, Parker JE, Turner JG. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J 2005; 42:95-110; PMID:15773856; http://dx.doi.org/10.1111/j.1365-313X.2005.02356.x
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci U S A 2000; 97:3718-23; PMID:10737809; http://dx.doi.org/10.1073/pnas.97.7.3718
- Le Maout S, Welling PA, Brejon M, Olsen O, Merot J. Basolateral membrane expression of a K+ channel, Kir 2.3, is directed by a cytoplasmic COOH-terminal domain. Proc Natl Acad Sci U S A 2001; 98:10475-80; PMID:11504929; http://dx.doi.org/10.1073/pnas.181481098
- Orgil U, Araki H, Tangchaiburana S, Berkey R, Xiao S. Intraspecific genetic variations, fitness cost and benefit of RPW8, a disease resistance locus in Arabidopsis thaliana. Genetics 2007; 176:2317-33; PMID:17565954; http://dx.doi.org/10.1534/genetics.107.070565
- Kim H, O'Connell R, Maekawa-Yoshikawa M, Uemura T, Neumann U, Schulze-Lefert P. The powdery mildew resistance protein RPW8.2 is carried on VAMP721/722 vesicles to the extrahaustorial membrane of haustorial complexes. Plant J 2014; 79:835-47; PMID:24941879
- Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H, et al. Co-option of a default secretory pathway for plant immune responses. Nature 2008; 451:835-40; PMID:18273019; http://dx.doi.org/10.1038/nature06545
- Heese M, Gansel X, Sticher L, Wick P, Grebe M, Granier F, Jurgens G. Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J Cell Biol 2001; 155:239-49; PMID:11591731; http://dx.doi.org/10.1083/jcb.200107126
- Zhang Z, Feechan A, Pedersen C, Newman MA, Qiu JL, Olesen KL, Thordal-Christensen H. A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J 2007; 49:302-12; PMID:17241452; http://dx.doi.org/10.1111/j.1365-313X.2006.02961.x
- Wang W, Ma X, Zhang Y, Luo M, Wang G, Bellizzi M, Xiong X, Xiao S. PAPP2C interacts with the atypical disease resistance protein RPW8.2 and negatively regulates salicylic acid-dependent defense responses in Arabidopsis. Mol Plant 2012; 5:1125-37; PMID:22334594; http://dx.doi.org/10.1093/mp/sss008
- Yang X, Wang W, Coleman M, Orgil U, Feng J, Ma X, Ferl R, Turner JG, Xiao S. Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J 2009; 60:539-50; PMID:19624472; http://dx.doi.org/10.1111/j.1365-313X.2009.03978.x
