Abstract
Protein misfolding cyclic amplification (PMCA) is an in vitro simulation of prion replication, which relies on the use of normal brain homogenate derived from host species as substrate for the specific amplification of abnormal prion protein, PrPSc. Studies showed that recombinant cellular PrP, PrPC, expressed in Escherichia coli lacks N-glycosylation and an glycophosphatidyl inositol anchor (GPI) and therefore may not be the most suitable substrate in seeded PMCA reactions to recapitulate prion conversion in vitro. In this study, we expressed 2 PRNP genotypes of sheep, V136L141R154Q171 and A136F141R154Q171, and one genotype of white-tailed deer (Q95G96, X132,Y216) using the baculovirus expression system and evaluated their suitability as substrates in seeded-PMCA. It has been reported that host-encoded mammalian RNA molecules and divalent cations play a role in the pathogenesis of prion diseases, and RNA molecules have also been shown to improve the sensitivity of PMCA assays. Therefore, we also assessed the effect of co-factors, such as prion-specific mRNA molecules and a divalent cation, manganese, on protein conversion. Here, we report that baculovirus-expressed recombinant PrPC shows a glycoform and GPI-anchor profile similar to mammalian brain-derived PrPC and supports amplification of PrPSc and PrPCWD derived from prion-affected animals in a single round of seeded PMCA in the absence of exogenous co-factors. Addition of species-specific in vitro transcribed PrP mRNA molecules stimulated the conversion efficiency resulting in increased PrPSc or PrPCWD production. Addition of 2 to 20 μM of manganese chloride (MnCl2) to unseeded PMCA resulted in conversion of recombinant PrPC to protease-resistant PrP. Collectively, we demonstrate, for the first time, that baculovirus expressed sheep and deer PrP can serve as a substrate in protein misfolding cyclic amplification for sheep and deer prions in the absence of additional exogenous co-factors.
Introduction
Prionoses, or transmissible spongiform encephalopathies (TSEs), are rare infectious neurodegenerative diseases that affect a range of mammalian species and include scrapie in sheep and goats, bovine spongiform encephalopathy in cattle, chronic wasting disease (CWD) in cervids, as well as Creutzfeldt - Jakob disease, Gerstman Straussler-Scheinker syndrome, Fatal familial insomnia and kuru in humans.Citation1-3 Infectious prions are devoid of nucleic acid and seem to be composed exclusively of a modified isoform of PrPC, designated PrPSc or PrPres.Citation1 In prionoses, the pathological, protease-resistant PrPres, appears to propagate itself in infected hosts by inducing a conformational conversion of the normal host-encoded PrPC into PrPSc; the molecular details of this conversion are not clear.Citation1,4,5 PrPC is a monomeric, glycophosphatidylinositol (GPI)-linked glycoprotein that has low β-sheet content and is sensitive to proteases. In contrast, PrPSc is a polymer, is detergent insoluble, has an increased proportion of β-sheet and is partially resistant to proteases.Citation6-8
Protein misfolding cyclic amplification (PMCA) is an accelerated simulation of prion replication in vitro and involves using minute amounts of PrPSc/res, called the seed, and excess amounts of PrPC, called the substrate.Citation7,9,10,11 PMCA has been shown to be efficient at detecting very small amounts of scrapie PrPScCitation12 and is capable of detecting scrapie PrPSc in the blood or urine of scrapie-infected hamsters.Citation13-16 Using normal brain homogenate (NBH) as a substrate in PMCA, it has been shown that the newly generated PrPSc exhibits the same biochemical and structural properties as brain-derived PrPSc.Citation17,18 In contrast, the biological properties of recombinant PrPSC (rPrPSc) generated by PMCA using bacterially-expressed PrPC (rPrPC) as substrate were different from those of the PrPSc seed, despite identical amino acid sequence of rPrPC to that of the brain-derived PrPC.Citation19 Mammalian PrPC is variably glycosylated at 2 potentially N-glycosylation sites and has a GPI anchor.Citation8,20 Efforts to mimic prion protein conversion and prion propagation in cell-free conversion systems relied on the use of brain-derived PrPC for CWDCitation21,22 and brain-derived or bacterially-expressed recombinant prion proteins for scrapie.Citation23-26 However, bacterially expressed prion proteins are unglycosylated and lack the glycophasphatidylinositol anchor (GPI); therefore these recombinant proteins may not be ideal substrates for the PrPC conversion process. Importantly, the GPI anchor was found to be important for the amplification of PrPSc by PMCA.Citation20,27 Thus we hypothesize that glycosylated and GPI-anchored recombinant proteins produced in a baculovirus expression system can function efficiently as substrate in the in vitro propagation of PrPSc using PMCA.
Studies using in vitro conversion systems suggest that host-encoded factors other than PrPC may be required to propagate prions.Citation28-33 Specifically, several classes of negatively charged macromolecules and host-encoded RNA molecules have been shown to bind to and/or stimulate PrP conversion in vitro.Citation34-38 Citation34,39 In this study, we tested the hypothesis that the addition of species specific mRNA molecules to PMCA would stimulate the conversion of baculovirus-expressed sheep and cervid PrPC. Herein, we report that baculovirus-expressed recombinant PrPC supports the amplification of PrPSc and PrPCWD in seeded PMCA, respectively. The addition of species-specific mRNA molecules to PMCA of sheep and cervid prions results in increased conversion efficiency to PrPSc and PrPCWD, whereas the addition of the divalent cation Manganese (Mn) to unseeded PMCA induces conversion of baculovirus-expressed PrPC to protease resistant PrP.
RESULTS
Expression and characterization of recombinant PrPC
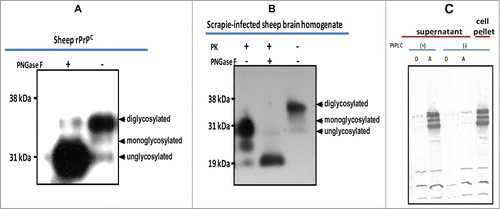
Using the baculovirus expression system we expressed recombinant PrPC of sheep () and white tailed deer (), in Sf9 or mimic Sf9 cells. Immunofluorescent antibody analysis showed high level of recombinant protein expression manifested by strong specific fluorescence around the cell surface (). Expression level of recombinant PrPC showed yield estimates ranging from 8 mg to 10 mg/L or 1.2 mg to1.5 mg per 2 × 107 cells. Western blot analysis of the expressed proteins showed that recombinant PrPC has a glycoform pattern similar to brain-derived PrPC from mammalian species (). The recombinant protein exhibited 3 glycoform bands: di-glycosylated band, mono-glycosylated and un-glycosylated band. Comparative quantitative analysis of rPrPC purification methods showed that a partial purification method yielded rPrP of sufficient quality for use in PMCA (). To examine whether rPrP expressed in insect cells was post-translationally modified with N-glycans, rPrPC as well as PrPSc (from PK-digested scrapie-infected sheep brain), were deglycosylated by treatment with the enzyme peptide-N-glycosidase F (PNGase F), which specifically cleaves N-linked glycans from glycoproteins. Digestion resulted in a single unglycosylated band with both recombinant brain-derived PrPC (). To assess whether the rPrPC was GPI-anchored similar to mammalian-expressed PrPC, we treated insect cells expressing rPrPC with phosphatidylinositol-specific phospholipase C (PIPLC) and subjected the supernatant to Triton X-114 phase partitioning. Cleavage of the GPI anchor resulted in appearance of rPrPC in the aqueous phase of TX-114 partition and in the cell pellet of untreated samples (), indicating that the rPrPC is GPI-anchored.
Figure 1. (A and B) Baculovirus-expressed PrP of sheep and white-tailed deer, respectively, showing the characteristic glycoform profiles, diglycosylated, monoglycosylated and unglycosylated bands, similar to brain-derived PrP from normal brain homogenate (NBH) of sheep. (C) An IFA staining of recombinant baculovirus-infected Sf9 cells showing expression of PrP. The cells were stained with P4 monoclonal antibody (mAbP4). Sheep PrP genotypes, VLRQ and AFRQ, as well as white-tailed deer, show specific fluorescence. The negative control shows no specific fluorescence. (D and E) Preparation of sheep rPrPC by affinity column chromatography and by the partial purification methods, respectively. W = wash/waste fraction; F1 – F2 = eluted fractions (1 to 5). CL = non-purified lysate; AFRQ and VLRQ depict sheep PrP genotypes. M = molecular weight marker. Proteins were detected with the monoclonal antibody, mAbP4.
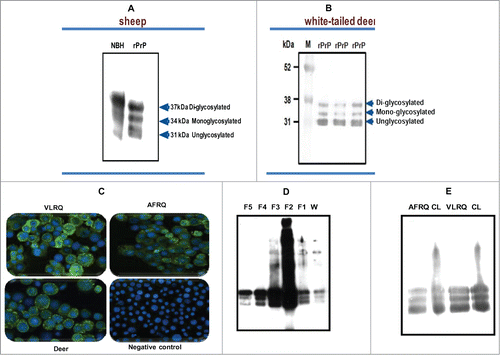
Figure 2. Characterization of baculovirus-expressed PrPC. (A) Cleavage of N-linked glycans from rPrPC by PNGaseF treatment. (B) Cleavage of N-linked glycans from sheep PrP derived from scrapie-infected sheep brain. A molecular weight shift confirms N-linked glycosylation of rPrPC characteristic of mammalian expressed PrPC. (C) GPI anchor assay using PIPLC and Triton X-114 phase partitioning. Cleavage by the enzyme releases the recombinant protein into the medium (supernatant), which upon partitioning was detected in the aqueous (A) phase. No recombinant protein could be detected in the supernatant of non-treated cells. The recombinant protein remained associated with the cell pellet in non-treated cells. Proteins were detected with the monoclonal antibody, mAbP4. PK = proteinase K.
PMCA
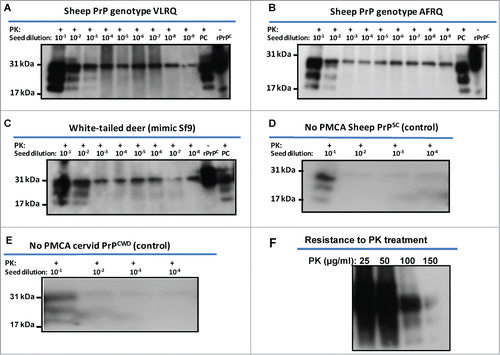
Using the recombinant PrPC proteins, PMCA was carried out according to the protocol described in materials and methods using baculovirus-expressed PrPC substrates. Using the proteins either partially or affinity purified in seeded PMCA as substrates showed no apparent differences in PMCA performance (data not shown). Given these results, subsequent PMCAs were carried out using partially purified recombinant PrP substrates. The reactions were seeded with PrPSc from the brain of scrapie-infected sheep or PrPCWD from a CWD-infected cervid. Both sheep rPrPC (from both genotypes) and white-tailed deer rPrPC supported seeded PMCA amplification of PrPSc and PrPCWD, respectively, without addition of exogenous cofactors (). Recombinant PrPC substrates of VLRQ genotype showed comparatively higher conversion efficiency than the AFRQ genotype, manifesting stronger band intensities and amplification of PrPSc inoculum in a 10–100 fold dilution above the baseline (). Recombinant PrPC of both genotypes of sheep supported amplification in a single round of PMCA of 48 cycles. Similarly, recombinant PrPC of white-tailed deer supported amplification of PrPCWD in a single round of PMCA (). Recombinant PrPC of sheep and white-tailed deer seeded with fold10- serially diluted PrPSc or PrPCWD as non-PMCA controls did not show amplification of the PrPSc or PrPCWD inoculum (). Unseeded negative controls were subjected to PMCA and have shown no specific amplification (data not shown). Using the baculovirus-expressed PrPC as substrate in PMCA, the resulting molecular weight of rPrPSc or rPrPCWD were similar to the brain-produced PrPSc or PrPCWD (the seed) (). The bands manifested the typical glycoform pattern, diglycosylated, monoglycosylated and unglycosylated isoforms of PrPSc. The rPrP seeded PMCA products manifested a fairly high degree of proteinase K resistance with resistance up to 100 μg/ml of PK digestion ().
Figure 3. Amplification of PrPSc or PrPCWD using sheep and white-tailed deer rPrPC as substrate. Recombinant sheep PrPC of both genotypes, VLRQ and AFRQ, and white-tailed deer PrPC support amplification of PrPSc (A and B) and PrPCWD (C), in seeded-PMCA; PMCA using white-tailed deer rPrPC was expressed in mimic Sf9 cells (Faint glycobands observed in 10−5 and 10−6 dilutions are attributed to carryover of unhomogenised residual inoculum, PrPCWD). (D) = No PMCA control for sheep PrPSc using sheep rPrPC (VLRQ) substrate; (E) = No PMCA control for cervid PrPCWD using white-tailed deer rPrPPC substrate; F = resistance of rPrPSc to PK treatment; PC = positive control (PK-treated scrapie or CWD-infected brain tissue from sheep or muntjac, respectively; PK = proteinase K.
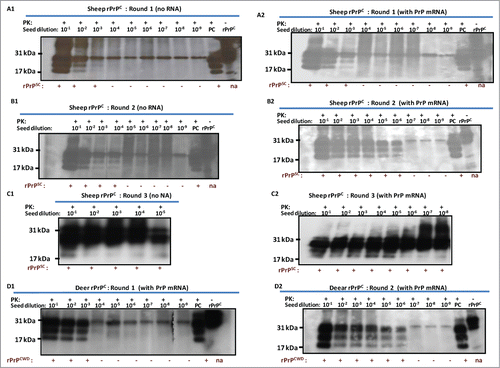
In vitro transcribed PrP mRNA enhances efficiency of seeded PMCA
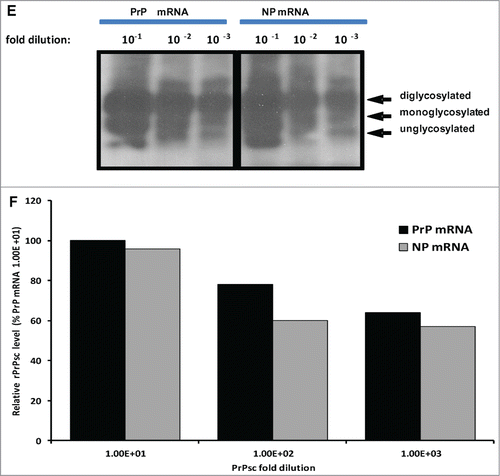
Previous experiments showed that RNA cofactors that stimulate PrPSc amplification are species specific.Citation34 In vitro transcribed sheep and deer PrP mRNA molecules were used to examine the effect of RNA on conversion efficiency in seeded PMCA. For this purpose, 12 μg of each transcript were added to respective PMCAs using reaction cycles as described above. PrP mRNA molecules of both sheep and white-tailed deer enhanced conversion of rPrPC to PrPSc or PrPCWD as manifested by increased signal intensity and increased detection signal ranging from 100–1000-fold (A1-D2). Addition of VLRQ PrP-specific mRNA transcripts to scrapie seeded PMCA, in the first round using VLRQ rPrPC as substrate, increased sensitivity and detection of the 3 glycoform bands of rPrPSc (). In a second round PMCA, addition of PrP-specific mRNA transcripts increased intensity of rPrPSc band intensity as well as detection sensitivity by a 100-fold; rPrPSc exhibiting the 3 glycoform bands, was amplified in 10−3 dilution of the inoculum, without PrP mRNA (), whereas with the addition of PrP-specific RNA, the efficiency of amplification increased 1,000-fold and rPrPSC was amplified in 10−6 dilution (). A notable increase in conversion efficiency manifested by enhanced band intensities occurred in the third round PMCA (). In this round, addition of PrP-specific mRNA increased detection sensitivity 1,000-fold resulting in amplification in up to10−8 dilution of the inoculums (). Similar results were obtained with AFRQ mRNA transcripts (data not shown). Addition of cervid PrP mRNA transcripts to CWD-seeded PMCA produced a similar effect of enhanced conversion efficiency and increased detection. Without mRNA, PrPCWD was amplified in a 10−3 dilution of the inoculum, whereas the addition of cervid PrP mRNA resulted in detection in up to 10−6 dilution (). To examine the comparative effect of species-specificity of mRNA molecules on seeded PMCA, we added 12 μg of in vitro mRNA transcripts of the Rift Valley fever virus (RVFV) nucleoprotein to seeded PMCA and subjected samples to 48 cycles of PMCA. No statistically significant difference (P > 0.05) in conversion or amplification efficiency was observed between the 2 RNA transcripts ().
Figure 4. Addition of 12 ug of in vitro transcribed sheep or white-tailed deer PrP mRNA molecules to seeded-PMCA resulted in enhanced PrPSc or PrPCWD amplification and increased sensitivity by at least a 100-fold in the second round PMCA. Panel A1 and A2 represent first round PMCA, without and with sheep PrP mRNA, respectively, of scrapie seeded PMCA using sheep rPrPC (VLRQ) as substrate. Panel B1 and B2, represent second round PMCA, without and with sheep PrP mRNA, respectively. Panel C1 and C2 show the third round of PMCA as indicated in the figure (negative and positive controls are on a separate gels and are not shown). Panel D1 and D2 show the effect of white-tailed deer PrP mRNA on the efficiency of first and second round CWD-seeded PMCA using white-tailed rPrPC as substrate. PMCA results (rPrPsC or rPrPCWD positivity) are indicated below each panel. Panel E shows the effect of specific (PrP mRNA) RNA on conversion efficiency in seeded PMCA. Rift Valley fever virus nucleoprotein (NP) mRNA is utilized as a non-specific RNA control. Densitometric analysis of the rPrPSC bands (Panel F). www.landesbioscience.
Manganese induces conversion of rPrPC to protease-resistant PrP
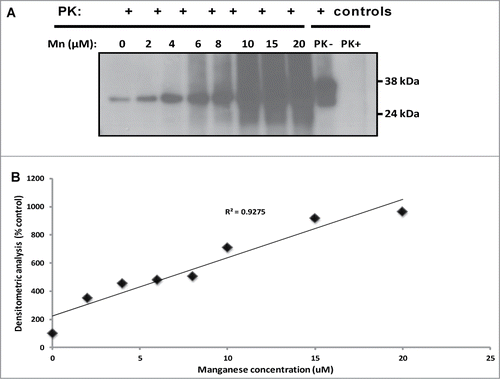
The effect of the divalent cation, manganese, on PrPSc formation was evaluated by addition of 2 – 20 μM of MnCl2 to PMCA using rPrPC of sheep () without seed and subjected them to 48 cycles of PMCA as described above. Manganese induced conversion of rPrPC to a protease resistant PrP, shown by the presence of a 32 kDa proteinase-K resistant band (up to 100 μg/ml). The level of protease-resistant PrP formation was dose-dependent (). Similar data was obtained with white-tailed deer PrPC (data not shown).
Figure 5. Manganese induces conversion of sheep rPrPC to a protease-resistant PrP in non-seeded PMCA. There is an increased formation of 32 kDa protease-resistant band in a dose –dependent manner (A); an increasing trend of protease-resistant PrP formation with increased manganese concentration is shown (B). Similar results were obtained with deer rPrPC (data not shown). Samples were digested with 100 ug/ml of proteinase K. Protease-resistant PrP was detected with the monoclonal antibody, mAbP4.
Discussion
The development of sensitive diagnostic assays to detect TSE infection in humans and animals is critical in addressing the challenges posed by prion diseases. In this study, we amplified, for the first time, PrPSc and PrPCWD by PMCA, with or without the addition of co-factors using baculovirus-expressed recombinant PrP of sheep and white-tailed deer as substrates. Herein, products derived from the seeded-PMCA were referred to as recombinant PrPSc (rPrPSc) or PrPCWD (rPrPCWD) since infectivity in a bioassay model is yet to be determined. A recent report, in contrast, shows that baculovirus-expressed PrPC substrate supports amplification of mouse scrapie prion but only in the presence of brain homogenate from Prnp knockout mouse (Prnp0/0), as a source of undefined co-factorsCitation27. Although the reasons for the divergent results are unknown, a host of factors could have contributed. The latter study used as seed the Chandler strain of mouse-adapted scrapie and a baculovirus-expressed PrPC substrate of mouse origin, which may contain subtle structural or conformational differences from the scrapie prion protein. There are reports describing utilization of different cellular co-factors by different prion strains or strains from different species,Citation40 including seeded conversion of recombinant prion protein expressed in E. coli by PMCA without the addition of co-factors.Citation4 The advantage of using baculovirus-expressed recombinant PrP as a substrate in seeded PMCA is that it seem to amplify and preserve the native structure of PrPSc (). Unlike E. coli-derived rPrP as substrates,Citation4,9,41 baculovirus-expressed rPrP preserved the typical protease-resistant core glycoform profile with diglycosylated, monoglycosylated and unglycosylated bands (). Given the fact that recombinant PrPC expressed in E. coli are unglycosylated and lack a GPI anchor,Citation4 the results suggest that that these posttranslational modifications are required for the preservation and amplification of a native PrPSc conformation. Indeed, the GPI anchor of PrPC has been shown to be important for the amplification of a native PrPSc conformation in vitro by PMCA.Citation20,27 Preservation of a native PrPSc conformation in seeded PMCA could be of significant importance for studies into the fundamental structure and conversion mechanism of PrPSc. Furthermore, the diglycosylated PrPres product vastly predominates over mono- and unglycosylated products; and in the starting substrate, the abundance of the 3 glycosylation forms appeared more balanced (). Such a pattern could be an intrinsic feature of baculovirus-expressed recombinant PrPC as substrate; similar results have been observed in animal models, in which the host has predominant influence over PrP glycoform patterns.
Recombinant PrPC of both sheep and white-tailed deer converted to rPrPSc and rPrPCWD, respectively, in seeded PMCA allowed detection of PrPSc or PrPCWD in 1000-fold dilutions of the inoculum. PrPC substrate of the VLRQ genotype resulted in a higher conversion efficiency (manifested by the intensity of protease-resistant bands) than the AFRQ substrate (data not shown); this appeared to correlate with the higher susceptibility of sheep of VLRQ genotype to scrapie prion disease. Additionally, with white-tailed deer rPrP expressed in regular Sf9 cells did efficiently efficiently support amplification of PrPCWD in CWD-seeded PMCA (data not shown); deer rPrPC expressed in mimic Sf9 cells. Mimic Sf9 cells are engineered to stably express human glycosyltransferases, and manifest a different pattern and complexity of glycosylation from the regular Sf9 cells.Citation42-44 This suggests that the structure and pattern of glycosylation of the PrPC substrate could influence the conversion efficiency of deer PrPC to the misfolded PrPCWD. Interestingly, glycosylation has been found to hinder conversion of protease-sensitive PrPC to protease-resistant PrPSc in a mouse-hamster model,Citation45 and has been attributed to partially obscuring the site in mouse PrPC bound by hamster PrPSc.Citation46 It is assumed that the conformational structure and glycosylation pattern of mimic Sf9-generated deer PrPC substrate presented less structural and steric hindrance to binding PrPCWD, thus facilitating conversion in PMCA.
RNA molecules have been reported to increase prion conversion efficiencyCitation39 in a species-specific manner.Citation34 Thus, in this study, the effect of in vitro transcribed prion-specific mRNA molecules on conversion efficiency of rPrPC to rPrPSc or rPrPCWD in seeded PMCA was examined. Indeed, PrP-specific mRNA transcripts of both sheep and white-tailed deer enhanced rPrPC conversion efficiency and increased sensitivity of seeded PMCA by at least a 100-fold in the second round (). However, this effect seems unspecific for PrP or species-specific mRNA as signals generated by the addition of RVFV nucleocapsid mRNA transcripts were similar when compared to those in the presence of PrP-specific mRNAs (P > 0.05). Several studies have demonstrated the stimulatory effect of various polyanionic compounds, including host-encoded RNA, on prion seeded conversion of PrPC into PrPSc molecules.Citation33,34,39,47-49 Although, the molecular mechanisms involved in the stimulation remain to be elucidated, it is postulated that endogenous polyanionic cofactors participate in prion propagation in vivo, either as catalysts or scaffolds complexed to PrPSc molecules.Citation39 Citation39,50
To improve our understanding of the role of divalent cations as cofactors in PrPSc formation or prion pathogenesis, the effect of manganese on rPrPC conversion was examined in unseeded PMCA. Micromolar amounts of manganese induced conversion of rPrPC to a protease-resistant PrP; the efficiency of conversion is dose-dependent and correlates with the amount of manganese in the reaction. Studies suggest that interactions between PrPC molecules and endogenous manganese may contribute to the pathogenesis of prion diseases.Citation39,51,52 Interestingly, the octapeptide repeat region in the N-terminus of the PrPC structure has been established as the target for metal binding,Citation51,53-55 and recent studies show that binding of manganese in this region causes conformational changes within the proteinCitation56,57 as well as structural reorganization,Citation58 which may promote misfolding and aggregation of the PrPC protein,Citation52 a hallmark of prion diseases.
In conclusion, this is the first report describing in vitro amplification of PrPSc or PrPCWD in PMCA using recombinant baculovirus-expressed PrP as substrates in the presence or absence of exogenous co-factors. It is also the first reported amplification of PrPCWD using recombinant substrates and could be an important contribution to efforts of developing a practical and sensitive diagnostic assay for the detection of CWD. The preservation of the native structure of PrPSc/PrPCWD in seeded PMCA using the recombinant baculovirus expressed PrPC substrates is an essential feature that could facilitate studies into the fundamental structure and conversion mechanism of PrPSc or PrPCWD. Collectively, this represents an important step toward the development of sensitive and practical means of detection of prion diseases using recombinant PrPC substrates derived from eukaryotic expression systems, which could have broader application in the detection of other protein misfolding diseases, such as Alzheimer, Parkinson and Huntington's diseases.
Materials and Methods
Cloning and expression of recombinant sheep and white-tailed deer PrP genes
A DNA fragment encoding amino acid residues 1 to 256 of sheep PrP from 2 prion protein gene (PRNP) genotypes, V136L141R154Q171 and A136 F141R154Q171, and white-tailed deer of genotype, Q95, G96, X132, Y216 was amplified by PCR using high fidelity pfx50 DNA polymerase (Invitrogen/Life Technologies, Carlsbad, CA). The primers used to amplify the PRNP genes are listed in . Genomic DNA purified from sheep brain tissue of 2 different sheep PrP genotypes, VLRQ and AFRQ, respectively, were used as templates in PCR. Genomic DNA purified from a lymph node obtained from a white-tailed deer was used as template to amplify the deer PrP coding sequence. The PCR fragments were cloned into pFastBac/CT-TOPO vector by blunt-end TOPO cloning according to the manufacturer's instruction. The cloning reaction was transformed into One Shot Mach1 T1 chemically competent E. coli (Invitrogen/Life Technologies) to create the following donor plasmids: pRF-VLRQ, pRF-AFRQ and pRF-QGXY for VLRQ, AFRQ genotypes of sheep and white-tailed deer, respectively. The correct size and sequence of the inserts in the donor clones was confirmed by PCR, restriction enzyme analysis and DNA sequencing. Each donor plasmid was transformed into MAX Efficiency DH10Bac competent E. coli (Invitrogen/Life Technologies) to generate recombinant bacmids by site-specific transpositioning. Transpositioning of the gene of interest in the recombinant bacmid was confirmed by PCR using M13F and M13R primers (). The purified recombinant bacmid DNA was used to transfect Spodoptera frugiperda 9 (Sf9) insect cells using Cellfectin reagent (Invitrogen/Life Technologies). These cells were maintained at 27°C, in order to generate the initial recombinant viral stock (P1 viral stock). In the case of white-tailed deer, the recombinant bacmid was also transfected into “mimic Sf9” cells (Invitrogen/Life Technologies), which stably expressed human N-glycosyltransferases. The rescued P1 virus stock was used to infect Sf9 or mimic Sf9 cells to increase the viral titer (P2 viral stock). Expression of recombinant PrPC was carried out using viral stocks P2 and above. Infected cells were harvested after 72 to 96 hours post infection and recombinant PrPC was purified and used as substrate in PMCA.
Table 1 Primers used for cloning PRNP genes and in vitro transcription of PrP mRNA molecules Primers used for cloning PRNP genes and amplification of PrP mRNA transcription templates
Immunofluorescence antibody assay (IFA)
To demonstrate PrPC expression by recombinant viruses and to identify the site of protein localization, we carried out indirect fluorescent antibody tests. Briefly, a suspension of recombinant baculovirus-infected Sf9 cells was cytospinned onto glass slides. The slides were fixed in 4% paraformaldehyde for 30 min at room temperature and then incubated in 1X PBS (pH 7.4) containing 1% BSA for 45 min at 37°C. Ten microliters of 1:100 dilution of monoclonal antibody (mAb) P4 (Ridascreen Biopharm, Darmstadt, Germany) was added and slides were incubated for 30 min at 37°C in a moist chamber. Then slides were soaked in 1X fluorescence antibody (FA) rinse buffer (2.85 g Na2CO3, 8.4 g NaHCO3, 2.125 g NaCl, distilled water to 1000 ml; pH 9 to 9.5) for 10 min. Ten microliters of FITC-conjugated anti-mouse IgG antibody was then added and incubated in a dark moist chamber at 37°C for 30 min. Slides were soaked in 1X FA rinse buffer for 10 min. Thereafter, one drop of anti-fade mounting buffer containing DAPI counter stain was added, the slide was covered with a cover slip and examined under a fluorescence microscope (Nikon, Eclipse 90i).
Peptide-N-Glycosidase deglycosylation assay
About 300 μl of baculovirus-expressed and column purified rPrPC was centrifuged at 100,000 x g for 30 min at 4°C. The pellet was resuspended in 2 μl of 10X denaturing buffer (New England Biolabs, Ipswich, MA) and 18 μl distilled water. As a control, brain-derived mammalian PrPC, and proteinase K-digested scrapie infected 10% brain homogenate from sheep was subjected to centrifugation and resuspension as described. The mixtures were denatured by heating at 100°C for 10 min. Thereafter, the following reagents were added to the reaction: 4 μl of 10x reaction buffer, 4 μl 10% NP40, 4 μl distilled water and 6 μl PNGase F (500,000 U/ml) (New England Biolabs, Beverly, MA). The mixture was incubated at 37°C for 1 hour. The reaction was stopped by adding SDS loading buffer. The samples were separated by SDS-PAGE in NuPAGE 12% Bis-Tris gels (Invitrogen/Life Technologies) and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were probed in western blot as described below.
Phosphatidylinositol-specific phospholipase C digestion and TritonX-114 phase partitioning
Triton X-114 phase partitioning analysis was performed as described previouslyCitation12 with some modifications. Cells were grown and infected with recombinant baculovirus in T75 flasks. The medium was discarded and the cells were washed twice with PBS (pH7.4) and resuspended in the same buffer. One batch of samples was treated with 0.4U PIPLC/ml and the other batch was mock-treated. Both samples were incubated at 37°C for 1 hr and centrifuged at 3000 x g for 3 min to collect the cell pellet. The supernatant of PIPLC-treated cells was re-centrifuged at 20,000 x g for 3 min. All pellets were resuspended in PBS containing 1X protease inhibitor. rPrPC in the supernatant from PIPLC-treated cells and non-treated cells was treated with 0.25 volume of TritonX-114 at 4°C. Samples were subsequently centrifuged at 3000 x g for 3 min at 4°C to remove any insoluble aggregates. The supernatant was heated at 37°C for 3 min to induce phase transitioning and then re-centrifuged at 3000 x g for 3 min at 25°C. Proteins in the upper (aqueous) and lower (detergent) phases were precipitated with 5 volumes of methanol. PrP in all fractions was analyzed by Western blot analysis.
Purification of recombinant PrPC (rPrPC) substrate
The use of rPrPC as substrate in PMCA requires a purification step that is laborious and time-consuming. Thus, 2 different methods were used to prepare and evaluate recombinant PrP for use as substrate in PMCA: i) partial purification by differential centrifugation and filtration and ii) affinity column purification using Ni-NTA superflow resin (Novagen-EMD Millipore, Billerica, MA). For partial purification, the cells were washed twice with PBS (pH 7.4) and resuspended in 1x PMCA conversion buffer composed of PBS pH 7.4 containing 150 mM NaCl, 4 mM EDTA, 1% Triton X-100 and Complete protease inhibitor (Roche Diagnostic, Rockland, MA). The suspension was incubated on ice for 10 min and passed through a 26.75 G needle twice for complete lysis. Benzonase (Novagen-EMD Millipore, Billerica, MA) was added to 10U per ml and incubated on ice for a further 15 min. The preparation was then centrifuged at 500 x g for 15 min at 4°C and the supernatant was collected and filtered through a 45 μm filter. The filtered protein was aliquoted in 500 μl aliquots and stored at -80°C until used. For Ni-NTA affinity column purification, cells were washed with PBS as described above. The pellet was resuspended in Ni-NTA binding buffer (300 mM NaCl, 50 mM Na3PO4, pH 8.0, and 10 mM imidazole) containing 1x Complete protease inhibitor; then the Insect Popculture Reagent (Novagen®) was added at 0.05 volumes of original culture volume. The lysate was incubated at room temperature for 15 min. The suspension was centrifuged at 500 x g at 4°C and the supernatant mixed with previously equilibrated Ni-NTA superflow resin (Novagen-EMD Millipore, Billerica, MA). The mixture was allowed to bind for 1 hour at 4°C. The suspension was loaded into a column and washed with 5 volumes of wash buffer (300 mM NaCl, 50 mM Na3PO4, pH 8.0, and 20 mM imidazole). Bound rPrPC was eluted with elution buffer (300 mM NaCl, 50 mM Na3PO4, pH 8.0, and 250 mM imidazole). Pooled fractions were dialysed overnight at 4°C against dialysis buffer (PBS pH 7.4, 150 mM NaCl, 4 mM EDTA). Recombinant proteins were analyzed by protein gel blot. Protein concentration was determined by bicinchinionic method using bovine serum albumin (BSA) as standard. Aliquots of the protein were stored at −80°C until used.
In vitro transcription of PrP mRNA molecules
In vitro transcription was carried out to generate mRNA molecules for VLRQ and AFRQ PRNP genotypes of sheep and for the white-tailed deer PRNP. To generate the transcription template for in vitro RNA synthesis, PCR was carried out on donor plasmids, pRF-VLRQ, pRF-AFRQ and pRF-QGXY to amplify the VLRQ, AFRQ and white-tailed deer PRNP transcription templates, respectively, using primers listed in . In vitro transcription was enabled by a T7 promoter cloned upstream of the transcription start site of each coding sequence. As a non-prion RNA control, the Rift Valley fever virus (RVFV) nucleoprotein mRNA was cloned and transcribed as described above. Briefly, PCR was carried out on plasmid pRF-N containing RVFV nucleoprotein coding sequence to generate the transcription template using primers listed in . All PCR reactions were carried out using high fidelity pfx DNA polymerase (Invitrogen/Life Technologies) which has a 3’ to 5’ exonuclease proof reading activity. The PCR products were purified using Qiagen PCR purification kit (Qiagen, Valencia, CA) and DNA concentrations were measured using the Nanodrop (Thermoscientific, Wilmington, DE). In vitro transcription was carried out in 200 μl reaction volumes containing 5X T7 transcription buffer (80 mM HEPES-KOH [pH 7.5], 24 mM MgCl2, 2 mM spermidine and 40 mM DTT) (Promega Corporation, Madison, WI), 7.5 mM of each of the NTPs (ATP, CTP, GTP, UTP) (Promega Corporation, Madison, WI), 9 μg of DNA template and 20 μl of enzyme mix (T7 RNA polymerase, recombinant RNasin Ribonuclease Inhibitor and Recombinant Inorganic Pyrophosphatase) (Promega Corporation, Madison, WI). The reaction was incubated at 37°C for 4 hrs. Afterwards, DNase was added to a concentration of 1 U/μg of template DNA and incubated for a further 15 min at 37°C. mRNA transcripts were extracted and cleaned using the RNeasy Clean-up protocol (Qiagen, Valencia, CA). RNA concentrations were measured at 260/280 nm as described previously. The integrity of RNA transcripts was analyzed by gel electrophoresis.
PMCA
PMCA was carried out in a total volume of 100 μl in 0.2 ml PCR tubes. For this, 90 μl of rPrPC resuspended in PMCA conversion buffer was added to each tube. Ten microliters of 10% scrapie-infected sheep brain or 10% CWD-infected muntjac brain homogenate serially diluted fold10- in conversion buffer was added to each substrate. To analyze the effect of in vitro synthesized mRNA molecules on the efficiency of PMCA, 12 μg of RNA molecules were added to each reaction. The tubes were positioned on an adaptor placed on the plate holder of a sonicator (Misonix Model 3000, Farmingdale, NY). For scrapie-seeded PMCA, each cycle consisted of 30 min incubation at 37°C, 30 sec pulse of sonication at 60% amplitude for a total of 48 cycles. For CWD-seeded PMCA, each cycle consisted of 29.50 min incubation at 37°C, 30 sec sonication at 40% amplitude for a total of 144 cycles. Thereafter 10 μl aliquot of the amplified material was diluted into 90 μl of recombinant PrPC substrate and a new round of cycles of PMCA as described above was performed. To evaluate the effect of manganese on rPrPC conversion, varying concentrations (2–20 μM) of Manganese chloride was added to rPrPC and subjected to 48 cycles of unseeded PMCA as described above for scrapie. All reactions were stopped by the addition of 2 mM of phenylmethylsufonyl fluoride (PMSF). After each round of PMCA, 25–50 μg/ml of proteinase K was added to aliquots of the reaction products and incubated at 37°C for 1 hr. Loading buffer was added and samples were boiled in a heating block at 100°C for at least 5 min and then analyzed by western blot.
Western blot
Proteins were fractionated by gel electrophoresis in 12% Bis-Tris Polyacrylamide gel using 1x MOPS running buffer (Invitrogen/Life Technologies) and then transferred onto PVDF membrane by electroblotting. The membrane was blocked in 0.1% Tween-20 in PBS (pH7.4) containing 3% bovine serum albumin (BSA). mAbP4 (Ridascreen Biopharma, Darmstadt, Germany), raised against PrP amino acid residues 89–104, diluted 1:3000 in blocking buffer, was used as the primary antibody. Horse-radish peroxidase-conjugated goat anti-mouse monoclonal antibody (Santa Cruz Biotechnology, Ca. USA) at a dilution of 1:5000 in blocking buffer was used as the secondary antibody. Immunoreactive bands were visualized by enhanced chemiluminescence using ECL detection kit (Amersham, Piscataway, NJ) and X-ray film. Densitometry to quantify signal intensity of reactive bands in PMCA was carried out using AlphaView software (ProteinSimple, Santa Clara, CA).
Prion isolates
The scrapie prion isolate used in this study originated from a sheep farm in Idaho, USA. Scrapie infection is maintained in the sheep herd and scrapie-infected brain homogenate was obtained from a clinical case of scrapie confirmed by protein gel blot and immunohistochemistry (IHC). The CWD prion isolate was kindly donated by Dr. Alan Young, South Dakota State University. The CWD-infected brain homogenate was obtained from a muntjac manifesting clinical signs of CWD, with CWD status confirmed by western blot and IHC.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
The authors would like to thank Dr. England of the University of Idaho for providing scrapie-infected brain material and Dr. Alan Young for providing CWD-infected muntjac brain material. We thank Dr. Bhupinder Bawa for the IFA imaging.
Funding
This study is supported by the National Institutes of Health (NIH) grants: P01 A1 77774–01 (JAR) and ES19267 (AGK).
References
- Prusiner SB. Prions. Proc Natl Acad Sci 1998; 95:13363-83; http://dx.doi.org/10.1073/pnas.95.23.13363
- Caughey B, Baron GS, Chesebro B, Jeffrey M. Getting a grip on prions: oligomers, amyloids, anchors and pathological membrane interactions. Annu Rev Biochem 2009; 78:177-204; PMID:19231987; http://dx.doi.org/10.1146/annurev.biochem.78.082907.145410
- Surewicz WK, Jones EM, Apetri AC. The emerging principles of mammalian prion propagation and transmissibility barriers: insight from studies in vitro. Acc Chem Res 2006; 39:654-62; PMID:16981682; http://dx.doi.org/10.1021/ar050226c
- Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, Onwubiko HA, Priola SA, Caughey B. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods 2007; 4:645-50; PMID:17643109; http://dx.doi.org/10.1038/nmeth1066
- Caughey B. Prions and spongiform encephalopathy (TSE) chemotherapeutics: a common mechanism for anti-TSE compounds? Acc Chem Res 2006; 39:646-53; PMID:16981681; http://dx.doi.org/10.1021/ar050068p
- Pan KM. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion protein. Proc Natl Acad Sci USA 1993; 90:10962-6; PMID:7902575; http://dx.doi.org/10.1073/pnas.90.23.10962
- Caughey B, Raymond GJ. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J Biol Chem 1991; 266:18217-23; PMID:1680859
- Rudd PM, Wormald MR, Wing DR, Prusiner SB, Dwek RA. Prion glycoprotein: structure, dynamics, and roles for the sugars. Biochemistry 2001; 40:3759-66; PMID:11300755; http://dx.doi.org/10.1021/bi002625f
- Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 2001; 411:810-3; PMID:11459061; http://dx.doi.org/10.1038/35081095
- Saa P, Castilla J, Soto C. Presymptomatic detection of prions in blood. Science 2006; 313:92-4; PMID:16825570; http://dx.doi.org/10.1126/science.1129051
- Soto C, Saborio GP, Anderes L. Cyclic amplification of protein misfolding: application to prion-related disorders and beyond. Trends Neurosci 2002; 25:390-4; PMID:12127750; http://dx.doi.org/10.1016/S0166-2236(02)02195-1
- Borchelt DR, Morales R, Stahl N, Telling G, Pruisner SB. Release of the cellular prion protein from cultured cells after loss of its glycoinositol phospholipid anchor. Glycobiology 1993; 3:319-29; PMID:7691278; http://dx.doi.org/10.1093/glycob/3.4.319
- Castilla J, Saa P, Soto C. Detection of prions in blood. Nat Med 2005; 11:982-5; PMID:16127436
- Gonzalez-Romero D, Barria MA, Leon P, Morales R, Soto C. Detection of infectious prions in urine. FEBS letters 2008; 582:3161-6; PMID:18706416; http://dx.doi.org/10.1016/j.febslet.2008.08.003
- Murayama Y, Yoshioka M, Okada H, Takata M, Yokoyama T, Mohri S. Urinary excretion and blood level of prions in scrapie-infected hamsters. J General Virol 2007; 88:2890-8; PMID:17872544; http://dx.doi.org/10.1099/vir.0.82786-0
- Thorne L, Terry LA. In vitro amplification of PrPSc derived from the brain and blood of sheep infected with scrapie. J Gen Virol 2008; 89:3177-84; PMID:19008409; http://dx.doi.org/10.1099/vir.0.2008/004226-0
- Castilla J, Morales R, Saa P, Barria M, Gambetti P, Soto C. Cell-free propagation of prion strains. EMBO J 2008; 27:2557-66; PMID:18800058; http://dx.doi.org/10.1038/emboj.2008.181
- Green KM, Castilla J, Seward TS, Napier DL, Jewell JE, Soto C, et al. Accelerated high fidelity prion amplification within and across prion species barriers. PLoS Pathog 2008; 4:e1000139; PMID:18769716; http://dx.doi.org/10.1371/journal.ppat.1000139
- Kim JI, Cali I, Surewicz K, Kong Q, Raymond GJ, Atarashi R, Race B, Qing L, Gambetti P, Caughey B, et al. Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J Biol Chem 2010; 285:14083-7; PMID:20304915; http://dx.doi.org/10.1074/jbc.C110.113464
- Kim J-I, Surewicz K, Gambetti P, Surewicz WK. The role of glycophosphatidylinositol anchor in the amplification of the scrapie isoform of prion protein in vitro. FEBS letters 2009; 583:3671-5; PMID:19854187; http://dx.doi.org/10.1016/j.febslet.2009.10.049
- Kurt TD, Perrott MR, Wilusz CJ, Wilusz J, Supattapone S, Telling GC, Zabel MD, Hoover EA. Efficient in vitro amplification of chronic wasting disease PrPRES. J Virol 2007; 81:9605-8; PMID:17553879; http://dx.doi.org/10.1128/JVI.00635-07
- Raymond GJ, Bossers A, Raymond LD, O’Rourke KI, McHolland LE, Bryant PK, 3rd, Miller MW, Williams ES, Smits M, Caughey B. Evidence of a molecular barrier limiting susceptibility of humans, cattle and sheep to chronic wasting disease. EMBO J 2000; 19:4425-30; PMID:10970836; http://dx.doi.org/10.1093/emboj/19.17.4425
- Kocisko DA. Cell-free formation of protease-resistant prion protein. Nature 1994; 370:471-4; PMID:7913989; http://dx.doi.org/10.1038/370471a0
- Kirby L, Birkett CR, Rudyk H, Gilbert IH, Hope J. In vitro cell-free conversion of bacterial recombinant PrP to PrPres as a model for conversion. J Gen Virol 2003; 84:1013-20; PMID:12655105; http://dx.doi.org/10.1099/vir.0.18903-0
- Caughey B. Formation of protease-resistant prion protein in cell-free systems. Curr Issues Mol Biol 2000; 2:95-101; PMID:11471561
- Wilham JM, Orru CD, Bessen RA, Atarashi R, Sano K, Race B, Meade-White KD, Taubner LM, Timmes A, Caughey B. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog 2010; 6:e1001217.
- Imamura M, Kato N, Yoshioka M, Okada H, Iwamaru Y, Shimizu Y, Mohri S, Yokoyama T, Murayama Y. Glycosylphosphatidylinositol Anchor-Dependent Stimulation Pathway Required for Generation of Baculovirus-Derived Recombinant Scrapie Prion Protein. J Virol 2011; 85:2582-8; PMID:21228241; http://dx.doi.org/10.1128/JVI.02098-10
- Telling GC. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell 1995; 83:79-90; PMID:7553876; http://dx.doi.org/10.1016/0092-8674(95)90236-8
- Saborio GP. Cell-lysate conversion of prion protein into its protease-resistant isoform suggests the participation of a cellular chaperone. Biochem Biophys Res Commun 1999; 258:470-5; PMID:10329411; http://dx.doi.org/10.1006/bbrc.1999.0660
- Stephenson DA, Chiotti K, Ebeling C, Groth D, DeArmond SJ, Prusiner SB, Carlson GA. Quantitative trait loci affecting prion incubation time in mice. Genomics 2000; 69:47-53; PMID:11013074; http://dx.doi.org/10.1006/geno.2000.6320
- Lloyd SE, Onwuazor ON, Beck JA, Mallinson G, Farrall M, Targonski P, Collinge J, Fisher EM. Identification of multiple quantitative trait loci linked to prion disease incubation period in mice. Proc Natl Acad Sci U S A 2001; 98:6279-83; PMID:11353827; http://dx.doi.org/10.1073/pnas.101130398
- Manolakou K, Beaton J, McConnell I, Farquar C, Manson J, Hastie ND, Bruce M, Jackson IJ. Genetic and environmental factors modify bovine spongiform encephalopathy incubation period in mice. Proc Natl Acad Sci U S A 2001; 98:7402-7; PMID:11404459; http://dx.doi.org/10.1073/pnas.121172098
- Deleault NR. Protease-resistant prion protein amplification reconstituted with partially purified substrates and synthetic polyanions. J Biol Chem 2005; 280:26873-9; PMID:15917229; http://dx.doi.org/10.1074/jbc.M503973200
- Deleault NR, Lucassen RW, Supattapone S. RNA molecules stimulate prion protein conversion. Nature 2003; 425:717-20; PMID:14562104; http://dx.doi.org/10.1038/nature01979
- Adler V, Zeiler B, Kryukov V, Kascsak R, Rubenstein R, Grossman A. Small, highly structured RNAs participate in the conversion of human recombinant PrP(Sen) to PrP(Res) in vitro. J Mol Biol 2003; 332:47-57; PMID:12946346; http://dx.doi.org/10.1016/S0022-2836(03)00919-7
- Gabus C, Derrington E, Leblanc P, Chnaiderman J, Dormont D, Swietnicki W, Swietnicki W, Morillas M, Surewicz WK, Marc D, et al. The prion protein has RNA binding and chaperoning properties characteristic of nucleocapsid protein NCP7 of HIV-1. J Biol Chem 2001; 276:19301-9; PMID:11278562; http://dx.doi.org/10.1074/jbc.M009754200
- Derrington E, Gabus C, Leblanc P, Chnaidermann J, Grave L, Dormont D, Swietnicki W, Morillas M, Marck D, Nandi P, et al. PrPC has nucleic acid chaperoning properties similar to the nucleocapsid protein of HIV-1. C R Biol 2002; 325:17-23; PMID:11862616; http://dx.doi.org/10.1016/S1631-0691(02)01388-4
- Nandi PK, Leclerc E, Nicole JC, Takahashi M. DNA-induced partial unfolding of prion protein leads to its polymerisation to amyloid. J Mol Biol 2002; 322:153-61; PMID:12215421; http://dx.doi.org/10.1016/S0022-2836(02)00750-7
- Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proceedings of the National Academy of Sciences 2007; 104:9741-6; http://dx.doi.org/10.1073/pnas.0702662104
- Deleault NR, Kascsak R, Geoghegan JC, Supattapone S. Species-dependent differences in cofactor utilization for formation of the protease-resistant prion protein in vitro. Biochemistry 2010; 49:3928-34; PMID:20377181; http://dx.doi.org/10.1021/bi100370b
- Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science 2010; 327:1132-5; PMID:20110469; http://dx.doi.org/10.1126/science.1183748
- Hollister J, Grabenhorst E, Nimtz M, Conradt H, Jarvis DL. Engineering the protein N-glycosylation pathway in insect cells for production of biantennary, complex N-glycans. Biochemistry 2002; 41:15093-104; PMID:12475259; http://dx.doi.org/10.1021/bi026455d
- Jarvis DL, Finn EE. Modifying the insect cell N-glycosylation pathway with immediate early baculovirus expression vectors. Nat Biotechnol 1996; 14:1288-92; PMID:9631095; http://dx.doi.org/10.1038/nbt1096-1288
- Jarvis DL, Kawar ZS, Hollister JR. Engineering N-glycosylation pathways in the baculovirus-insect cell system. Curr Opin Biotechnol 1998; 9:528-33; PMID:9821284; http://dx.doi.org/10.1016/S0958-1669(98)80041-4
- Priola SA, Lawson VA. Glycosylation influences cross-species formation of protease-resistant prion protein. EMBO J 2001; 20:6692-9; PMID:11726505; http://dx.doi.org/10.1093/emboj/20.23.6692
- Rudd PM, Merry AH, Wormald MR, Dwek RA. Glycosylation and prion protein. Curr Opin Struct Biol 2002; 12:578-86; PMID:12464308; http://dx.doi.org/10.1016/S0959-440X(02)00377-9
- Wong C. Sulfated glycans and elevated temperature stimulate PrPSc dependent cell-free formation of protease-resistant prion protein. EMBO J 2001; 20:377-86; PMID:11157745; http://dx.doi.org/10.1093/emboj/20.3.377
- Shaked GM, Meiner Z, Avraham I, Taraboulos A, Gabizon R. Reconstitution of prion infectivity from solubilized protease-resistant PrP and nonprotein components of prion rods. J Biol Chem 2001; 276:14324-8; PMID:11152454; http://dx.doi.org/10.1074/jbc.C100278200
- Ben-Zaken O. Cellular heparan sulfate participates in the metabolism of prions. J Biol Chem 2003; 278:40041-9; PMID:12871949; http://dx.doi.org/10.1074/jbc.M301152200
- Weissmann C. A unified theory of prion propagation. Nature 1991; 352:679-83; PMID:1876183; http://dx.doi.org/10.1038/352679a0
- Choi CJ, Anantharam V, Saetveit NJ, Houk RS, Kanthasamy A, Kanthasamy AG. Normal cellular prion protein protects against manganese-induced oxidative stress and apoptotic cell death. Toxicol Sci 2007; 98:495-509; PMID:17483122; http://dx.doi.org/10.1093/toxsci/kfm099
- Choi CJ, Anantharam V, Martin DP, Nicholson EM, Richt AJ, Kanthasamy A, Kanthasamy AG. Manganese upregulates cellular prion protein and contributes to altered stabilization and proteolysis: relevance to role of metals in pathogenesis of prion diseases. Toxicol Sci 2010; 115:535-46; PMID:20176619; http://dx.doi.org/10.1093/toxsci/kfq049
- Choi CJ, Kanthasamy A, Anantharam V, Kanthasamy AG. Interaction of metals with prion protein: possible role of divalent cations in the pathogenesis of prion diseases. Neurotoxicology 2006; 27:777-87; PMID:16860868; http://dx.doi.org/10.1016/j.neuro.2006.06.004
- Hooper NM, Taylor DR, Watt NT. Mechanism of the metal-mediated endocytosis of the prion protein. Biochem Soc Trans 2008; 36:1272-6; PMID:19021539; http://dx.doi.org/10.1042/BST0361272
- Todorova-Balvay D, Simon S, Creminon C, Grassi J, Srikrishnan T, Vijayalakshmi MA. Copper binding to prion octarepeat peptides, a combined metal chelate affinity and immunochemical approaches. J Chromatogr B Analyt Technol Biomed Life Sci 2005; 818:75-82; PMID:15722047; http://dx.doi.org/10.1016/j.jchromb.2004.10.011
- Brazier MW, Davies P, Player E, Marken F, Viles JH, Brown DR. Manganese binding to the prion protein. J Biol Chem 2008; 283:12831-9; PMID:18332141; http://dx.doi.org/10.1074/jbc.M709820200
- Brown DR, Hafiz F, Glasssmith LL, Wong BS, Jones IM, Clive C, Haswell SJ. Consequences of manganese replacement of copper for prion protein function and proteinase resistance. EMBO J 2000; 19:1180-6; PMID:10716918; http://dx.doi.org/10.1093/emboj/19.6.1180
- Brown DR. Brain proteins that mind metals: a neurodegenerative perspective. Dalton Trans 2009:4069-76; PMID:19452053; http://dx.doi.org/10.1039/b822135a