Abstract
The still elusive structural difference of non-infectious and infectious amyloid of the mammalian prion protein (PrP) is a major pending milestone in understanding protein-mediated infectivity in neurodegenerative diseases. Preparations of PrP-amyloid proven to be infectious have never been investigated with a high-resolution technique. All available models to date have been based on low-resolution data. Here, we establish protocols for the preparation of infectious samples of full-length recombinant (rec) PrP-amyloid in NMR-sufficient amounts by spontaneous fibrillation and seeded fibril growth from brain extract. We link biological and structural data of infectious recPrP-amyloid, derived from bioassays, atomic force microscopy, and solid-state NMR spectroscopy. Our data indicate a semi-mobile N‑terminus, some residues with secondary chemical shifts typical of α‑helical secondary structure in the middle part between ∼115 to ∼155, and a distinct β‑sheet core C‑terminal of residue ∼155. These findings are not in agreement with all current models for PrP-amyloid. We also provide evidence that samples seeded from brain extract may not differ in the overall arrangement of secondary structure elements, but rather in the flexibility of protein segments outside the β-core region. Taken together, our protocols provide an essential basis for the high-resolution characterization of non-infectious and infectious PrP-amyloid in the near future.
Introduction
Prion diseases are fatal neurodegenerative disorders including Creutzfeldt-Jakob disease (CJD) in humans, bovine spongiform encephalopathy (BSE) in cattle, and scrapie in sheep. These disorders are associated with the conformational conversion of the cellular prion protein (PrPC) into a misfolded isoform (PrPSc), the pathological and infectious prion agent.Citation1 Prions are formed via nucleation-dependent polymerization, in which pre-formed amyloid fibrils of PrPSc act as a template for PrPC-conversion. Infectious PrP-amyloid has also been formed spontaneously solely from recombinant (rec) PrP.Citation2 Differences in infectivity titers of PrPSc and recPrP-amyloid are explained by structural differences. X-ray diffraction spectra of recPrP-amyloids correspond to a stack of β‑sandwiches, while X-ray diffraction spectra of PrPSc lack a 10 nm equatorial reflection characteristic of β‑sandwiches.Citation3 Whereas solution-state NMR spectroscopy was used to determine the structure of soluble PrPC Citation4 and to identify segments involved in oligomerization,Citation5 the structure of PrP-amyloid is still elusive. Studies have so far been limited to relatively low-resolution techniques.Citation6-10 These studies have suggested a superpleated β‑sandwich model for recPrP-fibrils characterized by a parallel, in-register alignment of β‑strands within a core domain comprising residues 160‑220.Citation6 Alternatively, a left-handed parallel β‑helix with the β‑sheet core between residues 89‑175 has been suggested based on EM studies on 2D-crystals of PrP27‑30, a fragment of PrPSc resulting from proteinase K-digestion of the ∼90 N‑terminal residues.Citation11 Recent high-resolution solid-state NMR-studies of recPrP(23‑144)-fibrils suggested a β‑strand‑turn‑β‑strand motif for the ∼30 C‑terminal amino acid residues, whereas no distinct NMR resonances were observed for the highly dynamic and disordered N‑terminus.Citation12 A lack of β‑strands N-terminal of position 145 in full-length recPrP-fibrils as demonstrated by H/D exchange,Citation13 however, indicates that data on truncated PrP-constructs or PrP-fragments are difficult to be extrapolated to infer the full-length PrP-structure. For full-length recPrP(23‑231)-fibrils, solid-state NMR studies on selectively 13CO-labeled samples confirmed an in-register parallel arrangement of β‑strands.Citation14 In light of severely broadened lines in spectra of uniformly 13C,15N-labeled fibrils, however, only residue types could be partially assigned to cross peak clusters. Assignment probabilities predicted by a Monte Carlo/simulated annealing algorithm suggested a fibril core comprising the 173‑224 segment but could not rule out an involvement of the 95‑161 segment.Citation14 Scrapie prion-seeded Syrian hamster recPrP(90-231)-fibrils were confirmed by 1D-solid-state NMR studies on selectively 13CO-labeled samples to have a parallel in-register β‑sheet architecture in the C-terminal segment as well.Citation15
Taken together, there is a need for improved protocols and additional experimental data to discriminate among different models for amyloid fibrils of PrP and to understand which structural features can render proteins infectious. The lack of high-resolution data is not only due to the general challenge posed to structure analysis by insoluble, non-crystalline, and heterogeneous samples, but also to the impossibility to prepare NMR-sufficient amounts of homogeneous brain-purified samples. However, it is known that fibrils grown from seeds, i.e. short fibril fragments produced by sonication of brain-purified fibrils, retain the molecular structures of the seeds.Citation16-18 Consequently, seeded fibril growth can be exploited to amplify and label structures present in brain tissue.Citation15,19,20 Likewise, different strains of the yeast prion Sup35p were faithfully passed on to recombinantly expressed Sup35p monomers by seeding with yeast cell lysates.Citation21
In contrast to all other amyloid fibrils, in vitro-generated PrP-amyloid offers the crucial advantage that its physiological relevance to neurodegenerative diseases in mammals can be assessed directly. Only when in vitro-generated PrP-amyloid proves to be infectious in animals, it is relevant in vivo. Up to now, PrP-amyloid, proven to be infectious, has never been investigated with a high-resolution technique. Moreover, data of infectious samples are restricted to laboratory animals such as mouse and hamster. Species that are important from a veterinary point of view, such as cattle or sheep, have never been investigated.
Here, we demonstrate methodologies for reproducibly preparing infectious samples containing sheep full-length recPrP(25-233)-amyloid. Ovine recPrP(25‑233)-amyloid was formed by spontaneous conversion and by nucleation with scrapie brain seeds in high yield and purity. For the first time, we have investigated the biologically authentic system of amyloid fibrils of full-length recPrP (recPrP-amyloid) by a multidisciplinary combination of bioassays and biophysical characterization with solid-state NMR spectroscopy. Our present data are consistent with a β‑sheet core C‑terminal of residues ∼155 and a partially α‑helical middle segment between residues ∼115–155. Since NMR spectroscopy of our recPrP-amyloid samples investigates not only single molecules within the sample but the complete ensemble of all conformations present in oligomeric and fibrillar states, our data allow us to verify structural models for the dominating conformation. In addition, our data indicate that recPrP-amyloid after seeding with brain extracts may differ in the flexibility of the partially α‑helical middle segment. Based on our established protocols, a high-resolution characterization of infectious PrP-amyloid appears to be tangible in the near future.
Results
Concept of the study
We investigated infectious samples of ovine (ov) ARQ full-length recombinant (rec) PrP(25‑233)-amyloid for several reasons: (i) Only full-length recPrP (see Fig. S1) encompasses the entire brain PrPSc sequence containing all prion disease-related polymorphism sites. Only fibrils from full-length PrP closely resemble structures formed in vivo.Citation13 (ii) Scrapie is the biochemically most investigated as well as the most widespread prion disease. (iii) The well-defined set of sheep PrP-polymorphisms at positions 136 (A/V), 154 (R/H), and 171 (Q/R/H) governs scrapie susceptibility and allows further study of structure-infectivity relationships. The homozygous combination A136R154Q171 is associated with highest scrapie susceptibility and frequency.Citation22 (iv) RecPrP-fibrils can be generated either under partially denaturing conditions Citation2 or by protein misfolding cyclic amplification (PMCA).Citation23,24 Although PMCA generates highly infectious recPrP-samples, if only in some batches, its usefulness for NMR studies is limited because PMCA sonication introduces a high well-to-well variability in recPrPSc-yield. A gravely polymorphic mixture is the undesirable PMCA outcome.Citation25-27 This lack of homogeneity poses an insurmountable obstacle for latest state of the art solid-state NMR-spectroscopy of a 209-residue protein such as PrP. In contrast, uniform protein denaturation by guanidinhydrochloride (GdnHCl) and urea results in less infectious but homogeneous recPrP-samples (recPrP-amyloid). The less infectious character of recPrP-amyloid is explained by structural differences to brain-derived PrPSc 3.
Generation of NMR-sufficient amounts of ovrecPrP-amyloid
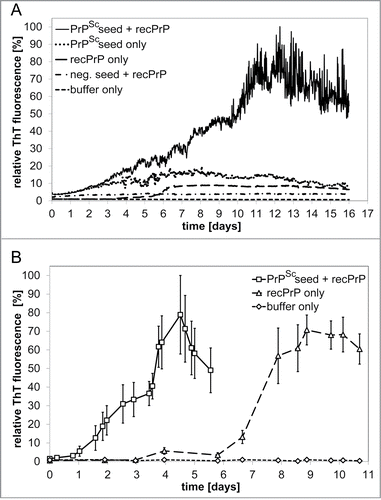
Reproducible fibrillation conditions were established in 96‑well plates for spontaneous conversion of ovrecPrP(25‑233). In a second approach, ovrecPrP(25‑233) was seeded with sequence-identical full-length PrPSc purified from scrapie sheep brain by PTA-precipitation (see Materials and Methods). PrPSc-seeded growth took place days before spontaneous conversion (). In light of this considerable difference in lag phases, PrPSc-seeded and spontaneous samples could be produced and analyzed independently of each other. Furthermore, this confirms that the fibrillation kinetics is dominated by a seeding reaction and not by spontaneous conversion. PTA-precipitation is not specific for PrPSc though. Therefore, normal sheep brain was subjected to the identical purification protocol and PTA-precipitated material (lacking PrPSc) was used as seed. A lack of any ThT-fluorescence increase, even after prolonged incubation times, confirmed that recPrP-fibrillation kinetics is dominated by propagation of specific seeds, namely PrPSc (). To further reduce the risk of generating polymorphic amyloid mixtures by pooling 96‑well plates, optimal conditions were scaled up to volumes of up to 10 ml (). Differential ultracentrifugation of samples from the fibrillation end point followed by densitometric quantification of Western blots indicated that about 95 % of ovrecPrP was always converted to a fibrillar form. In summary, NMR-sufficient fibrillar yields of 10 mg of both spontaneously generated and PrPSc-seeded ovrecPrP-amyloid could be prepared in 10 ml volumes each.
Figure 1. ThT fluorescence detection of amyloid formation during spontaneous and PrPSc-seeded fibrillation. (A) Amyloid formation of 100 ng μl‑1 ovrecPrP(25‑233) in volumes of 150 μl was analyzed in 96-well plates in absence or presence of PrPSc-seeds obtained by PTA-precipitation of scrapie sheep brain homogenate. As controls for PrPSc-seeded fibrillation, PTA-precipitated scrapie sheep brain homogenate in absence of ovrecPrP(25-233) (PrPSc seed only) and PTA-precipitated normal sheep brain homogenate in presence of ovrecPrP(25-233) (neg. seed + recPrP) demonstrate the specificity of the fibrillation assay. (B) Amyloid formation of 1000 ng μl−1 ovrecPrP(25‑233) in NMR-sufficient volumes of 10 ml was analyzed. Note that seeded fibril growth (PrPSc seed + recPrP) is completed before spontaneous conversion (recPrP only) starts. The decrease in ThT-fluorescence intensity after long times is attributed to lateral fibril association and blockage of ThT-binding sites. Values shown are means ± standard errors for up to 10 distinct experiments. The gradually increasing error bars reflect the fibril size distribution in a heterogeneous system.
Infectivity
Aliquots of 13C,15N-ARQ-ovrecPrP(25‑233)-samples were inoculated in transgenic mice, homozygously overexpressing ovine VRQ- or ARQ PrP(25‑233), respectively. Non-fibrillated ovrecPrP(25‑233) failed to induce noticeable prion disease symptoms in any mouse even 630 days after inoculation (), disproving a prion disease-inducing effect of the stress during sample injection. In contrast, 3 out of 12 mice inoculated with spontaneously fibrillated ovrecPrP (in the absence of seeding with PrPSc) succumbed to a prion disease, confirming the infectious character of our non-seeded samples. Because less than 100 % of mice developed a prion disease, the infectious titer can be considered to be lower than 102 ID50/ml.28 According to literature, the onset of neurological dysfunction in Syrian Gold hamsters or transgenic mice takes place between 380 and 750 days after inoculation of spontaneously generated recPrP-amyloid.Citation29-31 Although our incubation periods are at the shorter end of this range, their high standard deviation also indicates the presence of low infectivity titers. Whether our relatively short incubation periods are a feature of the details of our preparation conditions, the ovine recPrP-sequence, or the associated conformational stability remains uncertain.Citation29-31 After inoculation of the brain homogenates of these diseased mice into the same mouse line, 100 % of mice developed a prion disease with incubation periods characteristic for high prion infectivity. The incubation periods in transgenic ARQ mice even indicate prion infectivity as high as in brain-derived PrPSc (see ). Small standard deviations in incubation periods also indicate a highly efficient transmission. Taken together, these findings confirm the infectious character of our non-seeded samples. After one passage our low infectivity recPrP-amyloid was transformed into highly infectious PrPSc.Citation32 It is also noteworthy that the incubation periods observed in these second passages differ from incubation periods of PrPSc-seeds and PrPSc-seeded ovrecPrP-amyloid (see below). This makes a contamination of spontaneously generated ovrecPrP-amyloid with PrPSc-seeds highly unlikely.
Table 1. Incubation periods of in vitro-fibrillated ovrecPrP(25‑233) and PrPSc purified from scrapie-infected sheep brains
In order to confirm the presence of PrPSc in mice, brain extracts were analyzed for partial proteinase K (PK)-resistance which is the biochemical marker in routine prion tests.Citation33 A partial PK-resistance is evident from a molecular weight shift of the di-, mono-, and unglycosylated PrP‑forms caused by the digestion of about 70 N‑terminal residues. All mice inoculated with non-fibrillated ovrecPrP(25‑233) failed to show any PK-resistant PrPSc (). Although samples of spontaneously fibrillated ovrecPrP(25‑233) used for inoculation did not show any PK-resistance either (), the 3 partially PK-resistant PrPSc‑forms were present in brain extracts of all symptomatical mice (), confirming prion diseases.
Figure 2. Immunoblots of samples inoculated into transgenic mice and of brain material from transgenic mice. (A) PrPSc in brains of tgshpXI (ARQ) and tg338 (VRQ) mice, inoculated with scrapie PrPSc, after spontaneous fibrillation of ovrecPrP(25‑233) (SP), or with non-fibrillated ovrecPrP(25‑233) (NON), respectively, with (+) or without (−) digestion with 50 μg/ml PK for 2 h at 37°C. Only mice inoculated with fibrillated ovrecPrP(25‑233) show PK-resistant PrP27‑30 identical to scrapie PrP27‑30. (B) Spontaneously fibrillated ovrecPrP(25‑233) after digestion with increasing concentrations of PK for 1 h at 37°C does not show any PK-resistance. (C) For comparison, PrPSc-seeds are depicted as characterized by a partial PK-resistance even at high PK-concentrations. (D) PrPSc-seeded ovrecPrP(25‑233) after digestion with increasing concentrations of PK for 1 h at 37°C does not show any PK-resistance. In light of the 300-fold excess of ovrecPrP(25‑233) during seeded fibrillation (see Materials and Methods for details), the undigested PrPSc-specific bands are too weak to show any PK-resistance after digestion. Any atypical PrP-bands were not observed. Apparent molecular masses based on migration of protein standards are indicated in kDa.
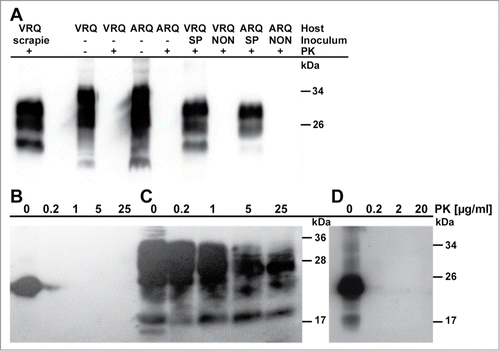
In contrast to spontaneously generated recPrP-amyloid, PrPSc-seeded samples induced prion diseases in 100 % of the inoculated mice at the first passage (). It is well known that animals inoculated with in vitro-propagated PrPSc exhibit an altered relationship between incubation period and infectivity titer Citation25. A comparison with the incubation periods induced by brain-derived PrPSc () is thus not valid. Samples of PrPSc-seeded ovrecPrP(25‑233) before inoculation also did not show any PK-resistance ().
It was beyond the scope of this manuscript to determine whether infectivity is associated with ovrecPrP-fibrils or pre-fibrillar aggregates. Solid-state NMR spectroscopy, however, allowed us to investigate the complete ensemble of conformations present after ovrecPrP-conversion. Measured line widths indicate structurally very similar conformations (see below).
Secondary and ultrastructural properties
A circular dichroism (CD) spectrum of the fibrillation starting point, i.e., monomeric ovrecPrP(25‑233), is indicative of low β‑sheet content as known for both PrPC and natively refolded recPrP.Citation33 Satisfactory CD spectra of recPrP-amyloid were only obtained after gentle sonication in presence of small concentrations of SDS. This mild treatment does not influence the secondary structure of recPrP-amyloid (see Materials and Methods for details) but allows for CD-analysis. OvrecPrP-conformations after spontaneous conversion or PrPSc-seeding combine a β-sheet structure with additional α‑helical/random coil segments and are thus completely different from natively folded monomeric ovrecPrP(25‑233) (). Although CD spectra after spontaneous conversion or PrPSc-seeding are characterized by identical minima, zero crossings, and maxima, the measured mean residue molar ellipticities of both sample types differ considerably. The higher mean residue molar ellipticity intensities (which are values normalized by the respective protein concentration) after PrPSc-seeding indicate an increased proportion of ovrecPrP-amyloid amenable to CD analysis.
Figure 3. Secondary structural properties. As revealed by CD-spectroscopy, the secondary structural transition of monomeric ovrecPrP(25 233) due to spontaneous and PrPSc-seeded fibrillation results in spectral minima at 218 nm typical for β-sheets. Zero crossings at 204 nm and spectral maxima at 193 nm are probable to indicate additional α-helical/random coil elements. For comparison, a spectrum of α‑helical monomeric ovrecPrP(25-233) is also presented. All given values are mean residue molar ellipticities ([θ]MRW).
![Figure 3. Secondary structural properties. As revealed by CD-spectroscopy, the secondary structural transition of monomeric ovrecPrP(25 233) due to spontaneous and PrPSc-seeded fibrillation results in spectral minima at 218 nm typical for β-sheets. Zero crossings at 204 nm and spectral maxima at 193 nm are probable to indicate additional α-helical/random coil elements. For comparison, a spectrum of α‑helical monomeric ovrecPrP(25-233) is also presented. All given values are mean residue molar ellipticities ([θ]MRW).](/cms/asset/bc2adc88-d230-41b7-ba35-f367b5811833/kprn_a_983754_f0003_b.gif)
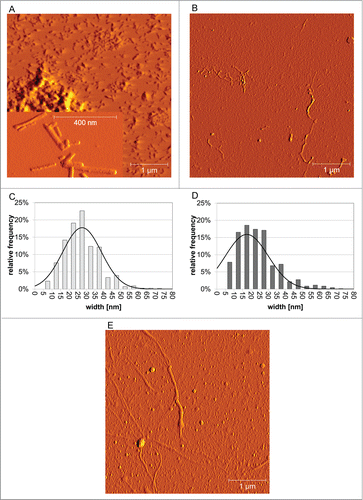
AFM analysis demonstrated that spontaneously generated and PrPSc-seeded ovrecPrP-fibrils substantially differ in morphology, but each fibril type is morphologically homogeneous. After spontaneous fibrillation (), we observed short amyloid fibrils similar to so-called prion rods. Prion rods are purified from infectious brain material using detergents and limited proteolytic digestion.Citation34 Our short fibrils are in agreement with the literatureCitation9,35 as they appear straight, up to 400 nm long, between 5 and 20 nm high, and exhibit a strong tendency to associate into higher order aggregates. OvrecPrP-fibrils longer than 400 nm or twisted fibrils were never observed, even when pre-formed fibrils were used for seeded fibrillation. Pre-fibrillar aggregates were never observed either. The histogram in showing the fibril widths distribution illustrates that the fibril species have an average width of 30 ± 11 nm, which is identical to literature values.Citation9 In contrast, ovine PrPSc-fibrils () are characterized by lengths of several μm and a far wider width distribution resulting in an average width of 56±31 nm. The presence of such long PrPSc-fibrils is reminiscent of full-length PrPSc in living cells which was observed by light, atomic force, and scanning electron microscopy to form thin, ≤5 μm-long structures on the cell surface.Citation36,37 Regarding the average width of PrPSc-fibrils, their appearance in our AFM images was obscured by co-purified brain matter in the form of extraneous non-fibrillar material. Also the natural presence of GPI-anchor and un-, mono, and diglycosylated PrPSc forms obscured the underlying proteinaceous parts of PrPSc-fibrils causing high width standard deviations. When ovrecPrP-amyloid was grown in presence of PrPSc-seeds [<0.5 % (vol/vol)], the resulting amyloid fibrils combined the PrPSc-like curvature and the length of several μm with an average fibrillar width of 25±13 nm (). In contrast to spontaneous preparations, short fibrils were never observed. Spontaneously generated and PrPSc-seeded ovrecPrP-fibrils have different lengths but indiscernible average fibrillar widths. The narrowest fibrillar structures were observed to be 6 to 8 nm in width, which is in agreement with diameters for single PrP-filaments as determined by negative stain EM (4.8 to 5.7 nm for PrP27‑30, 7.6 to 7.9 nm for recPrP(90-231)-fibrils).Citation3 Taken together, spontaneously generated ovrecPrP-fibrils differ morphologically from brain-purified PrPSc and PrPSc-seeded ovrecPrP-fibrils.
Figure 4. Ultrastructural properties. Low magnification amplitude AFM-images (A and B) and corresponding fibril width distributions (C and D) obtained after (A and C) spontaneous or (B and D) PrPSc-seeded fibrillation of ovrecPrP(25-233) display variability in fibrillar topology. The black curves in (C and D) are fits with a Gaussian function illustrating that spontaneous and PrPSc-seeded ovrecPrP-fibrils have identical average widths within the margin of error. However, spontaneously generated ovrecPrP-fibrils have only a length of up 400 nm (A), whereas PrPSc-fibrils in scrapie-infected sheep brain homogenate (E) and PrPSc-seeded fibrils (B) are several μm long. In (A), an inset of higher magnification shows the morphology of spontaneously generated ovrecPrP-fibrils to higher details. Immediately before AFM-measurement, all samples were subjected to mild sonication for 10 sec. Thus, (E) depicts PrPSc-fibrils as they were used for seeding of ovrecPrP(25‑233).
Absence of strongly mobile segments
NMR spectroscopy is sensitive to molecular motions on the μs to ms timescale. It is well-known that solid-state NMR spectra, which employ dipolar couplings for CP-excitation and magnetisation transfer, often display only peaks from a minor fraction of all residues. Only residues in rigid segments, i.e. in the rigid amyloid core, are characterized by strong dipolar couplings resulting in observable solid-state NMR signals. On the other hand, highly flexible protein segments, undergoing motions in the sub-microsecond range, may be discriminated easily from the rigid core by INEPT-based excitation.Citation21,38-41 In contrast to rigid and highly flexible residues though, signals from semi-flexible regions, such as the fuzzy coat of amyloid fibrils, may be completely missing due to intermediate range molecular motion resulting in severe line broadening and/or disappearance of signals.Citation12,21,38,42 In order to compare the amounts of rigid and mobile protein segments, we recorded 1D-13C-CP- and 1D-13C-INEPT-NMR spectra. CP-spectra, which selectively detect rigid protein segments, revealed identically strong and appropriately resolved resonances in samples after spontaneous conversion or PrPSc-seeding of ovrecPrP(25‑233). The absence of any protein signal in all INEPT-spectra, even after rehydration with 10 μl of H2O and measurement at 20°C, indicated a lack of highly flexible residues in any protein segment. When CP-excitation was combined with a transverse spin echo delay, leading to dipolar dephasing of magnetisation in rigid protein segments, all signals disappeared already after delays as short as 300 μs. In 2D-spectra, obtained by direct 13C-excitation followed by 100 ms PDSD-mixing, no additional cross peaks were visible, confirming that all NMR-visible residues had been observed in the spectra obtained with CP excitation. Taken together, all segments in ovrecPrP-conformers after spontaneous conversion or PrPSc-seeding are rigid on a sub-microsecond timescale.
Homogeneity of samples
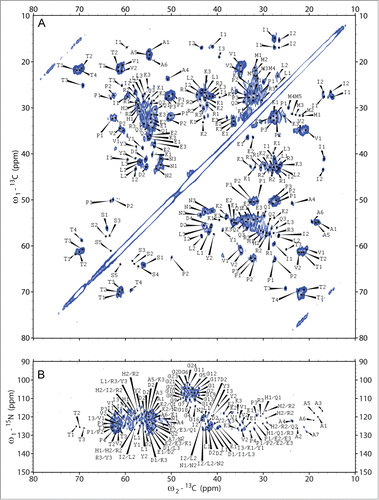
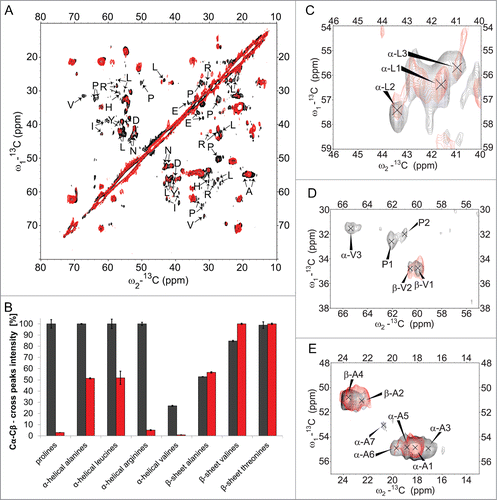
In order to identify spin systems visible in solid-state NMR spectra, we recorded sets of 2D-(13C‑13C)- and (15N‑13C)-correlation experiments (see Materials and Methods) at 0°C. Essentially identical spectra were obtained for 2 independently prepared samples, thus indicating the reproducibility of the experiments. In , 2D-(13C-13C)- and (15N-13C)-spectra are depicted exemplarily. Solid-state NMR spectroscopy investigates the complete ensemble of conformations present in pre-fibrillar and fibrillar states. 13C-line widths of isolated cross peaks of about 120 Hz thus indicate that all conformations present in our samples, be they infectious or non-infectious, need to be structurally very similar. A conformational ensemble comprising a continuous distribution of slightly varying conformations cannot be completely excluded though. We did neither observe any peak doubling nor find any other indication for the presence of completely different conformations.
Figure 5. 2D-(13C‑13C)- and (15N‑13C)-solid-state NMR correlation spectra. For identification of spin systems, homo- and heteronuclear correlation spectra were analyzed jointly. The aliphatic regions of (A) (13C‑13C)- and (B) (15N‑13C)-correlation spectra after spontaneous fibrillation of ovrecPrP(25‑233) are depicted, acquired with (A) PDSD-mixing for 20 ms at 11 kHz MAS and (B) DREAM-mixing at 23 kHz MAS, to obtain intraresidue correlations. All cross peaks are labeled according to Tables S1, S2, and S3. Note that in (B) several tentative assignments are present more than one time, reflecting the ambiguity as summarized in Table S3.
Despite careful control of sophisticated fibrillation conditions in one tube per sample and 13C and 15N line widths for resolved peaks of about 120 Hz and 450 Hz, respectively, average line widths could not be reduced further. Neither did hydration by addition of 10 μl H2O nor measurements at higher temperatures up to 20°C increase the spectral resolution. Also, 2 times seeding with seeds prepared from spontaneously generated ovrecPrP-amyloid did not render the conformational ensemble more homogeneous. Seeding with PrPSc, however, did result in samples whose 2D-correlation spectra exhibit both reduced intensities and absence of some cross peaks (see below).
Assignment process
In light of the spectral overlap, particularly in highly populated chemical shift regions (e.g. at 55 ppm × 35 ppm or 30 ppm × 25 ppm in ), the challenge of obtaining residue type-specific assignments could only be met by comparing peak maxima in a given spectrum with the complete set of all other homonuclear and heteronuclear 2D-spectra. A cross peak was only regarded to be genuine and residue type-specific, when (i) it was visible using different apodisation functions, (ii) a clear peak was visible in the corresponding 1D-traces, and most importantly (iii) cross peaks at corresponding chemical shifts were observed in all other correlation spectra. Using these stringent criteria, we could differentiate among distinct spin systems of nearly every amino acid type. For most of the amino acid types, however, we do not observe separated cross peaks indicative of different residues but clusters of merging cross peaks with a width of several hundred Hz. This is depicted exemplarily in Figure S2 for the threonine and asparagine regions of 2D-(13C-13C)-spectra.
Only by comparing peak maxima in a given spectrum with the complete set of all other homonuclear and heteronuclear spectra, peaks within the cross peak clusters could be assigned to different residues. This is demonstrated exemplary in Figures S3–S5 for the identification of alanine, leucine, and glycine spin systems. Alanine cross peaks, for example, could only be distinguished using the N‑Cβ-correlations resolved nicely in NCACB-type spectra (Fig. S3F). However, NMR assignments are only considered to be trustworthy, when there is just one assignment per separated cross peak. As more residues do contribute to the observed signals, Tables S1 and S2 summarize only a reduced set of average chemical shifts. These correspond to sub-clusters of cross peaks and stand up to close scrutiny. E.g. for threonine and asparagine residues, only 4 and 3 assignments are given, respectively, although the combination of information of all homonuclear and heteronuclear spectra indicates the presence of more residues (see ). Consequently, the numbers of given assignments (59 after PrPSc-seeded conversion and 73 after spontaneous conversion, respectively) are a lower estimate of visible residues. Standard deviations and numbers of assignments are provided in Tables S1 and S2 as a measure for the accuracy of our chemical shifts.
Sequence specific assignments can in principle be obtained by linking NCACX-cross peaks of individual residues via backbone 15N chemical shifts with NCOCX-cross peaks of the preceding residue in the amino acid sequence. Our heteronuclear spectra (see e.g., ), however, suffer from severe spectral overlap in the 15N-dimension. Since several residue-type assignments are conceivable for nearly every cross peak in (15N-13C)-correlation spectra (except alanine and threonine N‑Cβ- and glycine N‑Cα-cross peaks), no unambiguous sequential assignments could be obtained yet. Table S3 summarizes all possible residue type–specific 15N-assignments. To improve resolution and thereby allow for sequential assignments, heteronuclear 3D-spectra of selectively labeled samples are necessary. This analysis is currently underway in our laboratory but is beyond the scope of this manuscript.
Secondary structure distribution, comparison of amyloid types, and flexibility
In contrast to earlier studies of recPrP-samples, our solid-state NMR-data of infectious recPrP-amyloid allow us to draw the following conclusions:
The secondary chemical shifts of ovrecPrP(25‑233) after spontaneous conversion or PrPSc-seeding are similar though not identical (), indicating similar underlying conformational motifs. An overlay of solid-state NMR spectra of both preparations, however, visualises slightly different cross peak patterns and intensities suggesting the presence of deviating structural features. Since site-specific information is missing, however, it cannot be decided whether slightly different cross peak positions indicate local structural differences only.
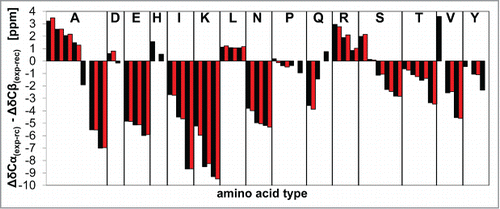
Figure 6. Secondary chemical shifts. A comparison of secondary chemical shifts of ovrecPrP(25‑233) after spontaneous (black) or PrPSc-seeded (red) conversion suggests that both amyloid types share a similar overall arrangement of secondary structure elements. All amino acid residues are listed in alphabetic order.
The number of identified residues in solid-state NMR spectra is apparently smaller than the number of amino acid residues in the ovrecPrP(25‑233)-sequence. This is mainly due to a severe spectral overlap, particularly in highly populated chemical shift regions. The numbers given in Tables S1 and S2 are a lower limit of visible residues because it is highly probable that more residues contribute to overlapping cross peaks. In addition, we cannot exclude that line broadening effects further reduce the number of identifiable residues. The complete lack of INEPT-signals (see above) confirms that all segments in ovrecPrP-conformers after spontaneous conversion or PrPSc-seeding are rigid on a sub-microsecond time scale. Apart from spectral overlap, a smaller than expected number of rigid residues may additionally be due to exchange broadening by molecular motions on the NMR-time scale or due to extensive inhomogeneous line broadening beyond detectability as a result of a high degree of disorder in completely rigid segments.
The segment C‑terminal of sequence position 100 contains only 10 out of 43 glycine residues. We observed, however, the up to twofold number of glycine cross peaks (see Fig. S5). A number of 10 glycine residues represent only 7.5 % of all 133 residues C‑terminal of sequence position 100. The integrals of the glycine-specific N‑Cα-region in 2D-(15N‑13Cα)-spectra (without any following mixing) and the glycine-specific CO‑Cα-region in 2D-(13C‑13C)-correlation spectra, however, constitute on average 14.6 ± 1.5 % of the entire Cα-region in the respective spectrum. This indicates that either a small fraction of the N-terminus does contribute to solid-state NMR-spectra or that glycine-containing regions C‑terminal of sequence position 100 are disordered to some extent.
The secondary chemical shifts () are in agreement with our CD measurements confirming a dominance of residues in a β‑sheet conformation. According to FTIR- and CD-data, PrPSc comprises 34–43 % β‑sheets, whereas PrP27‑30, obtained by proteolytic removal of the flexible N‑terminal PrPSc-segment, is characterized by 43–54 % β‑sheet contents.Citation43-45 When we differentiate our secondary chemical shifts into the categories >1 ppm α‑helical, <1 ppm/ >‑1 ppm random coil, and <‑1 ppm β‑sheet content, then β‑sheet contents of 60 % (PrPSc-seeded conversion) and 53 % (spontaneous conversion) match best with the values for PrP27‑30. This supports the view that it is mainly signals from a rigid C‑terminal fibril segment that contribute to our solid-state NMR spectra. Values for α‑helical contributions are not compared because FTIR- and CD-based data have been criticized recently not to support the presence of residual α‑helices in PrPSc and PrP27‑30.Citation46
A sequentially assigned minimum of 3 consecutive β‑sheet residues or 4 consecutive α‑helical residues are required to define a β‑strand or an α‑helix, respectively, at a distinct sequence position.Citation47 Although we lack site-specific assignments, we found some hints for the position of secondary structure elements by combining the secondary structure information for individual amino acid types which information about the primary sequence. The 3 ovrecPrP(25‑233)-leucine residues are at positions 128, 133, and 141, i.e., clustered in the middle region. All of them are characterized by α‑helical chemical shift signatures. Seven out of 9 alanine residues in ovrecPrP(25‑233) are clustered at positions 116, 118, 119, 120, 121, 123, and 136. At least 4 alanine residues identified in our spectra have chemical shifts indicative of α‑helical secondary structure. Eight out of 11 arginine residues in ovrecPrP(25‑233) are N‑terminal of position 168. All signals assigned to arginine residues have chemical shifts indicative of α‑helical secondary structure. These observations suggest that this middle region of fibrillated ovrecPrP(25‑233) contains either several closely adjacent β‑turns and/or a residual α‑helical secondary structure. All resonances with an α‑helical chemical shift signature can be assigned to this region, either unambiguously (Leu128, Leu133, Leu141) or tentatively (Ala116, Ala118, Ala119, Ala120, Ala121, Ala123, Ala136, Asp147, Asp150, Arg139, Arg151, Arg154, His114, His143, Gln101, Ser135, Ser138, Val124, and Val125).
All other secondary chemical shifts are indicative of amino acid residues in a β-sheet conformation. A semi-mobile N‑terminus and residual α‑helical segments between residues ∼115 and ∼155 imply that the majority of these β‑strand residues are confined to the segment C-terminal of residue ∼155.
Higher mean residue molar ellipticity intensities after PrPSc-seeding indicated an increased proportion of ovrecPrP-conformers amenable to CD analysis which implies higher flexibility of at least some segments of PrPSc-seeded conformations (). In 2D-(13C‑13C)-solid-state NMR spectra using homonuclear transfer based on dipolar recoupling, cross peak intensities of specific amino acid residues, e.g. of proline and leucine residues, are weakened when samples are generated by PrPSc-seeding. To account for possibly deviating hydration states, samples were rehydrated by adding 10 μl of H2O each. No change in the respective intensities or line widths of cross peaks was observed. A comparison of identically recorded and processed 2D-(13C‑13C)-DREAM spectra of identically prepared (except for the presence or absence of PrPSc-seeds during fibrillation) 10‑mg samples of both amyloid types confirmed an increased flexibility after PrPSc-seeding. An overlay of the aliphatic regions of these spectra is depicted in , whereas display some regions of interest in higher magnification. DREAM spectra display cross peaks only for correlations between directly bound 13C atoms and thus allowed for quantitative intensity analysis. Since only some cross peaks of lysine, glutamine, glutamate, aspartate, histidine, isoleucine, asparagine, serine, and tyrosine residues differed in intensity, a quantitative conclusion could hardly be drawn for them without heteronuclear 3D-spectra reducing the number of overlapping cross peaks. However, we determined the volumes of all Cα‑Cβ‑/Cβ‑Cα‑cross peaks with α‑helical or β‑sheet chemical shift signatures, respectively, for the spectrally separated proline, valine, arginine, leucine, alanine, and threonine residues.(). For threonine residues (not present between positions 115–155; 9 of 11 threonine residues are C‑terminal of position 185), no difference in the normalized cross peak intensities was observed comparing spontaneously generated and PrPSc-seeded samples. Cross peak intensities of β‑sheet alanine and β‑sheet valine residues also did not differ within an error margin of 10%. In contrast, all proline cross peaks almost completely vanished in the spectra of PrPSc-seeded samples. The most C-terminal of 15 ovrecPrP(25‑233) proline residues is located at sequence position 168. The substantially reduced proline intensities therefore suggest that PrPSc-seeding results in enhanced flexibility of the N‑terminal fibril segment. The cross peaks of α‑helical alanine, α‑helical leucine, α‑helical arginine, and α‑helical valine residues are also considerably weakened after PrPSc-seeding (). Seven of 9 alanine residues are clustered between positions 116 and 136. All 3 leucine residues are at positions 128, 133, and 141. Eight of 11 arginine residues are located N‑terminal of position 168. Valine residues with α‑helical chemical shift signatures are believed to be located between residues 115 to 155 (see above). Taken together, only cross peak intensities of residues N‑terminal of Pro168 seem to be affected indicating again that PrPSc-seeding induces a more flexible fibril region N-terminal of Pro168.
Figure 7. Structure and dynamics details. Solid-state NMR comparison of ovrecPrP(25‑233) after spontaneous (black) or PrPSc-seeded (red) conversion reveals an increased flexibility after PrPSc-seeding. (A) Overlay of 2D-(13C‑13C)-correlation spectra acquired with the identical number of transients and increments and processed identically. DREAM mixing at 23 kHz was performed to obtain only direct correlations. Spin systems after PrPSc-seeded conversion for which cross peaks are weakened up to almost completely vanished on both sides of the diagonal are indicated by arrows. (B) For intensity analysis, Cα‑Cβ/Cβ‑Cα-peak volumes in α‑helical and β‑sheet chemical shift ranges of 2D-(13C-13C)-DREAM spectra were calculated. Peak volumes were normalized 2-fold: (1) To account for deviating protein amounts in the samples, all values were standardized to the peak volume of the corresponding spectrum diagonal. (2) The highest peak volume of each amino acid type (α‑helical black or β‑sheet black or α‑helical red or β‑sheet red) was set to 100 %. Note that α‑helical and β‑sheet spin systems are not present for all amino acid types. Whereas all threonines are located in β‑strands, all arginines and leucines are characterized by α‑helical secondary structure signatures. Prolines are not known to be present in any secondary structure. Valines are located in α‑helical as well as β‑strand conformations. (C–E) High magnification overlays of some exemplary regions of (C) leucine, (D) proline and valine, and (E) alanine residues show that cross peak intensities for prolines (all N‑terminal of position 168), α‑helical valines, α‑helical leucines (at positions 128, 133, and 141), and α‑helical alanines (clustered between positions 116 and 136) are reduced or even completely vanished, whereas β‑strand alanines and β‑strand valines do not display a decrease in cross peak intensities.
Discussion
Several structural models of amyloid fibrils have been published.Citation48,49 Recent data indicate that aberrantly folded proteins in other neurodegenerative diseases share self-propagating infectious properties similar to those in spongiform encephalopathies caused by misfolded PrP.Citation50,51 This emphasizes the importance of investigating physiologically relevant conformations. Proving that in vitro-fibrillated structures are physiologically relevant is still a challenge though. Since no functional test is available, only infectious samples or brain-seeded fibrils have so far been regarded to reflect in vivo-structures.Citation19 Earlier high-resolution studies have never investigated samples proven to be infectious. Here, for the first time we have linked structural and biological data by applying a high-resolution technique to infectious samples of recPrP-amyloid from sheep, an important species from the veterinary point of view. Our use of solid-state NMR spectroscopy allows for direct probing of subunit structure and dynamics of the complete ensemble of conformations present after conversion of ovrecPrP(25‑233). Although our data are not high-resolution, our protocols pave the way for obtaining atomic resolution data in the near future.
As both samples prepared either in absence or presence of brain-purified PrPSc-seeds display similar, although not identical, cross peak positions in solid-state NMR spectra, they are likely to have many structural features in common. However, they differ in crucial aspects. Only after seeding of ovrecPrP(25‑233) with brain-purified PrPSc did we observe a morphological appearance of ovrecPrP-fibrils similar to PrPSc (). Considerably shortened lag phases during seeded ThT-assays () are also in agreement with a specific propagation of PrPSc. A higher proportion of ovrecPrP-conformers amenable to CD analysis after PrPSc-seeding () agrees with an increased flexibility in PrPSc-seeded samples as suggested by solid-state NMR spectra (). It is tempting to speculate that these findings are in agreement with an increased degree of disorder as known for high prion infectivity.Citation3,52 However, as the infectivity of our PrPSc-seeded ovrecPrP-amyloid is unknown yet (see above), we cannot derive any conclusions for PrPSc but for infectious recPrP-amyloid.
Natively folded ovine PrPC comprises a flexible unstructured N‑terminus, 3 α‑helices Asn146‑Tyr158, Asn174‑Thr196, and Glu203‑Tyr228, and a small β‑sheet formed by Gly129‑Gly134 and Gln163‑Arg167.Citation53 According to our solid-state NMR-data, PrPSc-seeded conversion results in (i) a semi-rigid N‑terminus, (ii) a partially α‑helical/β‑turn middle segment approximately between residues ∼115 to ∼155, and (iii) a β‑sheet-core C‑terminal of residue ∼155. No hints were found for an α‑helical region in the C‑terminus.
An unstructured semi-rigid N‑terminus agrees with biochemical literatureCitation33 and earlier solid-state NMR studiesCitation12 of non-infectious recPrP-fibrilsCitation54 concluding that the N‑terminus up to position 131 is not as mobile as a fully disordered polypeptide in solution.
An α‑helical/β‑turn middle segment would be divided by the α‑helix-breaking Pro140. An α‑helical segment C-terminal of Pro140 may correspond to the remainder of the PrPC α‑helix Asn146‑Tyr158. An α‑helical segment N-terminal of Pro140, however, would imply a rearrangement of the β‑strand Gly129‑Gly134 to an α‑helix upon fibrillation. Since such a conversion has never been observed, it is doubtful whether all α‑helical chemical shift signatures N‑terminal of Pro140 do indicate a stable α‑helical segment. At least 3 other explanations are conceivable. Firstly, α‑helical chemical shift signatures can indicate β‑turn motifs Citation55 which might adjoin to a more or less preserved PrPC β‑strand Gly129‑Gly134. Additional β‑turn motifs within the C‑terminal β‑sheet core cannot be excluded. Secondly, we prepared our recPrP-amyloid under denaturing conditions. For natively unstructured proteins it is known that transient α‑helical states can be sampled prior to transition to amyloid fibrils.Citation56,57 Thirdly, secondary chemical shifts in unfolded proteins were found to almost be in the range expected for α‑helical residues.Citation58 Whatever the structure of this middle segment is, it is incompatible with the adoption of a uniform in-register β‑sheet structure as demonstrated by 1D-solid-state NMR studies on selectively 13CO-labeled scrapie prion-seeded Syrian hamster recPrP(90-231)-fibrils.Citation15
A β‑sheet-core C‑terminal of ∼155 is consistent with the finding that Tyr152‑Arg154 remains flexible during fibrillation, whereas Tyr165‑Arg167 is part of the rigid core.Citation59 Although we cannot determine the precise location of β‑strands, a β‑sheet-core C-terminal of residue ∼155 implies a refolding of the 2 C‑terminal PrPC α‑helices. This location of β-strands is in agreement with that proposed for the β‑sandwich modelCitation6,15 but not with that suggested for the spiral modelCitation60 or the published β‑helix model.Citation11 An alternative β‑helix model Citation61 comprising the C‑terminal segment would be consistent with our solid-state NMR-data though.
In addition to morphological influences, PrPSc-seeding increases the flexibility of some protein regions. Although we cannot completely exclude an increased flexibility of C‑terminal β‑strands, our data suggest that PrPSc-seeding induces an enhanced flexibility of the α‑helical/β‑turn-rich segments between residues ∼115 to ∼155 (). This is in agreement with the observation that PrPSc-seeded recPrP(90‑231)-fibrils consist of subpopulations with highly accessible disordered as well as protected ordered segments in the regions 117‑133 and 145‑168.Citation62 Furthermore, it was demonstrated that residues 117‑119 and 140‑143 are more exposed to PK-treatment and, in turn, more likely to be in more flexible turns or loops with exposure to the fibril surface.Citation10 It also agrees with 1D-PITHIRDS-CT solid-state NMR-measurements demonstrating a parallel in-register β‑sheet structure for residues Ile182, Ile184, Ile197, Ile203 and 2 of 3 of the residues Phe140, Phe174, and Phe197, but a more disordered structure for residues Leu125 and Leu130 in scrapie prion-seeded Syrian hamster recPrP(90-231)-fibrils.Citation15 The presence of the segments N-terminal of position 165 is mandatory for fibrillation,Citation59 though it is not necessarily incorporated in the β‑sheet coreCitation6 but rather at the fibril surface. Taken together, we hypothesize that the flexibility of this segment might act as a sort of “gatekeeper,” subtly influencing the β‑interface between monomeric units within fibrils and, in turn, stability, length, and infectivity of recPrP-amyloid.
In summary, we have established protocols for the preparation of infectious samples of full-length ovine recPrP-amyloid in solid-state NMR-sufficient amounts uniformly labeled with 13C and 15N. Our biophysical and solid-state NMR characterization of several samples is consistent with recPrP-amyloid in a single conformation. Our previous data indicate (1) a unique structure characteristic in line with a distinct C‑terminal β‑sheet core and a partially α‑helical middle part as the common motif of infectious ovine recPrP-amyloid; and (2) deviating flexibility of non‑β‑core segments in different PrP-amyloid types. Our established protocols demonstrate potential for a high-resolution structural characterization of non-infectious and infectious preparations of recPrP-amyloid by solid-state NMR spectroscopy in the near future.
Materials and Methods
Biological safety
Prion material was handled in biosafety hoods in a BSL-3**-laboratory. For NMR-experiments, samples were transferred into deformable, unbreakable, and hermetically sealable polyformaldehyde rotor inserts that were custom-built and are NMR-silent in the chemical shift range typical for proteins (13C signals are at 89.1 ppm and 1.0 ppm).
Fibrillation
PrP expression in Escherichia coli and purification was performed as describedCitation63 and adapted to full-length ovine ARQ recPrP (25–233). Typical yields of uniformly 13C,15N isotope-labeled ovrecPrP(25‑233) of 10‑15 mg per liter M9 minimal medium were achieved. In vitro-conversion was carried out in presence of 1 M GdnHCl, 2.4 M urea, and 167 mM NaCl in 20 mM sodium acetate pH 5.0 with continuous shaking at 600 r.p.m. and 37°C with 100 ng μl‑1 ovrecPrP(25‑233) (150 μl in 96-well plates) or 1000 ng μl‑1 ovrecPrP(25‑233) (1 ml in 1.5 ml microcentrifuge tubes or 10 ml in 15 ml conical tubes), respectively. For seeded fibrillation, PrPSc-seeds were prepared by precipitating 700 μl of 10 % (vol/vol) scrapie-infected homozygous ARQ sheep brain homogenate with sodium phosphotungstic acid (NaPTA) adapted to ovine PrPSc Citation64 resulting in sequence-identical 30 μg-ARQ PrPSc-seeds and sonicated for 10 sec with a Misonix sonicator 3000 at 200 W and <25°C. As a control, brain homogenate of non-infected ARQ sheep was treated identically. At the conversion endpoint, the fibrillation buffer including residual non-fibrillar material was removed by differential ultracentrifugation in a TL‑100 ultracentrifuge (Beckman Instruments) with a TLA-45 rotor for 1 h at 100,000 × g and 4°C. Each pellet was washed with sterile bidest. water, centrifuged for 1 h at 100,000 × g and 4°C, washed and centrifuged again. NMR samples were dried until any solution was evaporated and immediately packed in 3.2-mm rotors and stored at 4°C.
Thioflavin T–assay
Fibrillation kinetics were monitored by thioflavin T (ThT)-fluorescence assays in 96-well, black-bottom Nunc plates in a microplate reader (Infinite M200 Pro, Tecan Trading AG) at 37°C in 150 μl fibrillation buffer and 5 μM ThT. The recording parameters were: 442 nm excitation wavelength, 475 nm-555 nm spectral range, 2 nm step resolution, 10 nm excitation bandwidth, 20 nm emission bandwidth, gain of 80, z-position of 18300±500 μm. For analysis, intensity integrals from 475 nm to 555 nm were calculated.
Mouse bioassay
Fibril preparations were inoculated in ovine PrP(25‑233) transgenic mice tgshpXI and tg338 homozygously overexpressing the ARQ- or VRQ-allele, respectively, which are both associated with highest scrapie susceptibility and frequency.Citation22 Before inoculation, each sample was suspended in sterile PBS pH 7.4, quick-frozen in liquid nitrogen, and stored at −80°C. For each determination of mean incubation periods, 6 tgshpXI mice and 6 tg338 mice aged 6 to 8 weeks were inoculated intracerebrally with 50 μl of a given sample, examined for clinical signs of mouse prion disease at least twice a week, and euthenized at the point of neurologic dysfunction. Only coded information was displayed on mice boxes to avoid observer bias. Bioassays were terminated 630 days after inoculation. Incubation periods were measured as the time from inoculation to onset of clinical symptoms. Crude brain homogenates [10 % (w/v)] were prepared from diseased mice inoculated with spontaneously generated ovrecPrP(25‑233)-amyloid and used for a second passage into the same mouse line.Citation65 PrPSc in the brains of animals was detected by digestion of 10 % brain homogenates with 50 μg/ml PK (recombinant grade; Roche Applied Science) for 2 h at 37°C followed by Western blot analysis.
Circular dichroism (CD) spectroscopy
Ten CD spectra per sample were collected with a Jasco J-815 spectropolarimeter at room temperature in 10 mM sodium phosphate pH 7.4 over the spectral range of 185 nm-260 nm at a scan speed of 50 nm/min and a step resolution of 1 nm. Only CD signals with a HT-value below 600 were regarded to be reliable. Below a wavelength of 185 nm, excessive buffer absorption impeded further readings. A blank spectrum for cuvette and buffer was subtracted from each spectrum. Satisfactory CD ellipticities of recPrP-amyloid were only obtained after increasing amyloid solubility by gentle sonication of 1 μg μl‑1 PrP in presence of 0.12 % (vol/vol) SDS with a Misonix sonicator 3000 at 240 W for 1 min at <25°C.Citation66 Neither sonication only nor SDS-treatment only did result in measurable ellipticity. An influence of sonication or SDS-treatment on the secondary structure of amyloid fibrils is assumed to be negligible. A SDS-concentration as high as 0.2 % (vol/vol) only solubilises prion fibrils.Citation67 Prion infectivity, which depends crucially on structural features, remains even after boiling in 5 % SDS.Citation68 Sonication of monomeric ovrecPrP(25‑233) for 45 min at 240 W changes neither ellipticity intensity nor spectral features. Sonication for 1 min at 240 W in 0.12 % (vol/vol) SDS resulted in an increase in ellipticity intensity, only accompanied by slight shifts in spectral features of ± 1nm. PTA-precipitated PrPSc was not measured because brain homogenate impurities, glycosylations, and GPI-anchor prevent a valid quantitative analysis.
Atomic force microscopy (AFM) and image processing
All AFM measurements were carried out with the sample dried on the mica surface. Liquid cell AFM could not be performed because of prion safety limitations, to avoid interference of the cantilever movement with the solvent, and because a soft coat formed in the presence of water by the disordered N-terminal segment of PrP-fibrils may hide fibrillar topology details.Citation9 Immediately before AFM measurement, samples were briefly sonicated for 10 sec with a Misonix sonicator 3000 at 200 W and <25°C. Sample volumes of 40 μl containing 0.44 – 4.4 μM ovrecPrP from the fibrillation end point or PrPSc-seeds purified from brain homogenate by precipitation with sodium phosphotungstic acid (NaPTA) were adhered for 30 min onto a freshly cleaved mica crystal, washed 30 times with 40 μl water drops, and dried at room temperature for 45 min. Samples were imaged with a NanoWizard II AFM (JPK Instruments AG) using Olympus silicon tips on a silicon cantilever (OMCL‑160TS‑R3) with a typical tip size of 7 nm, a spring constant of 26 N m−1, a drive frequency of 300±100 kHz, and a scan rate of 0.5 – 1 lines sec−1. Data were collected using intermittent current AFM-mode from up to 4 different samples and up to 3 different scanning sessions as 512 × 512 or 1024 × 1024 pixel images and processed using Gwyddion software 2.22. All images were levelled by mean plane subtraction to correct for tilting of the sample stage and servo range errors. Each scan line was corrected for streaks, scratches, and noise related to the mica surface. Fibrillar heights were measured perpendicular to the fibrillar axis using Gwyddion's extract profile tool, but not quantified in detail. In contrast to widths, the heights of soft objects are usually underestimated with AFMCitation69,70 because of the mechanical force applied, which results in elastic deformation of polymers such as nucleic acids and protein fibrils. Mean fibril widths and lengths were quantified at half of the maximal heights. Width distribution histograms were obtained by classifying the cross section widths of >700 ovrecPrP-fibrils into groups of 5 nm size. To avoid measurement errors from stacked fibrils, only isolated fibrils were considered. To account for tip convolution, all measured widths and lengths were corrected for the tip size.
Solid-state NMR spectroscopy
All NMR-experiments were conducted using 3.2 mm triple-resonance (1H, 13C, 15N) probe heads at static magnetic fields of 14.1 and 18.8 T (Agilent Technologies) and MAS frequencies of 11 kHz±3 Hz or 12.5 kHz±3 Hz, respectively, for (13C, 13C) PDSD‑Citation71 and NCACX-/NCOCX-type spectraCitation72 or 23.0 kHz±5 Hz for DREAM mixing,Citation73 respectively. Temperature was calibrated for each probe head and each spinning speed using nickelocene.Citation74 The cooling gas was set to a temperature that resulted in an actual sample temperature of 0°C ± 2.5°C. 13C and 15N chemical shifts were referenced externally to adamantane. Typical 1H, 13C, and 15N 90° pulse lengths were 3.0 μs, 5.5 μs, and 6 μs, respectively. For heteronuclear through-bond transfer, the INEPT schemeCitation75 was applied. For heteronuclear through-space transfer, chemical shift-selective (15N, 13C) and broad-band (1H, 13C)/(1H, 15N) SPECIFIC-CPCitation72 schemes were applied. (13C, 13C) mixing was performed using PDSD-mixing times of 5, 10, 20, and 80 ms for intraresidue correlations or 80, 150, and 500 ms for sequential correlations. (15N, 13C) spectra were recorded using SPECIFIC-CP contact times between 1500 and 3500 ms followed by homonuclear PDSD-mixing of 50 to 100 ms or DREAM mixing. During indirect chemical shift evolution periods and FID acquisition, 83 kHz SPINAL decouplingCitation76 was applied. All spectra were recorded in form of successive one-day data sets, which were recorded and added until no further improvement of signal-to-noise ratios was visible. Data sets were processed with NMRPipeCitation77 by routinely using different apodisation functions, primarily shifted sine-bell squared functions, with or without linear prediction in the indirect dimension and analyzed with Sparky version 3.114 (T. D. Goddard and D. G. Kneller, University of California, San Francisco). For peak intensity analysis, square chemical shift areas were integrated manually using Sparky's sum over box integration method. For every amino acid type, the identical chemical shift areas were integrated 10 times, to ensure that peak volumes were not biased by slight variations in the areas selected for integration. Standard deviations were calculated according to [∑(x-ℜ)2/(n-1)]1/2 with ℜ the sample mean and n the sample size.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
KPRN_A_983754_Supplemental_Figures.zip
Download Zip (4.6 MB)KPRN_A_983754_Supplemental_Tables.zip
Download Zip (111.5 KB)Acknowledgments
We thank Bernd Esters for recPrP(25‑233) expression and the precision engineering workshop of Waldemar Seidel for MAS-rotor inserts. We are deeply indebted to Detlev Riesner, Filipp Oesterhelt, Eva Birkmann, and Christian Dumpitak for fruitful discussions.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Prusiner S. Prions. Proc Natl Acad Sci U S A 1998; 95:13363-83; PMID:9811807; http://dx.doi.org/10.1073/pnas.95.23.13363
- Legname G, Baskakov I, Nguyen H, Riesner D, Cohen F, DeArmond S, Prusiner S. Synthetic mammalian prions. Science 2004; 305:673-6; PMID:15286374; http://dx.doi.org/10.1126/science.1100195
- Wille H, Bian W, McDonald M, Kendall A, Colby D, Bloch L, Ollesch J, Borovinskiy A, Cohen F, Prusiner S, Stubbs, G. Natural and synthetic prion structure from X-ray fiber diffraction. Proc Natl Acad Sci U S A 2009; 106:16990-5; PMID:19805070; http://dx.doi.org/10.1073/pnas.0909006106
- Wuthrich K, Riek R. Three-dimensional structures of prion proteins. Adv Protein Chem 2001; 57:55-82; PMID:11447697; http://dx.doi.org/10.1016/S0065-3233(01)57018-7
- Schlepckow K, Schwalbe H. Molecular mechanism of prion protein oligomerization at atomic resolution. Angew Chem Int Ed Engl 2013; 52:10002-10005; PMID:23934741; http://dx.doi.org/10.1002/anie.201305184
- Cobb N, Sonnichsen F, McHaourab H, Surewicz W. Molecular architecture of human prion protein amyloid: a parallel, in-register beta-structure. Proc Natl Acad Sci U S A 2007; 104:18946-51; PMID:18025469; http://dx.doi.org/10.1073/pnas.0706522104
- Bocharova O, Breydo L, Parfenov A, Salnikov V, Baskakov I. In vitro conversion of full-length mammalian prion protein produces amyloid form with physical properties of PrP(Sc). J Mol Biol 2005; 346:645-59; PMID:15670611; http://dx.doi.org/10.1016/j.jmb.2004.11.068
- Smirnovas V, Baron G, Offerdahl D, Raymond G, Caughey B, Surewicz W. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat Struct Mol Biol 2011; 18:504-6; PMID:21441913; http://dx.doi.org/10.1038/nsmb.2035
- Anderson M, Bocharova O, Makarava N, Breydo L, Salnikov V, Baskakov I. Polymorphism and ultrastructural organization of prion protein amyloid fibrils: an insight from high resolution atomic force microscopy. J Mol Biol 2006; 358:580-96; PMID:16519898; http://dx.doi.org/10.1016/j.jmb.2006.02.007
- Vazquez-Fernandez E, Alonso J, Pastrana MA, Ramos A, Stitz L, Vidal E, Dynin I, Petsch B, Silva CJ, Requena JR. Structural organization of mammalian prions as probed by limited proteolysis. PLoS One 2012; 7:e50111; PMID:PMID:23185550; http://dx.doi.org/10.1371/journal.pone.0050111
- Govaerts C, Wille H, Prusiner S, Cohen, F. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci U S A 2004; 101:8342-7; PMID:15155909; http://dx.doi.org/10.1073/pnas.0402254101
- Helmus J, Surewicz K, Nadaud P, Surewicz W, Jaroniec C. Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proc Natl Acad Sci U S A 2008; 105:6284-9; PMID:18436646; http://dx.doi.org/10.1073/pnas.0711716105
- Lu X, Wintrode P, Surewicz W. Beta-sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc Natl Acad Sci U S A 2007; 104:1510-5; PMID:17242357; http://dx.doi.org/10.1073/pnas.0608447104
- Tycko R, Savtchenko R, Ostapchenko V, Makarava N, Baskakov I. The alpha-helical C-terminal domain of full-length recombinant PrP converts to an in-register parallel beta-sheet structure in PrP fibrils: evidence from solid state nuclear magnetic resonance. Biochemistry 2010; 49:9488-97; PMID:20925423; http://dx.doi.org/10.1021/bi1013134
- Groveman BR, Dolan MA, Taubner LM, Kraus A, Wickner RB, Caughey B. Parallel in-register intermolecular beta-sheet architectures for prion-seeded prion protein (PrP) amyloids. J Biol Chem 2014; 289:24129-42; PMID:25028516; http://dx.doi.org/10.1074/jbc.M114.578344
- Kodali R, Williams AD, Chemuru S, Wetzel, R. Abeta(1-40) forms five distinct amyloid structures whose beta-sheet contents and fibril stabilities are correlated. J Mol Biol 2010; 401:503-17; PMID:20600131; http://dx.doi.org/10.1016/j.jmb.2010.06.023
- Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's beta-amyloid fibrils. Proc Natl Acad Sci U S A 2008; 105:18349-54; PMID:19015532; http://dx.doi.org/10.1073/pnas.0806270105
- Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science 2005; 307:262-5; PMID:15653506; http://dx.doi.org/10.1126/science.1105850
- Paravastu A, Qahwash I, Leapman R, Meredith S, Tycko R. Seeded growth of beta-amyloid fibrils from Alzheimer's brain-derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci U S A 2009; 106:7443-8; PMID:19376973; http://dx.doi.org/10.1073/pnas.0812033106
- Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular structure of beta-amyloid fibrils in Alzheimer's disease brain tissue. Cell 2013; 154:1257-68; PMID:24034249; http://dx.doi.org/10.1016/j.cell.2013.08.035
- Frederick KK, Debelouchina GT, Kayatekin C, Dorminy T, Jacavone AC, Griffin RG, Lindquist S. Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics. Chem Biol 2014; 21:295-305; PMID:24485763; http://dx.doi.org/10.1016/j.chembiol.2013.12.013
- Baylis M, Goldmann W. The genetics of scrapie in sheep and goats. Curr Mol Med 2004; 4:385-96; PMID:15354869; http://dx.doi.org/10.2174/1566524043360672
- Castilla J, Saa P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell 2005; 121:195-206; PMID:15851027; http://dx.doi.org/10.1016/j.cell.2005.02.011
- Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science 2010; 327:1132-5; PMID:20110469; http://dx.doi.org/10.1126/science.1183748
- Deleault N, Harris B, Rees J, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci U S A 2007; 104:9741-6; PMID:17535913; http://dx.doi.org/10.1073/pnas.0702662104
- Kim J, Cali I, Surewicz K, Kong Q, Raymond G, Atarashi R, Race B, Qing L, Gambetti P, Caughey B, Surewicz W. Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J Biol Chem 2010; 285:14083-7; PMID:20304915; http://dx.doi.org/10.1074/jbc.C110.113464
- Cosseddu G, Nonno R, Vaccari G, Bucalossi C, Fernandez-Borges N, Di Bari M, Castilla J, Agrimi U. Ultra-efficient PrP(Sc) amplification highlights potentialities and pitfalls of PMCA technology. PLoS Pathog 2011; 7:e1002370; PMID:22114554; http://dx.doi.org/10.1371/journal.ppat.1002370
- Prusiner S, Safar J, DeArmond S. Bioassays of prions. In: Prion Biology and Diseases. Eds. Prusiner, S.B. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2004; 143-186.
- Colby D, Giles K, Legname G, Wille H, Baskakov I, DeArmond S, Prusiner S. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci U S A 2009; 106:20417-22; PMID:19915150; http://dx.doi.org/10.1073/pnas.0910350106
- Colby D, Wain R, Baskakov I, Legname G, Palmer C, Nguyen H, Lemus A, Cohen F, DeArmond S, Prusiner S. Protease-sensitive synthetic prions. PLoS Pathog 2010; 6:e1000736
- Makarava N, Kovacs G, Bocharova O, Savtchenko R, Alexeeva I, Budka H, Rohwer R, Baskakov I. Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol 2010; 119:177-87; PMID:20052481; http://dx.doi.org/10.1007/s00401-009-0633-x
- Makarava N, Baskakov IV. The evolution of transmissible prions: the role of deformed templating. PLoS Pathog 2013; 9:e1003759; PMID:24339773; http://dx.doi.org/10.1371/journal.ppat.1003759
- Riesner D. The scrapie isoform of the prion protein PrPSc compared to the cellular isoform PrPC. In: PriOns and Prion Diseases. Eds. Hörnlimann B, Riesner D, Kretzschmar H. Berlin, New York: Walter de Gruyter, 2006, 505-15.
- McKinley M, Meyer R, Kenaga L, Rahbar F, Cotter R, Serban A, Prusiner S. Scrapie prion rod formation in vitro requires both detergent extraction and limited proteolysis. J Virol 1991; 65:1340-51; PMID:1704926
- Piro J, Wang F, Walsh D, Rees J, Ma J, Supattapone S. Seeding specificity and ultrastructural characteristics of infectious recombinant prions. Biochemistry 2011; 50:7111-6; PMID:21776987; http://dx.doi.org/10.1021/bi200786p
- Rouvinski A, Karniely S, Kounin M, Moussa S, Goldberg MD, Warburg G, Lyakhovetsky R, Papy-Garcia D, Kutzsche J, Korth C, Carlson GA, Godsave SF, Peters PJ, Luhr K, Kristensson K, Taraboulos A. Live imaging of prions reveals nascent PrPSc in cell-surface, raft-associated amyloid strings and webs. J Cell Biol 2014; 204:423-441; PMID:24493590; http://dx.doi.org/10.1083/jcb.201308028
- Wegmann S, Miesbauer M, Winklhofer KF, Tatzelt J, Muller DJ. Observing fibrillar assemblies on scrapie-infected cells. Pflugers Archiv-Eur J Physiol 2008; 456:83-93; PMID:18175144; http://dx.doi.org/10.1007/s00424-007-0433-x
- Heise H, Hoyer W, Becker S, Andronesi O, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci U S A 2005; 102:15871-6; PMID:16247008; http://dx.doi.org/10.1073/pnas.0506109102
- Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Am Chem Soc 2005; 127:12965-74; PMID:16159291; http://dx.doi.org/10.1021/ja0530164
- Siemer AB, Arnold AA, Ritter C, Westfeld T, Ernst M, Riek R, Meier BH. Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J Am Chem Soc 2006; 128:13224-8; PMID:17017802; http://dx.doi.org/10.1021/ja063639x
- Helmus JJ, Surewicz K, Surewicz WK, Jaroniec CP. Conformational flexibility of Y145Stop human prion protein amyloid fibrils probed by solid-state nuclear magnetic resonance spectroscopy. J Am Chem Soc 2010; 132:2393-403; PMID:20121096; http://dx.doi.org/10.1021/ja909827v
- Daebel V, Chinnathambi S, Biernat J, Schwalbe M, Habenstein B, Loquet A, Akoury E, Tepper K, Muller H, Baldus M, et al. beta-Sheet core of tau paired helical filaments revealed by solid-state NMR. J Am Chem Soc 2012; 134:13982-9; PMID:22862303; http://dx.doi.org/10.1021/ja305470p
- Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R, Cohen F, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A 1993; 90:10962-6; PMID:7902575; http://dx.doi.org/10.1073/pnas.90.23.10962
- Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry 1991; 30:7672-80; PMID:1678278; http://dx.doi.org/10.1021/bi00245a003
- Safar J, Roller PP, Gajdusek DC, Gibbs, CJ Jr. Conformational transitions, dissociation, and unfolding of scrapie amyloid (prion) protein. J Biol Chem 1993; 268:20276-84; PMID:8104185
- Requena JR, Wille H. The structure of the infectious prion protein: experimental data and molecular models. Prion 2014; 8:60-6; PMID:24583975; http://dx.doi.org/10.4161/pri.28368
- Wishart D, Sykes B. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR 1994; 4:171-80; PMID:8019132; http://dx.doi.org/10.1007/BF00175245
- Tycko R, Wickner RB. Molecular structures of amyloid and prion fibrils: consensus versus controversy. Acc Chem Res 2013; 46:1487-96; PMID:23294335; http://dx.doi.org/10.1021/ar300282r
- Heise H. Solid-state NMR spectroscopy of amyloid proteins. Chembiochem 2008; 9:179-89; PMID:18161737; http://dx.doi.org/10.1002/cbic.200700630
- Stohr J, Watts J, Mensinger Z, Oehler A, Grillo S, DeArmond S, Prusiner S, Giles K. Purified and synthetic Alzheimer's amyloid beta (Abeta) prions. Proc Natl Acad Sci U S A 2012; 109:11025-30; PMID:22711819; http://dx.doi.org/10.1073/pnas.1206555109
- Prusiner SB. Cell biology. A unifying role for prions in neurodegenerative diseases. Science 2012; 336:1511-3; PMID:22723400; http://dx.doi.org/10.1126/science.1222951
- Legname G, Nguyen H, Peretz D, Cohen F, DeArmond S, Prusiner S. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc Natl Acad Sci U S A 2006; 103:19105-10; PMID:17142317; http://dx.doi.org/10.1073/pnas.0608970103
- Eghiaian F, Grosclaude J, Lesceu S, Debey P, Doublet B, Treguer E, Rezaei H, Knossow M. Insight into the PrPC–>PrPSc conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants. Proc Natl Acad Sci U S A 2004; 101:10254-9; PMID:15240887; http://dx.doi.org/10.1073/pnas.0400014101
- Tateishi J, Kitamoto T. Inherited prion diseases and transmission to rodents. Brain Pathol 1995; 5:53-9; PMID:7767491; http://dx.doi.org/10.1111/j.1750-3639.1995.tb00577.x
- Shen Y, Bax A. Identification of helix capping and b-turn motifs from NMR chemical shifts. J Biomol NMR 2012; 52:211-32; PMID:22314702; http://dx.doi.org/10.1007/s10858-012-9602-0
- Williamson JA, Miranker AD. Direct detection of transient alpha-helical states in islet amyloid polypeptide. Protein Sci 2007; 16:110-7; PMID:17123962; http://dx.doi.org/10.1110/ps.062486907
- Kirkitadze MD, Condron MM, Teplow DB. Identification and characterization of key kinetic intermediates in amyloid beta-protein fibrillogenesis. J Mol Biol 2001; 312:1103-19; PMID:11580253; http://dx.doi.org/10.1006/jmbi.2001.4970
- Holtzer ME, Lovett EG, d'Avignon DA, Holtzer A. Thermal unfolding in a GCN4-like leucine zipper: 13C alpha NMR chemical shifts and local unfolding curves. Biophys J 1997; 73:1031-41; PMID:9251820; http://dx.doi.org/10.1016/S0006-3495(97)78136-0
- Kumar J, Sreeramulu S, Schmidt T, Richter C, Vonck J, Heckel A, Glaubitz C, Schwalbe, H. Prion protein amyloid formation involves structural rearrangements in the C-terminal domain. Chembiochem 2010; 11:1208-13; PMID:20458726; http://dx.doi.org/10.1002/cbic.201000076
- DeMarco M, Daggett V. Local environmental effects on the structure of the prion protein. C R Biol 2005; 328:847-62; PMID:16286076; http://dx.doi.org/10.1016/j.crvi.2005.05.001
- Kunes KC, Clark SC, Cox DL, Singh RRP. Left handed beta helix models for mammalian prion fibrils. Prion 2008; 2:81-90; PMID:19098440; http://dx.doi.org/10.4161/pri.2.2.7059
- Smirnovas V, Kim J, Lu X, Atarashi R, Caughey B, Surewicz W. Distinct structures of scrapie prion protein (PrPSc)-seeded versus spontaneous recombinant prion protein fibrils revealed by hydrogen/deuterium exchange. J Biol Chem 2009; 284:24233-41; PMID:19596861; http://dx.doi.org/10.1074/jbc.M109.036558
- Jansen K, Schafer O, Birkmann E, Post K, Serban H, Prusiner S, Riesner D. Structural intermediates in the putative pathway from the cellular prion protein to the pathogenic form. Biol Chem 2001; 382:683-91; PMID:11405232; http://dx.doi.org/10.1515/BC.2001.081
- Panza G, Stohr J, Dumpitak C, Papathanassiou D, Weiss J, Riesner D, Willbold D, Birkmann E. Spontaneous and BSE-prion-seeded amyloid formation of full length recombinant bovine prion protein. Biochem Biophys Res Commun 2008; 373:493-7; PMID:18585368; http://dx.doi.org/10.1016/j.bbrc.2008.06.059
- Torres JM, Espinosa JC, Aguilar-Calvo P, Herva ME, Relano-Gines A, Villa-Diaz A, Morales M, Parra B, Alamillo E, Brun A, et al. Elements modulating the prion species barrier and its passage consequences. PLoS One 2014; 9:e89722; PMID:24608126; http://dx.doi.org/10.1371/journal.pone.0089722
- Muller H, Stitz L, Wille H, Prusiner S, Riesner D. Influence of water, fat, and glycerol on the mechanism of thermal prion inactivation. J Biol Chem 2007; 282:35855-67; PMID:17878157; http://dx.doi.org/10.1074/jbc.M706883200
- Leffers KW, Schell J, Jansen K, Lucassen R, Kaimann T, Nagel-Steger L, Tatzelt J, Riesner, D. The structural transition of the prion protein into its pathogenic conformation is induced by unmasking hydrophobic sites. J Mol Biol 2004; 344:839-53; PMID:15533449; http://dx.doi.org/10.1016/j.jmb.2004.09.071
- Taylor DM, Fernie K, McConnell I, Steele PJ. Survival of scrapie agent after exposure to sodium dodecyl sulphate and heat. Vet Microbiol 1999; 67:13-6; PMID:10392773; http://dx.doi.org/10.1016/S0378-1135(99)00026-7
- Ukraintsev E, Kromka A, Kozak H, Remeš Z, Rezek B. Artifacts in Atomic Force Microscopy of Biological Samples. In: Force Microscopy Investigations into Biology – From Cell to Protein, Ed. Christopher F, InTech, 2012, 29-54. ISBN 978-953-51-0114-7.
- Rezek B, Shin D, Nebel C. Properties of hybridized DNA arrays on single-crystalline undoped and boron-doped (100) diamonds studied by atomic force microscopy in electrolytes. Langmuir 2007; 23:7626-33; PMID:17547423; http://dx.doi.org/10.1021/la0636661
- Szeverenyi N, Sullivan M, Maciel G. Observation of spin exchange by two-dimensional fourier-transform C-13 cross polarization-magic-angle spinning. J Magn Reson 1982; 47:462-475.
- Baldus M, Petkova A, Herzfeld J, Griffin R. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol Phys 1998; 95:1197-1207; PMID:16990042; http://dx.doi.org/10.1080/00268979809483251
- Verel R, Ernst M, Meier B. Adiabatic dipolar recoupling in solid-state NMR: the DREAM scheme. J Magn Reson 2001; 150:81-99; PMID:11330986; http://dx.doi.org/10.1006/jmre.2001.2310
- Heise H, Kohler F, Xie X. Solid-state NMR spectroscopy of paramagnetic metallocenes. J Magn Reson 2001; 150:198-206; PMID:11384181; http://dx.doi.org/10.1006/jmre.2001.2343
- Morris G, Freeman A. Enhancement of nuclear magnetic resonance signals by polarization transfer. J Am Chem Soc 1979; 101:760-762; http://dx.doi.org/10.1021/ja00497a058
- Bennett A, Rienstra C, Griffiths J, Zhen W, Lansbury P, Griffin R. Homonuclear radio frequency-driven recoupling in rotating solids. J Chem Phys 1998; 108:9463-9479; PMID:24313031; http://dx.doi.org/10.1063/1.476420
- Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. Nmrpipe – a multidimensional spectral processing system based on unix pipes. J Biomol NMR 1995; 6:277-293; PMID:8520220; http://dx.doi.org/10.1007/BF00197809
