Abstract
Proteases regulate a myriad of cell functions, both in normal and disease states. In addition to protein turnover, they regulate a range of signaling processes, including those mediated by Eph receptors and their ephrin ligands. A variety of proteases is reported to directly cleave Ephs and/or ephrins under different conditions, to promote receptor and/or ligand shedding, and regulate receptor/ligand internalisation and signaling. They also cleave other adhesion proteins in response to Eph-ephrin interactions, to indirectly facilitate Eph-mediated functions. Proteases thus contribute to Eph/ephrin mediated changes in cell-cell and cell-matrix interactions, in cell morphology and in cell migration and invasion, in a manner which appears to be tightly regulated by, and co-ordinated with, Eph signaling. This review summarizes the current literature describing the function and regulation of protease activities during Eph/ephrin-mediated cell signaling.
Abbreviations
| AD | = | Alzheimer's disease |
| ADAM | = | a disintegrin and metalloprotease |
| APP | = | Amyloid precursor protein |
| Arf1 | = | ADP rybosylation factor 1 |
| BACE | = | β-site APP cleaving enzyme |
| CAM | = | cell adhesion molecules |
| CTF | = | cytoplasmic fragment |
| CNC | = | cranial neural crest |
| ECM | = | extracelluar matrix |
| FLIM | = | fluorescence lifetime imaging microscopy |
| FN | = | fibronectin |
| iCLiP | = | intramembrane cleaving protease |
| LTP | = | long-term potentiation |
| MLC | = | myosin light chain |
| MMP | = | matrix-metalloprotease |
| MT1-MMP | = | membrane type-I matrix metalloproteinase |
| NCAM | = | neural cell adhesion molecule |
| NMDA | = | N-methyl-D-aspartate |
| NMJ | = | neuromuscular junction |
| NOX | = | NADPH oxidase |
| PDI | = | protein disulphide isomerise |
| PS | = | Presenilin |
| PTP | = | Protein Tyrosine Phosphatese |
| RIP | = | Regulated intramembrane proteolysis |
| ROS | = | reactive oxygen species |
| RTK | = | receptor tyrosine kinase |
| TACE | = | tumor necrosis factor-α-converting enzyme |
| TNFα | = | tumor necrosis factor-α |
| TGFβ | = | transforming growth factor β |
Introduction
Proteases in regulation of cell signaling in general
Proteases are enzymes that carry out the hydrolysis of peptide bonds within proteins and polypeptides. Apart from their obvious roles in digestion and protein turnover, proteases are important in tightly regulated signaling cascades which include cell cycle regulation, blood coagulation, apoptosis, antigen presentation and protein and organelle recycling.Citation1 The disruption of their strictly controlled equilibrium in the cell has been found in a number of pathologies, such as cardiovascular diseases, inflammation, neurodegenerative diseases, and cancer.Citation2,3 There are 5 major classes of proteases known in mammals including serine, cysteine, metallo, aspartic, and threonine proteases, which are categorised according to their different catalytic mechanisms.Citation4,5 The cellular localization of proteases is an important factor that affects their activity and substrate selection – the greater proportion are extracellular, followed by intracellular, with a minor proportion being localized in plasma and organelle membranes. Together, all these different properties and abilities of proteases tightly regulate cellular processes throughout the body.Citation4,5
In early years proteases were not regarded as traditional signaling molecules, although this view is dramatically changing. Nevertheless, there are significant differences between protease signaling and other types of cellular signaling. In protease-mediated signaling, the signal is transmitted through the cleavage of protein substrates with subsequent activation, inactivation, or modulation of function, which is irreversible.Citation6 Examples include activation of cytokines, inactivation of numerous repair proteins during apoptosis, exposure of cryptic sites,Citation6 and shedding of various transmembrane proteins such as RTK ligands and receptors.Citation7 A number of proteases act in cascades allowing more stringent regulation and amplification of the signal.Citation3 In addition to their catalytic domain, numerous additional domains or modules substantially increase the complexity of protease functions.Citation4,5
For many receptor families, including tyrosine kinases (RTKs), receptor activation is generally thought to require proteolytic shedding of ligands at a distance, either from the same cell or a different cell, facilitating autocrine or paracrine signaling, respectively. However, as described previously, and elsewhere in this issue, a defining feature of Eph/ephrin signaling is that interactions occur between membrane bound ligands and receptors on adjacent cells, mediating juxtacrine signaling at cell-cell contacts, and therefore a necessary role for proteolysis is not immediately obvious. These interactions involve clusters of receptors and ligands, leading to large receptor-ligand complexes, which combined with involvement of cadherins and other cell adhesion molecules, result in tethering cells together. Yet Eph-ephrin interactions can lead either to cell spreading and adhesion, or to cytoskeletal collapse and cell retraction or segregation, necessitating disruption of these complexes.Citation8 While trans-endocytosis of Eph-ephrin ligand complexes has been reported for EphBs,Citation9 a number of reports show an essential role of proteases in mediating a variety of Eph-ephrin functions.
Proteases regulating Eph/ephrin signaling
ADAMs
ADAMs (A Disintegrin And Metalloproteases) are transmembrane metalloproteases that process and shed the ectodomains of membrane-anchored growth factors, cytokines and receptors.Citation10 They belong to the metazincin superfamilyCitation11 which also includes the ADAM-TS, class III snake venoms and matrix metalloproteases (MMPs). ADAMs have essential roles in fertilization, angiogenesis, neurogenesis, heart development and cancerCitation10 shown by using ADAM loss- and gain-of-function mouse models. ADAMs are widely expressed in mammalian tissues, and the observed phenotypes of ADAM knockout mice are subsequently diverse,Citation12 although only ADAM10, 17 and 19 are essential for mouse development.Citation13-15 ADAM10 and ADAM17 knockout leads to abnormalities in almost all tissuesCitation13,14 compared to ADAM19 deletion which affects only the peripheral nervous system and heart development.Citation15 Additionally, several ADAMs play important roles in pathological situations, such as inflammation, carcinogenesis or stress-mediated angiogenic response.Citation12 Accordingly, ADAMs are up-regulated in a wide variety of cancers, both solid and hematological tumors. They are reported to target a variety of substrates directly implicated in human disease, including ligands for receptors, and pro-inflammatory cytokines (eg. TNFα for ADAM17/TACE), motivating strong interest in ADAMs as therapeutic targets in cancer and other diseases, as reviewed elsewhere.Citation16
A typical ADAM consists of conserved and characteristic protein domains in which an N-terminal signal sequence is followed by a pro-domain, a metalloprotease (MP) domain, a disintegrin domain (D), a cysteine-rich (C) region, an EGF-like domain (except ADAM10 and ADAM17), a transmembrane domain and a cytoplasmic domain ().Citation17 While the N-terminus signal sequence directs ADAMs into the secretory pathwayCitation18,19 the prodomain is thought to function as both a molecular chaperone, and an inhibitor of the metalloprotease domain.Citation20,21 It was thought that removal of the prodomain at a conserved Rx(R/K)R motif by proprotein convertases was necessary to activate ADAMs, occurring in the trans-Golgi network.Citation22-24 However, while cleavage of the ADAM10 prodomain can enhance protease activity, expression of a truncated ADAM10 lacking the pro-domain results in decreased activity that is rescued by addition of recombinant prodomain, suggesting the prodomain is also necessary to assist in the correct folding of the catalytic domainCitation19. In support, cleaved prodomain can remain attached to the mature proteaseCitation23 on the cell surface. Furthermore, removal of the prodomain is not necessary for activation, as unprocessed, cell surface ADAM17 exhibits rapid and reversible activation in response to stimuli, by alterations in catalytic site accessibility.Citation25 Initially, it was thought the inhibitory function of the ADAM prodomain operates via a cysteine switch mechanism where a conserved unpaired cysteine residue within the prodomain preferentially coordinates with the active site zinc atom of the metalloprotease domain. However, at least for ADAM10Citation26, ADAM12Citation27, ADAM9 and 17Citation25 this is not the case.
Figure 1. An illustration of ADAM domain structure. ADAMs consist of a prodomain (pro), metalloprotease domain (MP), disintegrin domain (D), cysteine-rich domain (C), EGF-like domain (except for ADAM10 and 17), a transmembrane domain and a cytoplasmic domain.
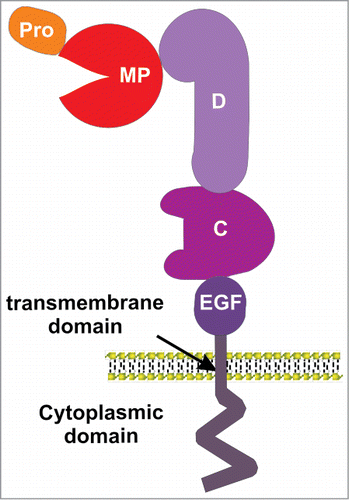
The best studied domain of the ADAM proteins is the metalloprotease domain, although this is not active in all ADAMs. Only 17 of the known 23 mammalian ADAMs contain the zinc-dependent metalloprotease catalytic site sequence,Citation11 which include consensus HEXGHXXGXXHD motif and a conserved methionine-turn in the active-site helix,Citation28 that confer proteolytic activity to ADAMs. Peptide libraries have been used to try to understand the selectivity of ADAMs for particular substrates. Although some sequence preferences exist for particular peptide substrates, there are no clear consensus cleavage motifs for ADAMs.Citation29 For example, while protein substrates for ADAM10 and ADAM17 are partially overlapping, each one appears to selectively target particular proteins in vivo.Citation30
Rather, control of substrate specificity resides in the D and C domains following the MP domains, which are involved in protein-protein interactions.Citation31 Using chimeric ADAM10-ADAM13 constructs it has been shown that ADAM13 D+C domains cannot be replaced by that of ADAM10 to induce hyperplasia in Xenopus laevis embryos, indicating alternate specificity.Citation32 ADAM13 C domain is also a major determinant for specific developmental events mediated by ADAM13 proteolytic shedding.Citation33 Similarly, a chimeric construct of ADAM17 with the D+C domains of ADAM10 could not reconstitute ADAM17 shedding of IL-1R-II.Citation34 As described below a substrate-binding pocket within the C domain of ADAM10 has been shown to define cleavage specificity in ephrin-Eph signaling.Citation44,45
Full length exodomain structures of mammalian ADAMs are not yet available, only for the isolated MP or D+C domains.Citation35,36 However they share an overall domain architecture similar to class-III snake venom metalloproteases, including conserved disulfide bond patterns, so these structures can provide insight into the likely structure of ADAMs. For example the VAP1 structure shares a curved D+C domain structure with ADAM10, which confers an overall C-shape structure with the MP domain in proximity to the substrate binding region of the C domain, consistent with the idea binding to the C domain controls specificity by positioning the substrate for cleavage.Citation37
Direct shedding of Ephs/ephrin ECDs by ADAMs
Fundamentally, a major outcome of ephrin-Eph signaling is cell-cell repulsion following engagement, which is important for neuronal axon guidance and also for establishment of the arterial and venous vascular networks.Citation38 Eph-induced retraction was first shown to require proteolytic cleavage of the Eph ligand ephrin-A2 on axons, following binding to its cognate receptor EphA3, through activity of ADAM10.Citation39 As outlined below a number of studies have now shown how ADAM10, and some other ADAM family members, regulate signaling by a number of different Eph/ephrins.
ADAM10
ADAM10 (Kuz) is one of the better characterized in the ADAM family with respect to proteolytic function. In addition to A-type ephrins (below), it has been identified as a sheddase of cell-surface bound proteins such as the epidermal growth factor receptor ligands EGF and betacellulin,Citation40 the erbB3/4 ligands neuregulins,Citation41,42 chemokines CX3CL1 and CXCL16,Citation43,44 E- and N-cadherin,Citation45,46 and adhesion molecule L1,Citation47 Amyloid precursor protein (APP),Citation48 the cellular prion precursor proteinCitation49 and many others. Nevertheless, many target substrates have been identified in vitro, using inhibitors that apply to a broad-range of metzincin family members.Citation50 The best characterized role of ADAM10 is in Notch signaling, where ADAM10 cleavage of both Notch receptorsCitation51 and ligands has been reportedCitation52,53 and, accordingly, phenotypes of ADAM10 and Notch1 deficient mice are very similar, including embryonic lethal defects in somitogenesis, neurogenesis and vasculogenesis.Citation54
In the first published study that demonstrates ADAM involvement in Eph signaling, ephrin-A2 was shown to be cleaved by ADAM10, disrupting Eph/ephrin cell contacts.Citation39 Here, ephrin-A2 was found in complex with ADAM10, interacting mainly via the non-catalytic regions and ephrin cleavage was triggered by binding to its receptor EphA3. Also, a motif search to identify sequences conserved between ADAM10 substrates, including all 8 vertebrate ephrins, the Notch ligand Delta, TNF-α, and APP, revealed a consensus sequence which lies not at the cleavage site but in the ligand-binding region of ephrins, indicating this region is mostly likely bound by ADAM10. A cleavage-inhibiting mutation in ephrin-A2 delayed axon withdrawal, highlighting the biological importance of protease recognition and regulation in EphA2/ephrin-A2 mediated axon detachment.Citation39
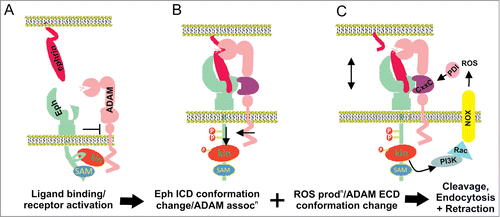
The mechanism by which ADAM10 recognizes and cleaves Eph-bound ephrin was clarified by structure-function studies interrogating the role of ADAM10 in EphA3/ephrin-A5 signaling.Citation35 ADAM10 demonstrated slight constitutive association with EphA3, rather than ephrin, but increased upon ephrin binding (). Determination of the crystal structure of the ADAM10 D+C domain identified an acidic pocket within the C domain which was required for binding to the Eph/ephrin complex, but did not effectively bind to either alone. This is consistent with the consensus recognition sequence identified by Hattori et al,Citation39 lying in the Eph-binding region of ephrins (above). Furthermore, as the Eph/ephrin interaction occurs between adjacent cells, ADAM10 on the Eph-expressing cell was found to cleave ephrin from the opposing cell, in a non cell autonomous manner (‘in trans’). Thus only receptor-bound ligand is cleaved, breaking the molecular tethers between the opposing cell surfaces, allowing internalization of the EphA3/ephrin-A5 complexes into the Eph-expressing cell.Citation55 The direct Eph-ADAM10 interaction was also subsequently shown using EphA2-expressing cells incubated on an artificial membrane displaying ephrin-A1.Citation56 Supporting the importance of the substrate recognition motif, monoclonal antibodies specifically recognizing this site are able to inhibit ephrin cleavage and ephrin-induced Eph receptor internalization, phosphorylation and Eph-mediated cell-cell migration.Citation57 This approach confirms the significance of the substrate-determining C domain as a potential target for therapeutic inhibition of ADAM10.
Figure 2. Proposed mechanisms of Eph/ephrin induced ADAM shedding. (A) Prior to Eph/ephrin interaction: Eph is in an inactive conformation, via interactions between the juxtamembrane and kinase domains; ADAM10 is poorly associated. (B) When Eph/ephrin interaction occurs between adjacent cells, the Eph/ephrin complex provide a new interface recognized by a substrate binding pocket in the C domain of ADAM10, positioning the protease domain to cleave ephrin from the opposing cell. Eph activation results in release of the inactive conformation and elongation of the EphA3 cytoplasmic domain, moving the kinase domain away from the plasma membrane, and facilitating ADAM10 association and ephrin cleavage. (C) Conformation changes in the ADAM C domain are proposed to occur via protein disulfide isomerase (PDI) activity, switching disufide bonds at a conserved CxxC motif. Upon activation of RTK receptors, reactive oxygen species (ROS) are produced by NOXs (NADPH-oxidases), which in turn inhibit PDI to favor an ‘active’ ADAM conformation, able to bind the Eph/ephrin complex, cleave ephrin and enable cell-cell retraction.
Regulation of ADAM-mediated ephrin cleavage
Importantly, Hattori et al found that ephrin cleavage by ADAM10 was also dependent on tyrosine kinase activity, as it was inhibited by treatment with the tyrosine kinase inhibitor genistein.Citation39 This is in line with a large body of work showing that elevated Eph kinase activity promotes cell-cell repulsion, while low kinase activity is associated with adhesion.Citation8 Similarly, cleavage of RTK ligands and receptors by various ADAMs increases in response to activation of PKC by phorbol esters, upon G protein coupled receptor activation, and following inhibition of protein tyrosine phosphatases (PTPs) (thereby promoting RTK activation), including ephrin-A2.Citation20,39 However, the mechanism through which receptor tyrosine kinase (RTK) signaling regulates ADAMs remained elusive. The ADAM10 or EphA3 cytoplasmic domains are not essential for ephrin shedding,Citation58 and similarly phorbol ester-stimulated shedding by ADAM17 is not effected by deletion of its cytoplasmic domain.Citation34 By performing fluorescence lifetime imaging microscopy (FLIM) and electron tomography on EphA3 with or without ephrin-A5 stimulation we have shown that a conformational change known to occur upon Eph activationCitation59 results in elongation of the EphA3 cytoplasmic domain, moving the kinase domain away from the plasma membrane, presumably relieving a steric hindrance or otherwise allowing productive association with ADAM10 upon activation by ephrin-A5 ().Citation58
Emerging data also suggests regulation of ADAM activity by reactive oxygen species (ROS) mediated conformational changes in the extracellular domain. In this model, highly conserved cysteine sulfhydryl (CxxC) motifs present in the ADAM17 (and ADAM10) D+C domain act as consensus sites for thiol disulphide exchange by protein disulphide isomerase (PDI), to induce change between inactive and active conformations. Thus, ADAM17 mutations replacing CxxC cysteine residues significantly decreased shedding of L selectin, while redox modulation by H2O2 treatment enhanced and reducing conditions inhibited ADAM17 activity in vitro and in vivo.Citation60 Subsequent studies revealed that inhibition or silencing of PDI also reduces PMA induced HB-EGF cleavage, suggesting that PDIs may control the conversion into an inactive form of ADAM17, and that PDI activity is modulated by oxidation.Citation61,62 ADAM10 contains a conserved CxxC motif (C594-x-x-C597) which lies adjacent to the substrate recognition site,Citation35 implying that similar to ADAM17, ADAM10 activity may also be regulated by ROS-mediated conformational changes, altering access to the substrate recognition site. During RTK signaling, cell membrane-resident NADPH oxidase generates ROSCitation63 which transiently activate RTKs by inhibiting regulatory PTPs, through oxidation of their catalytic cysteines.Citation64 Thus localized, NADPH oxidase–generated ROS may also trigger conformational changes controlling ADAM activity, suggesting a positive feedback loop where RTK activation causes ROS-mediated PTP inhibition and simultaneous activation of ADAM-mediated ligand shedding (). Considering that ROS levels are frequently elevated in tumors and other pathologies with elevated RTK and pro-inflammatory signaling,Citation65 and that ADAM activity is well-known to be upregulated in such environments,Citation66 this mechanism may be particularly relevant for diseases such as chronic inflammation and cancer.
In addition to ephrin-As, ADAM10 also associates with and cleaves ephrin-B2 during Xenopus development, in a manner regulated by interaction with flotillin-1.Citation67 Thus in the absence of flotillin-1, ephrin-B2 levels are dramatically reduced, leading to defects in neural tube closure, an effect dependent on function of ADAM10. Both ephrins and flotillins are known to co-locate to similar cholesterol-rich membrane microdomains,Citation68 while Ephs and ephrins have previously been shown to reside in distinct microdomains.Citation69 Given the association of ADAM10 with EphAs within a single membrane (above), this suggests that physical segregation of ADAM10 from ephrins may provide an extra level of regulation, a notion previously proposed for other targets.Citation66
Other ADAMs in Eph signaling
ADAM12 modulates intracellular signaling by cleaving various membrane bound signaling receptors and their ligands,Citation70 as well as degrading extracellular matrix proteins such as fibronectin, at least in vitro.Citation71 ADAM12 is highly expressed in glioblastoma multiforme, where it is linked to shedding of HB-EGF.Citation72 This tumor type also over-expresses EphA2 and shows high levels of shed ephrin-A1 suggesting a possible role of ADAM12 in ephrin shedding. Indeed, ADAM12 enhances ephrin-A1 shedding in primary tumors in response to transforming growth factorβ1 (TGF β1) activity, resulting in lung hyperpermeability that allows tumor cell migration into the lungs.Citation73 The receptors EphA1 and EphA2 both colocalise with ephrin-A1 in this setting, where they are thought to maintain cell-cell contacts, since loss of either contributes to lung permeability. Soluble ephrin-A1 disrupts these contacts, and inhibition of soluble ephrin-A1 by a neutralizing antibody significantly reduced lung metastasis.Citation73 This is consistent with the significant reduction in primary tumor growth and lung metastasis in ADAM12 KO mice.Citation74 Interestingly, EphA1 directly bound ADAM12 by yeast 2-hybrid screening, its co-expression in cells aided in ADAM12 translocation to the plasma membrane, and in co-culture experiments EphA1/ADAM12-expressing cells caused cleavage of ephrin-A1 on adjacent cells in trans,Citation73 in line with previous findings of ephrin-A5 trans cleavage by ADAM10 in complex with EphA3.Citation35
ADAM13 is also reported to cleave ephrin-B ligands,Citation75 in addition to a range of other substrates, including fibronectin (FN) and Cadherin-11.Citation76-78 ADAM13 is required for cranial neural crest (CNC) migration, a process where multipotent embryonic cells in the CNC migrate extensively to generate a large number of differentiated cell types in their target destinations, giving rise to craniofacial structures during embryonic development.Citation77-79 Ephrin-B1 acts as a guidance cue in segmental cranial/trunk neural crest migration, and ectopic expression of ADAM13 results in disoriented migration of both cranial and trunk neural crest similar to distorted ephrinB signaling.Citation80 Both ephrin-B1 and B2 were found to be substrates of ADAM13 cleavage in cells and embryos during CNC migration.Citation75 Interestingly, ADAM13 upregulated canonical Wnt signaling and early expression of the transcription factor snail2, whereas ephrin-B1 acts to inhibit this pathway, suggesting cleavage of ephrin-B1/2 by ADAM13 is required for derepression of canonical Wnt signaling and early CNC induction.Citation75 In addition, the cytoplasmic domain of ADAM13 is also shed and translocated to the nucleus where it controls expression of multiple genes in CNC.Citation79
While ADAM19 cleaves various ligands including neuregulins, HB-EGF, and TNF-α,Citation81 a different role has been suggested in Eph/ephrin signaling during development of the neuromuscular junction (NMJ). ADAM19 and ephrin-A5 both localize to the NMJ and participate in its formation, where ADAM19 interacts with EphA4 and blocks internalization of the ephrin-A5/EphA4 complex into the EphA4 expressing motor neurons, preventing repulsion of the axon terminal at the NMJ, independent of ADAM19 protease activity.Citation82 The cytoplasmic domain of EphA4 is postulated to be less accessible when it interacts with ADAM19, thus inhibiting binding of endocytosis regulators to the EphA4 SAM domain, preventing endocytosis.Citation82
Eph-dependent shedding of cell adhesion molecules by ADAMs
Eph receptors not only act directly to promote cellular activities (eg. adhesion, repulsion, migration and process extension) but also co-operate with other molecules like cell adhesion molecules (CAMs) to mediate such activities.Citation8 As discussed below, recently it has been shown that Eph signaling also instigates shedding of E-cadherin and the neural cell adhesion molecule (NCAM), further regulating cell adhesion and migration.Citation83,84
EphB signaling regulates the formation of E-cadherin based adhesions in epithelial cells to control cell migration in the intestinal epithelium. Thus, paneth cells in the intestinal crypt migrate downwards to localize at the crypt bottom resulting in compartmentalization that depends on the EphB2 and B3 expression levels of Paneth cellsCitation85 and ephrin-B1 expression in the transient amplifying compartment.Citation86 To determine ADAM10 involvement in this process, transgenic mice expressing dominant negative ADAM10 lacking its MP domain were generated. As a result of abrogated ADAM10 activity Paneth cell segregation was impaired, diminishing the boundary with the transient amplifying compartment.Citation83 This observation is reminiscent of EphB2/B3 knockout mice or intestinal ephrin-B1 null mice,Citation85 and implicates the likely involvement of ADAM10 in cellular boundary formation and compartmentalization. Further experiments reveal that monolayers of MDCK (Madin-Darby canine kidney) cells expressing dominant negative ADAM10 also have impaired segregation, promoting intermingling of EphB and ephrinB positive cells and blocking cell segregation. Interestingly, ADAM10, EphB2 and E-cadherin form a complex which initiates E-cadherin proteolytic shedding at sites of EphB/ephrin-B1 interactions.Citation83 However ephrin-B1 did not directly interact with ADAM10 or E-cadherin, which is compatible with a model where ADAM10 acts in trans to cleave E-cadherin from the opposing cells expressing ephrin-B1, upon substrate recognition via EphB2.
EphA/ephrinA signaling has an important role in axon guidance by controlling axon repulsion from inappropriate areas and constraining arborization in the neuronal cell populations.Citation87 NCAM, a potent promoter of axon growth and synaptic plasticity, interacts with EphA3 to stimulate growth cone collapse, reduce arborization, and limit the number of synapses of cortical GABAergic interneurons.Citation84 Ephrin-A5-induced growth cone collapse is ADAM10-dependent and is inhibited in cortical cultures from NCAM null mice and restored by the introduction of wild type NCAM.Citation88 NCAM, ADAM10, and EphA3 co-localize in cortical neurons and, analogous to E-cadnerin, EphA/ephrinA signaling triggers ADAM10 mediated NCAM shedding to promote growth cone collapse.Citation88
MMPs
Matrix-metalloproteases (MMPs) are a family of nearly 25 members that, like ADAMs, also belong to the metzincin superfamily. MMPs are secreted or remain membrane-anchored and subsequently cleave proteins on the membrane, secretary pathway or extracellular space.Citation89 The conserved pro-domain and the catalytic domain are the common structural features to all the MMP family members. The catalytic domain includes a zinc ion in the active site and interacts with 3 conserved histidine residues (HEXXHXXGXXH).Citation90 Most non-membrane bound MMPs also consist of a hemopexin domain at their C termini that mediates protein–protein interactions facilitating substrate recognition, activation of the enzyme, protease localization, internalization and degradation.Citation5 A subgroup of membrane bound MMPs, the MT-MMPs, are either type I transmembrane proteins or tethered to the plasma membrane by a GPI anchor making them important effectors in pericellular proteolysis. A typical function of MMPs is to degrade the structural components of the extracelluar matrix (ECM) to facilitate cell migration, including regulating signaling via ECM-associated membrane bound receptors such as integrins.Citation91 However MMPs also participate in the release of cell-membrane-bound precursor forms of many growth factors and their receptors (eg. FGFR1, ErbBs).Citation92 Thus, MMPs are critical in regulation of cell migration, especially in disease settings such as cancer metastasis and angiogenesis, and in inflammation and arthritis.Citation93,94
Direct effects of MMPs on Ephs/ephrins
A number of MMPs are reported to cleave Eph receptors and ephrins. Ephrin-A1, which is overexpressed in multiple human malignancies along with its corresponding receptor EphA2, was reported to be released from cells by proteolytic activity, present in conditioned medium and serum, and sensitive to broad MMP and serine-protease inhibitors.Citation95 Mass-spectrometry analysis and use of cleavage site mutants showed 3 cleavage sites on ephrin-A1 for MMPs. Soluble cleaved ephrin-A1 acted similar to homodimeric ephrin-A1 by activating EphA2 receptor and facilitating its internalization over time. Co-incubation of various recombinant MMPs in a non-cell-based assay showed ephrin-A5 was best cleaved by MMP1, 2, 9 and 13, although the biological relevance of this was not verified.Citation95 However, MMP-specific inhibitor treatment of melanoma cells does not alter EphA2-induced invasive capacity, which is mediated rather by Rho-mediated cytoskeletal changes, conferring a shift from mesenchymal to amoeboid-like cell motility,Citation96 although the same study confirmed the role of MMPs in invasion in response to other pathways, such as TNF signaling. Thus it is likely various proteases co-operate to promote cancer cell invasion, including ADAMs (as reviewed above), and interestingly, ADAM12Citation73, and MMP2 and 9Citation79 are all upregulated by TGF-β1 signaling in tumor metastasis.
EphBs and ephrin-Bs have also been reported to undergo cleavage by MMPs. EphB2-stimulated ephrin-B2 cleavage was first described in the context of subsequent intramembrane proteolysis by Presenilin (see below), and was sensitive to a broad metalloprotease inhibitor (GM6001),Citation98 although as this inhibits both MMPs and ADAMs it is unclear which is responsible. However there is evidence MMP8-dependent cleavage of ephrin-B1 occurs, causing ectodomain release into the culture medium of pancreatic cancer cell lines, and indeed recombinant ephrin-B1.Fc is cleaved preferentially by MMP8, compared to the other metalloproteases tested.Citation99 As with ephrin-B2, cleavage of ephrin-B1 was enhanced by interaction with the EphB2 receptor, which activated secretion of MMP8. This was dependent on the C-terminus of ephrin-B1, and stimulation of ephrin-B1 by EphB2 activated the ADP rybosylation factor 1 (Arf1) GTPase, a critical regulator of membrane trafficking, via the C-terminus of ephrin-B1.Citation99 Therefore it is speculated that ephrin-B1 regulates the exocytosis of MMP-8 by activation of Arf1, a process implicated in invasion of collagen by prostate cancer cellsCitation99 and in scirrhous gastric carcinoma cells.Citation100 Moreover, there was also dependence on ephrin-B1 association with the adapter protein Dishevelled, which activates the small GTPase RhoA, mediating cell-cell repulsion in EphB reverse signaling.Citation101,102 Transient expression of ephrin-B1 increased the level of GTP-bound RhoA activation, which was reduced by introduction of an ephrin-B1 C-terminus peptide.Citation100 Thus the ephrin-B1 C-terminus is suggested to activate both RhoA, to regulate cytoskeletal changes, and Arf1, to facilitate MMP8 mediated invasion ().
Figure 3. MMPs in Eph and/or ephrin shedding. (A) Ephrin-B1 ectodomain cleavage by MMP8, which is enhanced upon interaction with the EphB2 receptor.Citation99,100 Stimulation of ephrin-B1 by EphB2 activates the Arf1 GTPase, a critical regulator of membrane trafficking, most likely increasing the exocytosis of MMP8. The ephrin-B1 C-terminus also activates RhoA by binding to the adapter protein Dishevelled (Dsh), facilitating cell-cell repulsion and invasion. (B) Ephrin-B1-induced EphB2 receptor activation and its cleavage by MMP 2/9 cause RhoA activity via recruitment and subsequent activation of FAK, regulating growth cone withdrawal and collapse.Citation103 (C) Clusters of overexpressed EphA2 recruit MT1-MMP via its non-catalytic domains, mediating cleavage of EphA2 in cis at the fibronectin repeat type-III domain, stimulated by binding to ephrin-A1, and activating Src/Rho-mediated invasion.Citation107
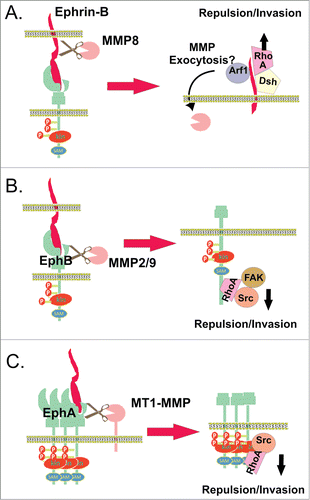
The EphB2 receptor has also been reported to be cleaved by MMPs in response to ligand stimulation in primary hippocampal neuron cultures.Citation103 EphB2-Fc fusion protein is cleaved in vitro by MMP9, and mutation of a potential MMP cleavage site in EphB2 inhibited cleavage, and prevented cell-cell repulsion upon interaction with ephrin-B2 stably expressing cells. EphB2 mediated growth cone collapse and withdrawal in hippocampal neurons was also significantly inhibited by mutation of the EphB2 MMP cleavage site and partially reduced by an MMP 2/9 inhibitor.Citation103 Interestingly, inhibition of MMP 2/9, or expression of the cleavage-resistant EphB2 mutant, reduced phosphorylation and recruitment of signaling proteins Src and FAK and activation of RhoA,Citation103 events previously described to promote ephrin-B/EphB-induced actin myosin contractility of the cytoskeleton ().Citation104 Interestingly, MMP 2/9 expression is elevated in EphB2 overexpressing breast cancer cells where MMP neutralising antibodies reduce EphB2 induced invasiveness,Citation105 suggesting a potential role in cancer invasion. In contrast, another study using fibroblasts and rat cortical neurons showed ligand-induced cleavage of EphB2 to be independent of metalloprotease activity, while ligand-independent cleavage was metalloprotease-dependent, probably via ADAM activity, since it was inhibited by the selective ADAM10 inhibitor GI254023X.Citation106 Both ligand and receptor cleavage events lead to subsequent intramembrane cleavage releasing the intracellular domain, described in more detail below.
The membrane type-I matrix metalloproteinase (MT1-MMP) also has a reported role in promoting cancer cell migration and invasion via cleaving the EphA2 receptor. Analysis of a panel of breast carcinoma cell lines shows co-overexpression of MT1-MMP with EphA2.Citation107 EphA2 clusters recruit MT1-MMP via a mechanism independent of its catalytic domain,Citation107 reminiscent of recruitment of MT1-MMP to CD44H, a major hyaluronan receptor that plays an important role in migration.Citation108 Subsequently, EphA2 is proteolytically processed by MT1-MMP, as MT1-MMP depletion and inhibition resulted in reduced EphA2 receptor cleavage. Mass spectrometry studies indicate MT1-MMP mediated cleavage of EphA2 receptor occurs at the FN type-III domain, likely in cis on the cell surface, as there was some constitutive shedding in the absence of added ligand, which was enhanced by soluble ephrin-A1. SiRNA knockdown of MT1-MMP and expression of EphA2-cleavage site mutant effectively reduced intracellular accumulation of EphA2, cell invasion and Src signaling indicating interconnection of these processes. Moreover, EphA2 association with RhoA lead to cell retraction, which decreased in the presence of inactive EphA2 and EphA2 MT1-MMP cleavage-site mutant.Citation107 Thus MT1-MMP/Src activity-dependent intracellular translocation of EphA2 receptor and cytoskeleton contractility via RhoA activation contribute to Eph/ephrin-mediated cell repulsion and invasion ().
Effects of Ephs on MMPs and cell migration/invasion
In addition to the above mechanisms describing direct Eph/ephrin cleavage, Eph signaling can regulate other MMP functions such as degradation of extracellular matrix facilitating cell migration and invasion. In gastric carcinoma cells, silencing of EphA2 expression inhibits cell proliferation, invasion and expression of MMP 9 in vitro and in vivo, while active EphA2 promotes MMP9 expression so that it can cleave type-IV collagen, a major scaffolding molecule in the basement membrane, assisting invasion of metastatic cancer cells.Citation109 Overexpression of EphA2 in pancreatic adenocarcinoma also increases MMP2 expression and FAK phosphorylation, which in turn leads to cellular invasion, although stimulation with recombinant ephrin-A1-Fc decreased cell invasion.Citation110 Similarly, MMP2 is down-regulated in ephrin-B2-Fc mediated EphB4 signaling in breast cancer cells, inactivating an oncogenic pathway involving Abl family tyrosine kinases and the Crk adaptor protein, and exerting a tumor suppressive effect by restraining cell motility and invasion.Citation111 In this context MMP2 is most likely to degrade extracellular matrix facilitating cell invasion, rather than directly shedding ephrinB ligands. Conversely, a previous study on endothelial cells demonstrated that EphB4/ephrin-B2 signaling mediates migration via the PI3K/Akt pathway, in which case MMP2/9 expression was increased and thought to facilitate migration by degrading collagen within the basement membrane.Citation112
Regulated intramembrane proteolysis
Regulated intramembrane proteolysis (RIP) describes a processing event that often follows ectodomain shedding in which an integral membrane protein is cleaved within its transmembrane domain to yield a soluble protein fragment.Citation113 The initial shedding, generally mediated by MMPs or ADAMs, or by the aspartyl proteases BACE1 and BACE2 (β-site APP cleaving enzymes),Citation114-116 is a prerequisite for the second cleavage.Citation117 The proteases that catalyze RIP are known as intramembrane cleaving proteases (iCLiPs), and are currently represented by 3 distinct and evolutionarily conserved families: the aspartyl protease-like (including presenilin-dependent γ-secretase, signal peptide peptidase, and signal peptide peptidase-like), the zinc metalloproteinase site-2 protease, and the serine protease rhomboid family (rhomboid and PARL). There are 2 types of RIP: type 1 RIP utilizes the aspartyl protease-like family enzyme (eg. γ-secretase) to processes proteins whose carboxy terminus is in the cytosol; whereas type 2 RIP employs a zinc metalloproteinase site-2 protease to process proteins with their N-terminus in the cytosol.Citation118
RIP is employed by a large number of membrane proteins, implicated in a wide range of biological processes. To date, over 60 substrates have been identified, including growth factors, cytokines, receptors, cell adhesion proteins, signal peptides and viral proteins. These substrates include Notch, EpCAM, N-cadherin, E-cadherin, and the p75 neurotrophin receptor.Citation114,119 The canonical Notch signaling pathway is a prime example of RIP, where ligand binding to the ectodomain of the Notch receptor on neighboring cells triggers sequential cleavage of the ectodomain and the transmembrane domain by metalloproteases and γ-secretase, respectively.Citation113 The released intracellular domain is then translocated to the nucleus where it regulates gene transcription by binding to transcription factors.Citation120 RTK signaling can also involve RIP, including the ErbB4 receptor.Citation121 There is some evidence suggesting Eph receptors/ligands undergo intramembrane processing, although sometimes atypical of standard, ligand-induced RIP.
γ-secretase
γ-secretase is a membrane-embedded proteolytic complex consisting of 4 subunits; Presenilin (PS), Nicastrin, Aph1, and Pen-2. Presenilin is endoproteolytically cleaved to form 2 fragments, one of which assembles with the other 3 subunits to make the γ-secretase complex, with Presenilin residing at the catalytic core of the protease complex.Citation122. Together this complex manages the intramembranous cleavage of transmembrane proteins such as APP and Notch, following ectodomain cleavage.Citation123
Ephrin-B2 was first identified to undergo RIP-like processing, where binding to EphB2 stimulates metalloprotease cleavage of ephrin-B2, yielding a membrane bound cytoplasmic fragment (CTF1) which is subsequently cleaved by PS1-dependent γ-secretases to produce a soluble CTF (CTF2) of ephrin-B2.Citation98 Thus PS1 Wt cells show less accumulation of membrane bound-ephrin-B2-CTF1 compared to PS1 knockout cells, and inhibiting γ-secretase accumulates membrane bound ephrin-B2 CTF. Stimulation of ephrin-B2 by EphB2 lead to detectable soluble ephrin-B2 CTF2 only in Wt fibroblasts, in which it also activated Src kinase, previously shown to mediate EphB-induced sprouting of endothelial cells, by inhibiting association with the inhibitory Csk kinase. This in turn facilitates phosphorylation of ephrin-B2 tyrosine residues.Citation98 A later study showed involvement of the Csk binding protein (PAG/Cbp), an adaptor protein that controls the activity of Src kinases.Citation124 Ephrin-B2-soluble CTF2 forms a complex with PAG/Cbp promoting Src activation,Citation124 which in other contexts has been shown to control various cellular events such as cell proliferation, survival, and migrationCitation125 most likely via remodelling of the actin cystoskeletonCitation126 ( ). Furthermore, PS1 mutants found in familial Alzheimer's disease (AD) inhibit ephrin-B2 CTF cleavage as well as activation of Src, suggesting a potential role in disease.Citation98 Indeed, Src kinases play an important role in neuronal function and degeneration since their loss results in neuronal defects, abnormalities in the hippocampus, and long-term potentiation (LTP) impairment.Citation127,128
Table 1. Summary of protease function in Eph/ephrin signaling
Figure 4. Regulated intramembrane proteolysis of Ephs/ephrins. (A) EphB/ephrin-B interaction can lead to sequential ectodomain and γ-secretase-mediated intramembrane proteolysis of both ligand and receptor in interacting cells. EphB2 bound ephrin-B2 ectodomain is cleaved by metalloprotease activity, generating a membrane bound cytoplasmic fragment (ephrin-B2-CTF1) which is further processed by γ-secretase, yielding a soluble cytoplasmic fragment (ephrin-B2-CTF2) (top cell). Inside the cell, ephrin-B2-CTF promotes tyrosine dephosphorylation of (PAG/Cbp), and relieves Csk-inhibition of Src kinases, which in turn facilitates tyrosine phosphorylation of ephrin-B2.Citation98,124 Ligand-binding to EphB2 also promotes receptor ectodomain cleavage, endocytosis, and γ-secretase cleavage, to produce soluble Eph cytoplasmic domain (bottom cell).Citation106 This regulated intramembrane proteolysis in both cells contributes to cytoskeletal regulation, migration, proliferation and survival. (B) EphA/B receptor ectodomain shedding also occurs in the apparent absence of ligand, via MMP/ADAM activity, stimulated by NMDA receptor signaling or calcim influx. The plasma membrane bound Eph cytoplasmic fragment (Eph-CTF) then undergoes γ-secretase cleavage. Both ligand-independent and -dependent EphB pathways may regulate NMDA receptor signaling and synaptic plasticity.Citation106 In addition, calcium-stimulated EphA4 shedding at synapses promotes dendritic spine formation via Rac signaling.Citation133
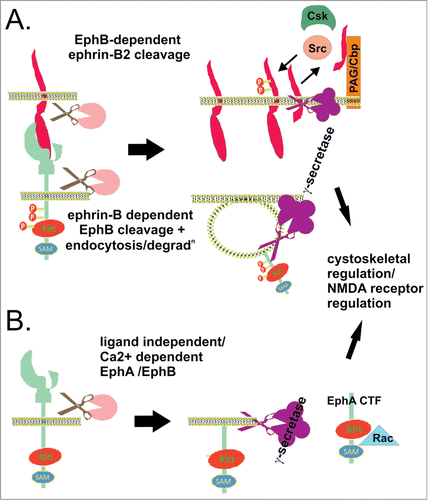
The same group also found the EphB2 receptor to be processed by γ-secretase, via both ligand dependent and independent pathways. Interestingly, during ligand induced signaling (), while the ephrin is sequentially processed by metalloprotease and γ-secretase (above), the receptor ectodomain cleavage occurs in endosomes, leads to receptor degradation, and is metalloprotease-independent. Cleavage was blocked by the BACE inhibitor peptide ZVLL but not other BACE inhibitors, so it is not clear what protease activity is responsible. In contrast, the ligand independent pathway () occurs in response to calcium influx and NMDA receptor activation, where broad metalloprotease and more selective ADAM10 inhibition prevents generation of the EphB2 CTF.Citation106 Analagous to ephrin-B2, brain extracts from PS1 knockout mice have increased accumulation of membrane-bound EphB2-CTF1, as do EphB2 transfected fibroblasts from PS1 -/- versus Wt mice, while the soluble EphB2 CTF2 is only detectable in Wt fibroblasts.Citation106 Thus ligand-independent and -dependent EphB2 signaling pathways involve distinct EphB2 ectodomain cleavage mechanisms, leading to cleaveage by γ-secretase in distinct compartmentsCitation106. Since neuronal ephrin-B/EphB interactions promote tyrosine phosphorylation of the NMDA receptor (NMDAR) and regulate NMDAR-dependent calcium influx and synaptic plasticity,Citation129 the authors suggest the EphB2-CTF may function in signaling cascades initiated both by EphB/ephrin-B binding or by activation of the NMDAR. Interestingly, unlike ephrin-B2, the effect of EphB2 CTF on NMDAR phosphorylation is apparently direct and independent of Src,Citation130 NMDA receptor signaling is also implicated in Alzheimer's disease (AD), and these potential functions may be impaired by familial AD mutations in PS1 that inhibit production of both ephrin-B and EphB-soluble CTFs.Citation131,132
EphA4 was also identified as a γ-secretase target, by mass spectrometry analysis of proteins associated with γ-secretase purified from synaptic lipid raft membranes.Citation133 Indeed, EphA4 also contains a lysine/arginine motif preferred by γ-secretase, and production of EphA4-soluble CTF was blocked by γ-secretase inhibitors or siRNA depletion. γ-secretase and EphA4 co-localized at the synaptic lipid raft membrane, where synaptic activity increased formation of EphA4-soluble CTF. Like EphB2, the intramembrane cleavage followed EphA4 ectodomain cleavage which was ligand independent, but stimulated by calcium influx, and dependent on metalloprotease activity (). In this situation, processing of EphA4 CTF by γ-secretase activates the Rac signaling pathway, contributing to the formation and maintenance of dentritic spines,Citation133 typical of a ligand-independent adhesive response.Citation8 Although soluble CTF was found in the nucleus, nuclear localization was not required for Rac signaling.Citation133 Importantly, this process is also affected by PS1 mutation found in familial AD, reducing EphA4 CTF cleavage, and inhibiting dentritic spine formation.Citation133 Interestingly, the occipital lobes of AD patients show significantly attenuated EphA4-soluble CTF levels and Rac1 signaling,Citation134 indicating likely physiological importance of γ-secretase EphA4 cleavage.
Serine Proteases
Members of the serine protease super family have a conserved serine residue in their active site. Originally they were recognized for the presence of the Asp−His−Ser “charge relay” system or “catalytic triad,”Citation135,136 although new catalytic triads and dyads have been discovered.Citation135 Serine proteases reside in a wide range of tissues and biological fluids and participate in diverse biological activities such as blood coagulation, wound healing, digestion and immune responses. In addition, deregulated activation of this protease family contributes to disease, including tumor growth, invasion and metastasis.Citation137
Rhomboids
Rhomboids are intramembrane serine proteases that have been reported throughout the eukaryotes. Three major groups of Rhomboids have been described: Rhomboid-like proteases (active rhomboids), iRhoms (inactive rhomboids) and other inactive rhomboid-like proteins.Citation138 Unlike most of the other serine proteases, Rhomboids use a catalytic dyad consisting of serine and histidine.Citation139 Drosophila Rhomboid-1 cleaves the EGF-like growth factor Spitz,Citation140 and similarly, human EGF is a substrate of the mammalian rhomboid RHBDL2, cleaving just outside its transmembrane domain, thereby facilitating the activation of the EGF receptor.Citation141 Increased activity of RHBDL2 in tumors overexpressing the EGF receptor implicates its involvement in RTK receptor signaling and tumor growth.Citation141
Based on a sequence motif at the luminal face of the membrane domain of Spitz, which determines its susceptibility to cleavage by RHBDL2, analysis of other type-I membrane proteins was performed to determine potential substrates. This identified ephrin-B1, with ephrin-B2 and B3 also potential candidates due to abundance of alanines or glycines in the luminal region, preferred by RHBDL2. Spitz/ephrin-B chimeric protein cleavage assays suggested that only ephrin-B3 is cleaved by RHBDL2, and overexpression of mammalian Rhomboid-1 or Rhomboid-2 together with ephrin-B3 resulted in efficient ephrin-B3 cleavage.Citation142 The authors suggest that RHBDL2 cleavage of ephrin-B3 may represent an additional mechanism of Eph/ephrin mediated cell signaling.Citation142
Neuropsin
Neuropsin is a sectreted, multi-domain serine protease that belongs to the kallikrein-related endopeptidase 8 (KLK8) family,Citation143 present in the limbic areas of the brain. Neuropsin exhibits the catalytic triad typical of other serine proteases and the overall structure is homologous to that of the chymotrypsin-type.Citation144 Enzymatically active neuropsin has important functions in maintaining synaptic plasticity, by establishing the early phase of long-term potentiation (LTP). Since neuropsin deficiency completely inhibits the early phase of LTP, associations of Neuropsin between early and persistent-LTP synapses are speculated to be related to mammalian working memory and consequently integration in learning and memory.Citation145
A recent report shows that neuropsin in the mouse brain is important for stress-related plasticity in the amygdala (a region involved in processing emotions).Citation146 Stress initiates neuropsin-dependent cleavage of EphB2 in the amygdala, resulting in dissociation of EphB2 from the NR1 subunit of the NMDA receptor, thus inducing membrane turnover of EphB2 receptors. Usually, EphB2–NR1 interaction increases NMDA receptor signals, inducing the expression of the glucocorticoid receptor sensitivity controlling protein Fkbp5, which in turn leads to behavioral signatures of anxiety. Neuropsin-deficient mice do not show EphB2 cleavage and EphB2 dissociation from the NR1 subunit under stress, resulting in static EphB2-NR1 interaction that attenuates production of Fkbp5 and anxiety. Anxiety can be restored by introduction of Neuropsin into the amygdala of Neuropsin deficient mice. In addition, anxiety levels of wild-type mice can be suppressed by silencing the Fkbp5 gene in the amygdale, or remarkably, by injection of anti-EphB2 antibodies.Citation146
Conclusion
It is clear from the reviewed literature that protease activity is becoming increasingly recognized as an important aspect in the regulation of Eph/ephrin signaling, beyond classical signal termination by proteosomal degradation. Thus different proteases contribute in diverse ways to facilitate both the opposing responses of cell-cell adhesion and retraction mediated by Eph/ephrin signaling. How these functions are regulated is only emerging, but it is clear that ligand-receptor interaction and ensuing bi-directional signaling are key to coordinate protease activity with changes in morphology and migration that follow Eph-driven cytoskeletal remodelling. ADAM10 is a prime example, where regulation likely occurs at multiple levels. Thus formation of a ligand-receptor complex across cell-cell junctions enables recognition by the ADAM10 substrate-binding domain, while the ensuing receptor activation likely leads to conformational changes in both Eph and ADAM that promote their association. This results in cleavage of ephrins and associated adhesion molecules from the opposing cell, breaking the tether between cells, while at the same time allowing internalisation of signaling complexes driving cytoskeletal collapse and cell retraction. These co-ordinated events are likely de-regulated in cancer and inflammatory diseases, where ADAMs and Ephs are often over-expressed, and high RTK and inflammatory signaling leads to oxidative conditions that favor protease activity. Similarly Eph/ephrin interaction appears to regulate their cleavage by MMPs, as well as cleavage of the extracellular matrix, although in the latter case ligand stimulation of Ephs can either promote or inhibit MMP-mediated cancer cell invasion. Lastly, ectodomain cleavage is commonly followed by intramembrane cleavage by gamma-secretase, releasing the cytoplasmic domain. This can occur in response to Eph/ephrin interaction, or to other stimuli, for example in neuronal cells where this function is implicated in Alzheimer's disease. Thus protease regulation of Eph/ephrin signaling occurs at multiple levels and has broad significance for disease, and as these mechanisms become better understood, targeted inhibition of specific protease activities may provide a useful approach for the development of new therapies to block aberrant Eph/ephrin function.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This review is dedicated to the memory of Martin Lackmann, an inspirational mentor and colleague who contributed much to the understanding of Eph/ephrin signaling, including key insights into the role and regulation of protease function.
References
- Serim S, Haedke U, Verhelst SHL. Activity-based probes for the study of proteases: recent advances and developments. ChemMedChem 2012; 7:1146-59; PMID:22431376; http://dx.doi.org/10.1002/cmdc.201200057
- Drag M, Salvesen GS. Emerging principles in protease-based drug discovery. Nat Rev Drug Discov 2010; 9:690-701; PMID:20811381; http://dx.doi.org/10.1038/nrd3053
- Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Dis 2006; 5:785-99; PMID:16955069; http://dx.doi.org/10.1038/nrd2092
- Lopez-Otin C, Bond JS. Proteases: multifunctional enzymes in life and disease. J Biol Chem 2008; 283:30433-7; PMID:18650443; http://dx.doi.org/10.1074/jbc.R800035200
- Overall CM, Blobel CP. In search of partners: linking extracellular proteases to substrates. Nat Rev Mol Cell Biol 2007; 8:245-57; PMID:17299501; http://dx.doi.org/10.1038/nrm2120
- Turk B, Turk D, Turk V. Protease signalling: the cutting edge. EMBO J 2012; 31:1630-43; PMID:22367392; http://dx.doi.org/10.1038/emboj.2012.42
- Adrain C, Freeman M. Regulation of receptor tyrosine kinase ligand processing. Cold Spring Harb Perspect Biol 2014; 6:pii: a008995; PMID:24384567; http://dx.doi.org/10.1101/cshperspect.a008995
- Nievergall E, Lackmann M, Janes PW. Eph-dependent cell-cell adhesion and segregation in development and cancer. Cell Mol Life Sci 2012; 69:1813-42; PMID:22204021; http://dx.doi.org/10.1007/s00018-011-0900-6
- Marston DJ, Dickinson S, Nobes CD. Rac-dependent trans-endocytosis of ephrinBs regulates Eph-ephrin contact repulsion. Nat Cell Biol 2003; 5:879-88; PMID:12973357; http://dx.doi.org/10.1038/ncb1044
- Klein T, Bischoff R. Active metalloproteases of the A Disintegrin and Metalloprotease (ADAM) family: biological function and structure. J Proteome Res 2011; 10:17-33; PMID:20849079; http://dx.doi.org/10.1021/pr100556z
- Wolfsberg TG, White JM. 207–ADAM metalloproteinases. In: Barrett AJ, Rawlings ND, Woessner JF, eds. Handbook of Proteolytic Enzymes. Second Edition. London: Academic Press, 2004:709-14
- Weber S, Saftig P. Ectodomain shedding and ADAMs in development. Development (Cambridge, England) 2012; 139:3693-709; PMID:22991436; http://dx.doi.org/10.1242/dev.076398
- Reiss K, Saftig P. The “a disintegrin and metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin Cell Dev Biol 2009; 20:126-37; PMID:19049889; http://dx.doi.org/10.1016/j.semcdb.2008.11.002
- Tousseyn T, Jorissen E, Reiss K, Hartmann D. (Make) stick and cut loose–disintegrin metalloproteases in development and disease. Birth Defects Res C Embryo Today 2006; 78:24-46; PMID:16622847; http://dx.doi.org/10.1002/bdrc.20066
- Kurohara K, Komatsu K, Kurisaki T, Masuda A, Irie N, Asano M, Sudo K, Nabeshima Y, Iwakura Y, Sehara-Fujisawa A. Essential roles of Meltrin beta (ADAM19) in heart development. Dev Biol 2004; 267:14-28; PMID:14975714; http://dx.doi.org/10.1016/j.ydbio.2003.10.021
- Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer 2008; 8:929-41; PMID:19005493; http://dx.doi.org/10.1038/nrc2459
- Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol 2005; 6:32-43; PMID:15688065; http://dx.doi.org/10.1038/nrm1548
- Loechel F, Overgaard MT, Oxvig C, Albrechtsen R, Wewer UM. Regulation of human ADAM 12 protease by the prodomain. Evidence for a functional cysteine switch. J Biol Chem 1999; 274:13427-33; PMID:10224107; http://dx.doi.org/10.1074/jbc.274.19.13427
- Anders A, Gilbert S, Garten W, Postina R, Fahrenholz F. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J 2001; 15:1837-9; PMID:11481247
- Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev 2003; 17:7-30; PMID:12514095; http://dx.doi.org/10.1101/gad.1039703
- Fahrenholz F, Gilbert S, Kojro E, Lammich S, Postina R. Alpha-secretase activity of the disintegrin metalloprotease ADAM 10. Influences of domain structure. Ann N Y Acad Sci 2000; 920:215-22; PMID:11193153; http://dx.doi.org/10.1111/j.1749-6632.2000.tb06925.x
- Howard L, Maciewicz RA, Blobel CP. Cloning and characterization of ADAM28: evidence for autocatalytic pro-domain removal and for cell surface localization of mature ADAM28. Biochem J 2000; 348 Pt 1:21-7; PMID:10794709; http://dx.doi.org/10.1042/0264-6021:3480021
- Roghani M, Becherer JD, Moss ML, Atherton RE, Erdjument-Bromage H, Arribas J, Blackburn RK, Weskamp G, Tempst P, Blobel CP. Metalloprotease-disintegrin MDC9: intracellular maturation and catalytic activity. J Biol Chem 1999; 274:3531-40; PMID:9920899; http://dx.doi.org/10.1074/jbc.274.6.3531
- Lum L, Reid MS, Blobel CP. Intracellular maturation of the mouse metalloprotease disintegrin MDC15. J Biol Chem 1998; 273:26236-47; PMID:9748307; http://dx.doi.org/10.1074/jbc.273.40.26236
- Laisney JAGC, Mueller TD, Schartl M, Meierjohann S. Hyperactivation of constitutively dimerized oncogenic EGF receptors by autocrine loops. Oncogene 2013; 32:2403-11; PMID:22751127; http://dx.doi.org/10.1038/onc.2012.267
- Moss ML, Bomar M, Liu Q, Sage H, Dempsey P, Lenhart PM, Gillispie PA, Stoeck A, Wildeboer D, Bartsch JW, et al. The ADAM10 prodomain is a specific inhibitor of ADAM10 proteolytic activity and inhibits cellular shedding events. J Biol Chem 2007; 282:35712-21; PMID:17895248; http://dx.doi.org/10.1074/jbc.M703231200
- Loechel F, Gilpin BJ, Engvall E, Albrechtsen R, Wewer UM. Human ADAM 12 (meltrin alpha) is an active metalloprotease. J Biol Chem 1998; 273:16993-7; PMID:9642263; http://dx.doi.org/10.1074/jbc.273.27.16993
- Saftig P, Reiss K. The “A Disintegrin And Metalloproteases” ADAM10 and ADAM17: novel drug targets with therapeutic potential? Eur J Cell Biol 2011; 90:527-35; PMID:21194787; http://dx.doi.org/10.1016/j.ejcb.2010.11.005
- White JM. ADAMs: modulators of cell–cell and cell–matrix interactions. Curr Opin Cell Biol 2003; 15:598-606; PMID:14519395; http://dx.doi.org/10.1016/j.ceb.2003.08.001
- Caescu CI, Jeschke GR, Turk BE. Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem J 2009; 424:79-88; PMID:19715556; http://dx.doi.org/10.1042/BJ20090549
- Moss ML, Lambert MH. Shedding of membrane proteins by ADAM family proteases. Essays Biochem 2002; 38:141-53; PMID:12463167
- Smith KM, Gaultier A, Cousin H, Alfandari D, White JM, DeSimone DW. The cysteine-rich domain regulates ADAM protease function in vivo. J Cell Biol 2002; 159:893-902; PMID:12460986; http://dx.doi.org/10.1083/jcb.200206023
- Alfandari D. ADAM13 Function in Development. In: Hooper N, Lendeckel U, eds. The ADAM Family of Proteases: Springer US, 2005:147-69
- Reddy P, Slack JL, Davis R, Cerretti DP, Kozlosky CJ, Blanton RA, Shows D, Peschon JJ, Black RA. Functional analysis of the domain structure of tumor necrosis factor-α converting enzyme. J Biol Chem 2000; 275:14608-14; PMID:10799547; http://dx.doi.org/10.1074/jbc.275.19.14608
- Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E, Blobel CP, Himanen JP, Lackmann M, Nikolov DB. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 2005; 123:291-304; PMID:16239146; http://dx.doi.org/10.1016/j.cell.2005.08.014
- Maskos K, Fernandez-Catalan C, Huber R, Bourenkov GP, Bartunik H, Ellestad GA, Reddy P, Wolfson MF, Rauch CT, Castner BJ, et al. Crystal structure of the catalytic domain of human tumor necrosis factor-α-converting enzyme. Proc Natl Acad Sci U S A 1998; 95:3408-12; PMID:9520379; http://dx.doi.org/10.1073/pnas.95.7.3408
- Takeda S, Igarashi T, Mori H, Araki S. Crystal structures of VAP1 reveal ADAMs’ MDC domain architecture and its unique C-shaped scaffold. EMBO J 2006; 25:2388-96; PMID:16688218; http://dx.doi.org/10.1038/sj.emboj.7601131
- Kullander K, Klein R. Mechanisms and functions of eph and ephrin signalling. Nat Rev Mol Cell Biol 2002; 3:475-86; PMID:12094214; http://dx.doi.org/10.1038/nrm856
- Hattori M, Osterfield M, Flanagan JG. Regulated cleavage of a contact-mediated axon repellent. Science 2000; 289:1360-5; PMID:10958785; http://dx.doi.org/10.1126/science.289.5483.1360
- Sahin U, Weskamp G, Kelly K, Zhou H-M, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol 2004; 164:769-79; PMID:14993236; http://dx.doi.org/10.1083/jcb.200307137
- Luo X, Prior M, He W, Hu X, Tang X, Shen W, Yadav S, Kiryu-Seo S, Miller R, Trapp s, et al. Cleavage of neuregulin-1 by BACE1 or ADAM10 protein produces differential effects on myelination. J Biol Chem 2011; 286:23967-74; PMID:21576249; http://dx.doi.org/10.1074/jbc.M111.251538
- Pruessmeyer J, Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol 2009; 20:164-74; PMID:18951988; http://dx.doi.org/10.1016/j.semcdb.2008.09.005
- Abel S, Hundhausen C, Mentlein R, Schulte A, Berkhout TA, Broadway N, Hartmann D, Sedlacek R, Dietrich S, Muetze B, et al. The transmembrane CXC-chemokine ligand 16 is induced by IFN-gamma and TNF-alpha and shed by the activity of the disintegrin-like metalloproteinase ADAM10. J Immunol 2004; 172:6362-72; PMID:15128827; http://dx.doi.org/10.4049/jimmunol.172.10.6362
- Hundhausen C, Misztela D, Berkhout TA, Broadway N, Saftig P, Reiss K, Hartmann D, Fahrenholz F, Postina R, Matthews V, et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood 2003; 102:1186-95; PMID:12714508; http://dx.doi.org/10.1182/blood-2002-12-3775
- Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, de Strooper B, Hartmann D, Saftig P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and β-catenin translocation. Proc Natl Acad Sci U S A 2005; 102:9182-7; PMID:15958533; http://dx.doi.org/10.1073/pnas.0500918102
- Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell–cell adhesion and β-catenin nuclear signalling. EMBO J 2005; 24:742–52.
- Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Gast D, Joumaa S, Zentgraf H, Fogel M, Altevogt DP. ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J 2003; 17:292-4; PMID:12475894
- Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Rossner S, Lichtenthaler SF. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J 2010; 29:3020-32; PMID:20676056; http://dx.doi.org/10.1038/emboj.2010.167
- Vincent B, Paitel E, Saftig P, Frobert Y, Hartmann D, De Strooper B, Grassi J, Lopez-Perez E, Checler F. The disintegrins ADAM10 and TACE contribute to the constitutive and phorbol ester-regulated normal cleavage of the cellular prion protein. J Biol Chem 2001; 276:37743-6; PMID:11477090; http://dx.doi.org/10.1074/jbc.M003965200
- Ludwig A, Hundhausen C, Lambert MH, Broadway N, Andrews RC, Bickett DM, Leesnitzer MA, Becherer JD. Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Comb Chem High Throughput Screen 2005; 8:161-71; PMID:15777180; http://dx.doi.org/10.2174/1386207053258488
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 2006; 7:678-89; PMID:16921404; http://dx.doi.org/10.1038/nrm2009
- D’Souza B, Miyamoto A, Weinmaster G. The many facets of Notch ligands. Oncogene 2008; 27:5148-67; http://dx.doi.org/10.1038/onc.2008.229
- Zolkiewska A. ADAM proteases: ligand processing and modulation of the Notch pathway. Cell Mol Life Sci 2008; 65:2056-68; PMID:18344021; http://dx.doi.org/10.1007/s00018-008-7586-4
- Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lübke T, Lena Illert A, von Figura K, et al. The disintegrinmetalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet 2002; 11:2615-24; PMID:12354787; http://dx.doi.org/10.1093/hmg/11.21.2615
- Mancia F, Shapiro L. ADAM and Eph: how Ephrin-signaling cells become detached. Cell 2005; 123:185-7; PMID:16239135; http://dx.doi.org/10.1016/j.cell.2005.10.004
- Salaita K, Nair PM, Petit RS, Neve RM, Das D, Gray JW, Groves JT. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science 2010; 327:1380-5; PMID:20223987; http://dx.doi.org/10.1126/science.1181729
- Atapattu L, Saha N, Llerena C, Vail ME, Scott AM, Nikolov DB, Lackmann M, Janes PW. Antibodies binding the ADAM10 substrate recognition domain inhibit Eph function. J Cell Sci 2012; 125:6084-93; PMID:23108669; http://dx.doi.org/10.1242/jcs.112631
- Janes PW, Wimmer-Kleikamp SH, Frangakis AS, Treble K, Griesshaber B, Sabet O, Grabenbauer M, Ting AY, Saftig P, Bastiaens PI, et al. Cytoplasmic relaxation of active Eph controls ephrin shedding by ADAM10. PLoS Biol 2009; 7:e1000215; PMID:19823572; http://dx.doi.org/10.1371/journal.pbio.1000215
- Wybenga-Groot LE, Baskin B, Ong SH, Tong J, Pawson T, Sicheri F. Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region. Cell 2001; 106:745-57; PMID:11572780; http://dx.doi.org/10.1016/S0092-8674(01)00496-2
- Wang Y, Herrera AH, Li Y, Belani KK, Walcheck B. Regulation of mature ADAM17 by redox agents for L-selectin shedding. J Immunol 2009; 182:2449-57; PMID:19201900; http://dx.doi.org/10.4049/jimmunol.0802770
- Willems SH, Tape CJ, Stanley PL, Taylor NA, Mills IG, Neal DE, McCafferty J, Murphy G, et al. Thiol isomerases negatively regulate the cellular shedding activity of ADAM17. Biochem J 2010; 428:439-50; PMID:20345372; http://dx.doi.org/10.1042/BJ20100179
- Dusterhoft S, Jung S, Hung CW, Tholey A, Sonnichsen FD, Grotzinger J, Lorenzen I. Membrane-proximal domain of a disintegrin and metalloprotease-17 represents the putative molecular switch of its shedding activity operated by protein-disulfide isomerase. J Am Chem Soc 2013; 135:5776-81; PMID:23521534; http://dx.doi.org/10.1021/ja400340u
- Chiarugi P, Cirri P. Redox regulation of protein tyrosine phosphatases during receptor tyrosine kinase signal transduction. Trends Biochem Sci 2003; 28:509-14; PMID:13678963; http://dx.doi.org/10.1016/S0968-0004(03)00174-9
- Chiarugi P, Buricchi F. Protein tyrosine phosphorylation and reversible oxidation: two cross-talking posttranslation modifications. Antioxid Redox Signal 2007; 9:1-24; PMID:17115885; http://dx.doi.org/10.1089/ars.2007.9.1
- van Kempen LCL, de Visser KE, Coussens LM. Inflammation, proteases and cancer. Eur J Cancer 2006; 42:728-34; PMID:16524717; http://dx.doi.org/10.1016/j.ejca.2006.01.004
- Hartmann M, Herrlich A, Herrlich P. Who decides when to cleave an ectodomain? Trends Biochem Sci 2013; 38:111-20; PMID:23298902; http://dx.doi.org/10.1016/j.tibs.2012.12.002
- Ji YJ, Hwang YS, Mood K, Cho HJ, Lee HS, Winterbottom E, Cousin H, Daar IO. EphrinB2 affects apical constriction in Xenopus embryos and is regulated by ADAM10 and flotillin-1. Nat Commun 2014; 5:3516; PMID:24662724
- Gauthier LR, Robbins SM. Ephrin signaling: One raft to rule them all? One raft to sort them? One raft to spread their call and in signaling bind them? Life Sci 2003; 74:207-16; PMID:14607248; http://dx.doi.org/10.1016/j.lfs.2003.09.029
- Marquardt T, Shirasaki R, Ghosh S, Andrews SE, Carter N, Hunter T, Pfaff SL. Coexpressed EphA receptors and Ephrin-A ligands mediate opposing actions on growth cone navigation from distinct membrane domains. Cell 2005; 121:127-39; PMID:15820684; http://dx.doi.org/10.1016/j.cell.2005.01.020
- Asakura M, Kitakaze M, Takashima S, Liao Y, Ishikura F, Yoshinaka T, Ohmoto H, Node K, Yoshino K, Ishiguro H, et al. Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: metalloproteinase inhibitors as a new therapy. Nat Med 2002; 8:35-40; PMID:11786904; http://dx.doi.org/10.1038/nm0102-35
- Roy R, Wewer UM, Zurakowski D, Pories SE, Moses MA. ADAM 12 cleaves extracellular matrix proteins and correlates with cancer status and stage. J Biol Chem 2004; 279:51323-30; PMID:15381692; http://dx.doi.org/10.1074/jbc.M409565200
- Kodama T, Ikeda E, Okada A, Ohtsuka T, Shimoda M, Shiomi T, Yoshida K, Nakada M, Ohuchi E, Okada Y. ADAM12 is selectively overexpressed in human glioblastomas and is associated with glioblastoma cell proliferation and shedding of heparin-binding epidermal growth factor. Am J Pathol 2004; 165:1743-53; PMID:15509542; http://dx.doi.org/10.1016/S0002-9440(10)63429-3
- Ieguchi K, Tomita T, Omori T, Komatsu A, Deguchi A, Masuda J, Duffy SL, Coulthard MG, Boyd A, Maru Y. ADAM12-cleaved ephrin-A1 contributes to lung metastasis. Oncogene 2014; 33:2179-90; PMID:23686306; http://dx.doi.org/10.1038/onc.2013.180
- Frohlich C, Nehammer C, Albrechtsen R, Kronqvist P, Kveiborg M, Sehara-Fujisawa A, Mercurio AM, Wewer UM. ADAM12 produced by tumor cells rather than stromal cells accelerates breast tumor progression. Mol Cancer Res 2011; 9:1449-61; PMID:21875931; http://dx.doi.org/10.1158/1541-7786.MCR-11-0100
- Wei S, Xu G, Bridges LC, Williams P, White JM, DeSimone DW. ADAM13 induces cranial neural crest by cleaving class B Ephrins and regulating Wnt signaling. Dev Cell 2010; 19:345-52; PMID:20708595; http://dx.doi.org/10.1016/j.devcel.2010.07.012
- Gaultier A, Cousin H, Darribere T, Alfandari D. ADAM13 disintegrin and cysteine-rich domains bind to the second heparin-binding domain of fibronectin. J Biol Chem 2002; 277:23336-44; PMID:11967265; http://dx.doi.org/10.1074/jbc.M201792200
- Alfandari D, Cousin H, Gaultier A, Smith K, White JM, Darribere T, DeSimone DW. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr Biol 2001; 11:918-30; PMID:11448768; http://dx.doi.org/10.1016/S0960-9822(01)00263-9
- McCusker C, Cousin H, Neuner R, Alfandari D. Extracellular cleavage of cadherin-11 by ADAM metalloproteases is essential for Xenopus cranial neural crest cell migration. Mol Biol Cell 2009; 20:78-89; PMID:18946084; http://dx.doi.org/10.1091/mbc.E08-05-0535
- Cousin H, Abbruzzese G, Kerdavid E, Gaultier A, Alfandari D. Translocation of the cytoplasmic domain of ADAM13 to the nucleus is essential for Calpain8-a expression and cranial neural crest cell migration. Dev Cell 2011; 20:256-63; PMID:21316592; http://dx.doi.org/10.1016/j.devcel.2010.12.009
- Krull CE, Lansford R, Gale NW, Collazo A, Marcelle C, Yancopoulos GD, Fraser SE, Bronner-Fraser M. Interactions of Eph-related receptors and ligands confer rostrocaudal pattern to trunk neural crest migration. Cu Biol 1997; 7:571-80; PMID:9259560; http://dx.doi.org/10.1016/S0960-9822(06)00256-9
- Qi B, Newcomer RG, Sang Q-XA. ADAM19Adamalysin 19 structure, function, and role as a putative target in tumors and inflammatory diseases. Curr Pharm Des 2009; 15:2336-48; PMID:19601835; http://dx.doi.org/10.2174/138161209788682352
- Yumoto N, Wakatsuki S, Kurisaki T, Hara Y, Osumi N, Frisen J, Sehara-Fujisawa A. Meltrin betaADAM19 interacting with EphA4 in developing neural cells participates in formation of the neuromuscular junction. PloS One 2008; 3:e3322; PMID:18830404; http://dx.doi.org/10.1371/journal.pone.0003322
- Solanas G, Cortina C, Sevillano M, Batlle E. Cleavage of E-cadherin by ADAM10 mediates epithelial cell sorting downstream of EphB signalling. Nat Cell Biol 2011; 13:1100-7; PMID:21804545; http://dx.doi.org/10.1038/ncb2298
- Brennaman LH, Zhang X, Guan H, Triplett JW, Brown A, Demyanenko GP, Manis PB, Landmesser L, Maness PF. Polysialylated NCAM and EphrinAEphA regulate synaptic development of GABAergic interneurons in prefrontal cortex. Cereb Cortex 2013; 23:162-77; PMID:22275477; http://dx.doi.org/10.1093/cercor/bhr392
- Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphBephrinB. Cell 2002; 111:251-63; PMID:12408869; http://dx.doi.org/10.1016/S0092-8674(02)01015-2
- Cortina C, Palomo-Ponce S, Iglesias M, Fernandez-Masip JL, Vivancos A, Whissell G, Humà M, Peiró N, Gallego L, Jonkheer S, et al. EphB-ephrin-B interactions suppress colorectal cancer progression by compartmentalizing tumor cells. Nat Genet 2007; 39:1376-83; PMID:17906625; http://dx.doi.org/10.1038/ng.2007.11
- Chen Y, Fu AKY, Ip NY. Eph receptors at synapses: implications in neurodegenerative diseases. Cell Signal 2012; 24:606-11; PMID:22120527; http://dx.doi.org/10.1016/j.cellsig.2011.11.016
- Brennaman LH, Moss ML, Maness PF. EphrinAEphA-induced ectodomain shedding of neural cell adhesion molecule regulates growth cone repulsion through ADAM10 metalloprotease. J Neurochem 2014; 128:267-79; PMID:24117969; http://dx.doi.org/10.1111/jnc.12468
- Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol 2007; 8:221-33; PMID:17318226; http://dx.doi.org/10.1038/nrm2125
- Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol 2004; 4:617-29; PMID:15286728; http://dx.doi.org/10.1038/nri1418
- Streuli C. Extracellular matrix remodelling and cellular differentiation. Curr Opin Cell Biol 1999; 11:634-40; PMID:10508658; http://dx.doi.org/10.1016/S0955-0674(99)00026-5
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, et al. An essential role for ectodomain shedding in mammalian development. Science 1998; 282:1281-4; PMID:9812885; http://dx.doi.org/10.1126/science.282.5392.1281
- Palmisano R, Itoh Y. Analysis of MMP-Dependent Cell Migration and Invasion. Meth Molec Bio 2010; 622:379-92.
- Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 2010; 141:52-67; PMID:20371345; http://dx.doi.org/10.1016/j.cell.2010.03.015
- Beauchamp A, Lively MO, Mintz A, Gibo D, Wykosky J, Debinski W. EphrinA1 is released in three forms from cancer cells by matrix metalloproteases. Mol Cell Biol 2012; 32:3253-64; PMID:22688511; http://dx.doi.org/10.1128/MCB.06791-11
- Parri M, Taddei ML, Bianchini F, Calorini L, Chiarugi P. EphA2 reexpression prompts invasion of melanoma cells shifting from mesenchymal to amoeboid-like motility style. Cancer Res 2009; 69:2072-81; PMID:19244130; http://dx.doi.org/10.1158/0008-5472.CAN-08-1845
- Kim ES, Kim MS, Moon A. TGF-beta-induced upregulation of MMP-2 and MMP-9 depends on p38 MAPK, but not ERK signaling in MCF10A human breast epithelial cells. Int J Oncol 2004; 25:1375-82; PMID:15492828
- Georgakopoulos A, Litterst C, Ghersi E, Baki L, Xu C, Serban G, Robakis NK. MetalloproteinasePresenilin1 processing of ephrinB regulates EphB-induced Src phosphorylation and signaling. EMBO J 2006; 25:1242-52; PMID:16511561; http://dx.doi.org/10.1038/sj.emboj.7601031
- Tanaka M, Sasaki K, Kamata R, Sakai R. The C-terminus of ephrin-B1 regulates metalloproteinase secretion and invasion of cancer cells. J Cell Sci 2007; 120:2179-89; PMID:17567680; http://dx.doi.org/10.1242/jcs.008607
- Tanaka M, Kamata R, Yanagihara K, Sakai R. Suppression of gastric cancer dissemination by ephrin-B1-derived peptide. Cancer Sci 2010; 101:87-93; PMID:19804421; http://dx.doi.org/10.1111/j.1349-7006.2009.01352.x
- Lee HS, Bong YS, Moore KB, Soria K, Moody SA, Daar IO. Dishevelled mediates ephrinB1 signalling in the eye field through the planar cell polarity pathway. Nat Cell Biol 2006; 8:55-63; PMID:16362052; http://dx.doi.org/10.1038/ncb1344
- Ogawa K, Wada H, Okada N, Harada I, Nakajima T, Pasquale EB, Tsuyama S. EphB2 and ephrin-B1 expressed in the adult kidney regulate the cytoarchitecture of medullary tubule cells through Rho family GTPases. J Cell Sci 2006; 119:559-70; PMID:16443753; http://dx.doi.org/10.1242/jcs.02777
- Lin KT, Sloniowski S, Ethell DW, Ethell IM. Ephrin-B2-induced cleavage of EphB2 receptor is mediated by matrix metalloproteinases to trigger cell repulsion. J Biol Chem 2008; 283:28969-79; PMID:18713744; http://dx.doi.org/10.1074/jbc.M804401200
- Moeller ML, Shi Y, Reichardt LF, Ethell IM. EphB receptors regulate dendritic spine morphogenesis through the recruitmentphosphorylation of focal adhesion kinase and RhoA activation. J Biol Chem 2006; 281:1587-98; PMID:16298995; http://dx.doi.org/10.1074/jbc.M511756200
- Chukkapalli S, Amessou M, Dilly AK, Dekhil H, Zhao J, Liu Q, Bejna A, Thomas RD, Bandyopadhyay S, Bismar TA, et al. Role of the EphB2 receptor in autophagy, apoptosis and invasion in human breast cancer cells. Exp Cell Res 2014; 320:233-46; PMID:24211352; http://dx.doi.org/10.1016/j.yexcr.2013.10.022
- Litterst C, Georgakopoulos A, Shioi J, Ghersi E, Wisniewski T, Wang R, Ludwig A, Robakis NK. Ligand binding and calcium influx induce distinct ectodomaingamma-secretase-processing pathways of EphB2 receptor. JBiol Chem 2007; 282:16155-63; PMID:17428795; http://dx.doi.org/10.1074/jbc.M611449200
- Sugiyama N, Gucciardo E, Tatti O, Varjosalo M, Hyytiainen M, Gstaiger M, Gstaiger M, Lehti K. EphA2 cleavage by MT1-MMP triggers single cancer cell invasion via homotypic cell repulsion. J Cell Biol 2013; 201:467-84; PMID:23629968; http://dx.doi.org/10.1083/jcb.201205176
- Suenaga N, Mori H, Itoh Y, Seiki M. CD44 binding through the hemopexin-like domain is critical for its shedding by membrane-type 1 matrix metalloproteinase. Oncogene 2005; 24:859-68; PMID:15558018; http://dx.doi.org/10.1038/sj.onc.1208258
- Yuan W, Chen Z, Chen Z, Wu S, Guo J, Ge J, Yang P, Huang J. Silencing of EphA2 inhibits invasion of human gastric cancer SGC-7901 cells in vitro and in vivo. Neoplasma 2012; 59:105-13; PMID:22103904; http://dx.doi.org/10.4149/neo_2012_014
- Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. Ligation of EphA2 by Ephrin A1-Fc inhibits pancreatic adenocarcinoma cellular invasiveness. Biochem Biophys Res Commun 2004; 320:1096-102; PMID:15249202; http://dx.doi.org/10.1016/j.bbrc.2004.06.054
- Noren NK, Foos G, Hauser CA, Pasquale EB. The EphB4 receptor suppresses breast cancer cell tumorigenicity through an Abl-Crk pathway. Nat Cell Biol 2006; 8:815-25; PMID:16862147; http://dx.doi.org/10.1038/ncb1438
- Steinle JJ, Meininger CJ, Forough R, Wu G, Wu MH, Granger HJ. Eph B4 receptor signaling mediates endothelial cell migration and proliferation via the phosphatidylinositol 3-kinase pathway. J Biol Chem 2002; 277:43830-5; PMID:12235151; http://dx.doi.org/10.1074/jbc.M207221200
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 2000; 100:391-8; PMID:10693756; http://dx.doi.org/10.1016/S0092-8674(00)80675-3
- Lichtenthaler SF, Haass C, Steiner H. Regulated intramembrane proteolysis - lessons from amyloid precursor protein processing. J Neurochem 2011; 117:779-96; PMID:21413990; http://dx.doi.org/10.1111/j.1471-4159.2011.07248.x
- Murphy G, Murthy A, Khokha R. Clipping, shedding and RIPping keep immunity on cue. Trends Immunol 2008; 29:75-82; PMID:18182322; http://dx.doi.org/10.1016/j.it.2007.10.009
- Prox J, Rittger A, Saftig P. Physiological functions of the amyloid precursor protein secretases ADAM10, BACE1, and Presenilin. Exp Brain Res 2012; 217:331-41; PMID:22120156; http://dx.doi.org/10.1007/s00221-011-2952-0
- Strooper BD, Annaert W. Presenilins and the intramembrane proteolysis of proteins: facts and fiction. Nat Cell Bio 2001; 3:E221-5.
- Morohashi Y, Tomita T. Protein trafficking and maturation regulate intramembrane proteolysis. Biochim Biophys Acta 2013; 1828:2855-61; PMID:23770323; http://dx.doi.org/10.1016/j.bbamem.2013.06.001
- Rawson RB. Regulated intramembrane proteolysis: from the endoplasmic reticulum to the nucleus. Essays Biochem 2002; 38:155-68; PMID:12463168
- Nakayama K, Nagase H, Koh C-S, Ohkawara T. γ-secretase-regulated signaling: notch, APP, and Alzheimer's disease. Curr Psychopharmacol 2012; 1:155-66; http://dx.doi.org/10.2174/2211556011201020155
- Ni C-Y, Murphy MP, Golde TE, Carpenter G. γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 2001; 294:2179-81; PMID:11679632; http://dx.doi.org/10.1126/science.1065412
- Wolfe MS. Structure, mechanism and inhibition of gamma-secretase and presenilin-like proteases. Biol Chem 2010; 391:839-47; PMID:20482315; http://dx.doi.org/10.1515/bc.2010.086
- Bergmans BA, De Strooper B. γ-secretases: from cell biology to therapeutic strategies. Lancet Neurol 2010; 9:215-26; PMID:20129170; http://dx.doi.org/10.1016/S1474-4422(09)70332-1
- Georgakopoulos A, Xu J, Xu C, Mauger G, Barthet G, Robakis NK. Presenilin1gamma-secretase promotes the EphB2-induced phosphorylation of ephrinB2 by regulating phosphoprotein associated with glycosphingolipid-enriched microdomainsCsk binding protein. FASEB J 2011; 25:3594-604; PMID:21746865; http://dx.doi.org/10.1096/fj.11-187856
- Playford MP, Schaller MD. The interplay between Src and integrins in normal and tumor biology. Oncogene 2004; 23:7928-46; PMID:15489911; http://dx.doi.org/10.1038/sj.onc.1208080
- Kanou T, Oneyama C, Kawahara K, Okimura A, Ohta M, Ikeda N, Shintani Y, Okumura M, Okada M. The transmembrane adaptor CbpPAG1 controls the malignant potential of human non–small celllung cancers that have c-Src upregulation. Mol Cancer Res 2011; 9:103-14; PMID:21156787; http://dx.doi.org/10.1158/1541-7786.MCR-10-0340
- Grant SG, O’Dell TJ, Karl KA, Stein PL, Soriano P, Kandel ER. Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice. Science 1992; 258:1903-10; PMID:1361685; http://dx.doi.org/10.1126/science.1361685
- Lu YM, Roder JC, Davidow J, Salter MW. Src activation in the tnduction of long-term potentiation in CA1 hippocampal neurons. Science 1998; 279:1363-8; PMID:9478899; http://dx.doi.org/10.1126/science.279.5355.1363
- Henderson JT, Georgiou J, Jia Z, Robertson J, Elowe S, Roder JC, Pawson T. The receptor tyrosine kinase EphB2 regulates NMDA-dependent synaptic function. Neuron 2001; 32:1041-56; PMID:11754836; http://dx.doi.org/10.1016/S0896-6273(01)00553-0
- Ionides EL, Fang KS, Rivkah Isseroff R, Oster GF. Stochastic models for cell motion and taxis. J Math Biol 2004; 48:23-37; PMID:14685770; http://dx.doi.org/10.1007/s00285-003-0220-z
- Danysz W, Parsons CG. Alzheimer's disease, beta-amyloid, glutamate, NMDA receptors and memantine–searching for the connections. Br J Pharmacol 2012; 167:324-52; PMID:22646481; http://dx.doi.org/10.1111/j.1476-5381.2012.02057.x
- Snyder EM, Nong Y, Almeida CG, Paul S, Moran T, Choi EY, Nairn AC, Salter MW, Lombroso PJ, Gouras GK. Regulation of NMDA receptor trafficking by amyloid-beta. Nat Neurosci 2005; 8:1051-8; PMID:16025111; http://dx.doi.org/10.1038/nn1503
- Inoue E, Deguchi-Tawarada M, Togawa A, Matsui C, Arita K, Katahira-Tayama S, Sato T, Yamauchi E, Oda Y, Takai Y. Synaptic activity prompts gamma-secretase-mediated cleavage of EphA4 and dendritic spine formation. J Cell Biol 2009; 185:551-64; PMID:19414612; http://dx.doi.org/10.1083/jcb.200809151
- Matsui C, Inoue E, Kakita A, Arita K, Deguchi-Tawarada M, Togawa A, Yamada A, Takai Y, Takahashi H. Involvement of the gamma-secretase-mediated EphA4 signaling pathway in synaptic pathogenesis of Alzheimer's disease. Brain Pathol 2012; 22:776-87; PMID:22404518; http://dx.doi.org/10.1111/j.1750-3639.2012.00587.x
- Hedstrom L. Serine protease mechanism and specificity. Chem Rev 2002; 102:4501-24; PMID:12475199; http://dx.doi.org/10.1021/cr000033x
- Polgar L. The catalytic triad of serine peptidases. Cellular and molecular life sciences : CMLS 2005; 62:2161-72; PMID:16003488; http://dx.doi.org/10.1007/s00018-005-5160-x
- Netzel-Arnett S, Hooper JD, Szabo R, Madison EL, Quigley JP, Bugge TH, Antalis TM. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev 2003; 22:237-58; PMID:12784999; http://dx.doi.org/10.1023/A:1023003616848
- M. Santos J, Graindorge A, Soldati-Favre D. New insights into parasite rhomboid proteases. Mol Biochem Parasitol 2012; 182:27-36; PMID:22173057; http://dx.doi.org/10.1016/j.molbiopara.2011.11.010
- Lemberg MK, Menendez J, Misik A, Garcia M, Koth CM, Freeman M. Mechanism of intramembrane proteolysis investigated with purified rhomboid proteases. EMBO J 2005; 24:464-72.
- Lee JR, Urban S, Garvey CF, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in drosophila. Cell 2001; 107:161-71; PMID:11672524; http://dx.doi.org/10.1016/S0092-8674(01)00526-8
- Adrain C, Strisovsky K, Zettl M, Hu L, Lemberg MK, Freeman M. Mammalian EGF receptor activation by the rhomboid protease RHBDL2. EMBO Rep 2011; 12:421-7; PMID:21494248; http://dx.doi.org/10.1038/embor.2011.50
- Pascall JC, Brown KD. Intramembrane cleavage of ephrinB3 by the human rhomboid family protease, RHBDL2. Biochem Biophys Res Commun 2004; 317:244-52; PMID:15047175; http://dx.doi.org/10.1016/j.bbrc.2004.03.039
- Chen ZL, Yoshida S, Kato K, Momota Y, Suzuki J, Tanaka T, Ito J, Nishino H, Aimoto S, Kiyama H, et al. Expression and activity-dependent changes of a novel limbic-serine protease gene in the hippocampus. J Neurosci 1995; 15:5088-97; PMID:7623137
- Kishi T, Kato M, Shimizu T, Kato K, Matsumoto K, Yoshida S, Shiosaka S, Hakoshima T. Crystal structure of neuropsin, a hippocampal protease involved in kindling epileptogenesis. J Biol Chem 1999; 274:4220-4; PMID:9933620; http://dx.doi.org/10.1074/jbc.274.7.4220
- Shiosaka S, Ishikawa Y. Neuropsin—A possible modulator of synaptic plasticity. J Chem Neuroanat 2011; 42:24-9; PMID:21679765; http://dx.doi.org/10.1016/j.jchemneu.2011.05.014
- Attwood BK, Bourgognon J-M, Patel S, Mucha M, Schiavon E, Skrzypiec AE, Young KW, Shiosaka S, Korostynski M, Piechota M, et al. Neuropsin cleaves EphB2 in the amygdala to control anxiety. Nature 2011; 473:372-5; PMID:21508957; http://dx.doi.org/10.1038/nature09938
