 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
One of the most important features of malignant cells is their capacity to invade adjacent tissues and metastasize to distant organs. This process involves the creation, by tumor and stroma cells, of a specific microenvironment, suitable for proliferation, migration and invasion of tumor cells. The ADAM family of proteins has been involved in these processes. This work aimed to investigate the role of the recombinant disintegrin domain of the human ADAM9 (rADAM9D) on the adhesive and mobility properties of DU145 prostate tumor cells. rADAM9D was able to support DU145 cell adhesion, inhibit the migration of DU145 cells, as well as the invasion of this cell line through matrigel in vitro. Overall this work demonstrates that rADAM9D induces specific cellular migratory properties when compared with different constructs having additional domains, specially those of metalloproteinase and cysteine-rich domains. Furthermore, we showed that rADAM9D was able to inhibit cell adhesion, migration and invasion mainly through interacting with α6β1 in DU145 tumor cell line. These results may contribute to the development of new therapeutic strategies for prostate cancer.
Introduction
Prostate cancer is the second leading cause of death by cancer in men. It has become the most common cancer in men in the Western world, with approximately 40,000 new cases yearly in the UK. In Europe and North America, more than 500,000 cases are diagnosed per year. The disease incidence varies worldwide, being more common in Western countries compared to East Asian and developing countries. The disease incidence has increased in the past 2 decades, probably due the improvements in early detection and/or treatment, and also an increased public awareness.Citation11 Because of the high prostate cancer incidence, the development of new therapeutic tools should be created to combat this disease.
The ADAM (A Disintegrin And Metalloproteinase) family of proteins comprises a group of multifunctional proteins that play important roles in many biological processes, such as cell fusion, cell adhesion, proteolysis, and in some diseases as well, including cancer.Citation2,3 These type I transmembrane proteins are characterized by the presence of a prodomain, an N-terminal metallopeptidase domain, a disintegrin domain, a cysteine-rich domain, an EGF-like domain, a transmembrane region, and a cytoplasmic tail with signaling properties.Citation4
Among the 30 described members of the ADAM family, ADAM9 or Meltrin-γ, is a widely expressed, non-RGD-containing protein that has been shown to bind to the α6β1 integrin on fibroblasts,Citation5 αvβ5 on myeloma cellsCitation6 as well as osteoblasts.Citation7 Furthermore, the disintegrin domain of ADAM9 was demonstrated to bind directly to α6β4 and α2β1 integrins on the surface of colon carcinoma cells.Citation8 After binding to these integrins, the disintegrin domain of ADAM9 is able to promote different cell signaling responses, such as an increase in IL-6 production via the p38MAPK and cPLA2 pathways.Citation7 Recombinant disintegrin domain of ADAM9 (rADAM9D) was demonstrated to bind to MDA-MB-231 tumor breast cells through β1, α3, αvβ3, αvβ5 and α2 integrins and also to inhibit the adhesion of this cell line and platelets to collagen type I under dynamic flow conditions.Citation9
The adhesive properties of the different ADAMs domains are still a matter of controversy. Whereas some works attribute adhesive properties to the disintegrin domain,Citation5,7-9 others indicate the cysteine-rich domain as having adhesive functions.Citation10, 11 Recent data from different model systems suggest that ADAM9 is involved in tumor formation/progression. In many types of human cancers ADAM9, as well as other specific ADAMs, are up-regulated.Citation12-18
In the present work we demonstrated the pro- and anti-adhesive properties of the rADAM9D on DU145 prostate cancer cells. rADAM9D was able to promote adhesion of DU145 cells, therefore acting as an adhesion molecule, similarly to other molecules such as collagens, laminin and fibronectin. Furthermore, rADAM9D inhibited the adhesion of DU145 cells to laminin-coated wells, but not to collagen type I. The adhesion of rADAM9D to DU145 cells can occur through β1, α6, αvβ5 and αvβ3, since the incubation with anti-integrin blocker antibodies directed against these integrins, inhibited cell adhesion to rADAM9D. rADAM9D was also able to inhibit DU145 cell migration and invasion through matrigel. Overall, this study may contribute to the development of new therapeutic strategies for prostate cancer.
Results
rADAM9D promotes DU145 cell adhesion
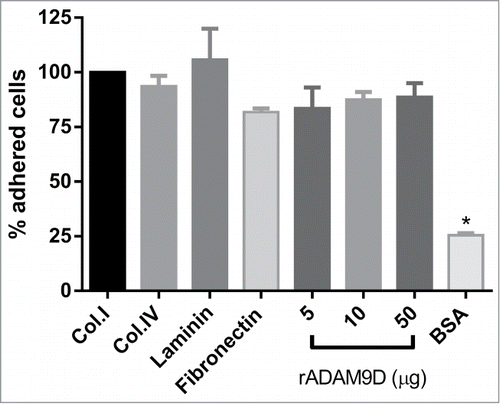
rADAM9D was able to promote adhesion of DU145 cell line, similarly to different extracellular matrix proteins (collagen type I and IV, laminin and fibronectin), therefore supporting cell adhesion as an extracellular matrix protein, probably by interacting with DU145 integrins. There was no difference among quantities (5, 10 or 50 µg) of rADAM9 plated on the wells to promote the adhesion of DU145 cells ().
Figure 1. rADAM9D promotes DU145 cell adhesion. Collagen type I (Col.I), collagen type IV (Col.IV), laminin, fibronectin (10 µg) (positive controls) or different quantities of ADAM9D (5, 10 and 50μg) were immobilized in the wells of a 96-well plate in adhesion buffer at 4°C. After blocking with 1% BSA (negative control), CMFDA-labeled DU145 cells (1 × 105 cells/well) were seeded in the wells. The plates were incubated at 37 °C for 30 min, washed, lysed and read for the release of fluorescence. BSA was used as negative control for cell adhesion. The results were obtained from 3 independent experiments in triplicate. The means that are significantly different from those of cells growing on collagen using ANOVA followed by post hoc Dunnett's test were shown by *(P ≤ 0.001). The results were normalized by the collagen type I values in each experiment. The error bars show the SE of three samples with less deviation from the mean.
ADAM9D interacts with DU145 prostate tumor cells through β1, α6, αvβ5 and αvβ3
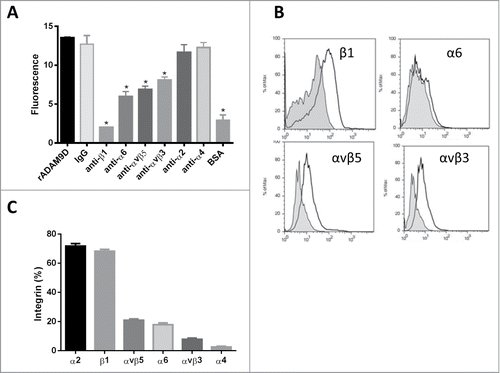
Since rADAM9D supported DU145 cell adhesion we verified which integrins in this prostate cancer cell line would be specific for this disintegrin. For that, we incubated DU145 cells with different anti-integrin blocking antibodies before plate on an rADAM9D (10 µg) coating. The antibodies against β1, α6, αvβ5 and αvβ3, inhibited DU145 cell adhesion to rADAM9D (), confirming that this recombinant protein could bind to these integrins on the surface of DU145 cells. On the other hand, antibodies against α2 and α4 integrins chains did not inhibit the adhesion of DU145 cells to rADAM9D (). To confirm this result, rADAM9D (1 µM) was previously incubated with DU145 cells and the mixture was then incubated with anti-integrin antibodies. Subsequently, cells were analyzed by flow cytometry () and the results obtained confirmed that rADAM9D inhibits the binding of anti-β1, anti-α6, anti-αvβ5 and anti-αvβ3 antibodies to DU145 cells, probably because rADAM9D binds to these integrins on these cell line, preventing previous binding of cited antibodies. The inhibition of anti-α6 antibody promoted by rADAM9D was lower compared to the other antibodies using cytometry analysis (). rADAM9D did not promote inhibition of the binding of anti-α2 and α-4 antibodies to DU145 cells. Integrin profile in DU145 cell line was measured by flow cytometry, using the antibodies mentioned above. This cell line presented higher levels of α2 and β1, moderate levels of α6, αvβ3 and αvβ5, and low levels of α4 integrin subunit ().
Figure 2. rADAM9D binds to DU145 through β1, α6, αvβ5 and αvβ3 integrins as demonstrated by an antibody competition assay (A) and by flow cytometry analysis (B). The integrin content of DU145 cell line was assessed by flow cytometry (C). (A) For antibody competition assay CMFDA-labeled cells were incubated with different anti-integrin antibodies (β1, α6, αvβ5, αvβ3, α2 and α4, at 10 µg/ml) and IgG control (10 µg/ml) before being plated on rADAM9D-coated (10 µg) wells. DU145 directly plated on rADAM9D or IgG-coated wells, without previous incubation with any antibody was used as positive control and BSA was used as negative control. (B) The integrin content of DU145 cells was determined by flow cytometry. Cells (1 × 105) were incubated for 40 min at 4°C with the specific antibodies cited earlier or control IgG. Cells were washed and incubated with secondary antibody labeled with FITC, at same conditions described before, washed and fixed with FACs buffer contaning 1% phormaldehyde overnight at 4°C. C. To verify the interaction with integrins, rADAM9D (1 µM) was previously incubated (30 min at room temperature) with DU145 cells, before the addition of antibodies. Cells were analyzed in FACSCanto. The results were obtained from 3 independent experiments in triplicate. The error bars show the SE of three samples with less deviation from the mean. The means that are significantly different from rADAM9D and IgG-coated wells using ANOVA followed of post hoc Dunnett's test were shown by *(P ≤ 0.001).
rADAM9D inhibits the adhesion of DU145 cells to laminin but not to collagen type I
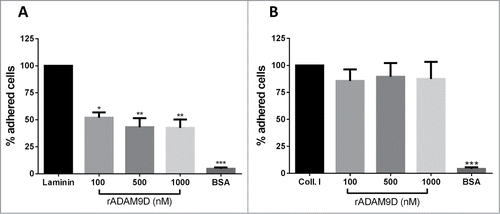
Since rADAM9D seemed to bind specifically α6β1 integrin on DU145 prostate cancer cell line, we further investigate whether this rADAM9 domain would inhibit tumor cell adhesion to laminin, a known α6β1 integrin ligand. Therefore, we tested the capacity of rADAM9D to inhibit the adhesion of DU145 cells to laminin and collagen type I, which has no specificity for α6β1 integrin. For that, DU145 cells were previously incubated with different rADAM9D concentrations and then plated on laminin or collagen type I-coated wells. Results indicate that incubation of rADAM9D with DU145 cells inhibited their adhesion to laminin () but not to collagen type I-coated wells (). In other words, the blocking of receptors on DU145 cells promoted by rADAM9D was specific to inhibit the adhesion of this cell line to laminin but not to collagen type I. rADAM9D at concentrations of 2000nM were also tested for the inhibition of DU145 cells to collagen type I, but there was no inhibition (data not shown).
Figure 3. rADAM9D inhibits the adhesion of DU145 to laminin but not to collagen type I. Ninety six-well plates were coated with laminin (A) or collagen type I (B) (10 µg/well) in adhesion buffer or 0.1% acetic acid, respectively, overnight at 4°C. After blocking with 1% BSA, CMFDA-labeled cells (1 × 105 cells/well) were incubated with different concentrations (100, 500, 1000nM) of rADAM9D and seeded in the wells. The plates were incubated at 37°C for additional 30 min. After washing, remaining cells were lysed, and the plate was read for the release of fluorescence. The results were obtained from 3 independent experiments and in triplicate. The results for rADAM9D were normalized by the collagen or laminin values in each experiment. The error bars show the SE of three samples with less deviation from the mean. The means for all rADAM9 concentrations were significantly different from the laminin using ANOVA followed by post hoc Dunnett's test: *(P ≤ 0.05), **(P ≤ 0.01) and ***(P ≤ 0.001).
rADAM9D inhibits invasion and migration of DU145 cells
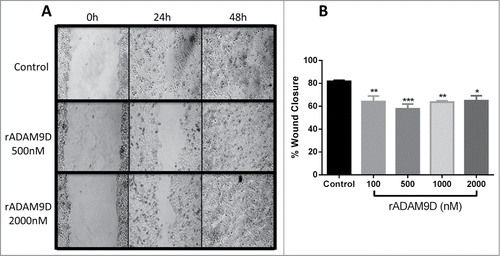
rADAM9D was tested to its ability to inhibit DU145 cell invasion and migration. rADAM9D, at concentrations ranging from 100 to 1000 nM, was able to significantly inhibit DU145 cell invasion (). In a wound healing migration assay, rADAM9D significantly inhibited DU145 cell migration at 100, 500, 1000 and 2000nM (). The effects were more striking after 24 hours of wound repopulation.
Figure 4. rADAM9D inhibits the invasion of DU145 cells through matrigel. DU145 cells (1.25 × 105 cells/ml) were seeded on the inserts (12 well-plate) of the invasion chamber in the presence or absence of rADAM9D (1 µM). Complete medium was used as a chemoattractant at the lower chamber. Plates were incubated for 22 h at 37°C and 5% CO2. Non-invading cells were removed with a cotton swab from the upper surface of the membrane and invading cells were fixed and 10 random fields from microscope slides were photographed and cells were counted using Image J software. Positive control (+ control) was made in the presence of chemoattractant (10% FBS) at the lower chamber and negative control (− control) was FBS free medium at the lower chamber. The results were obtained from 3 independent experiments in triplicate. The error bars show the SE of three samples with less deviation from the mean. The means that are significantly different from the positive control using ANOVA followed by post hoc Dunnett's test: *(P ≤ 0.001).
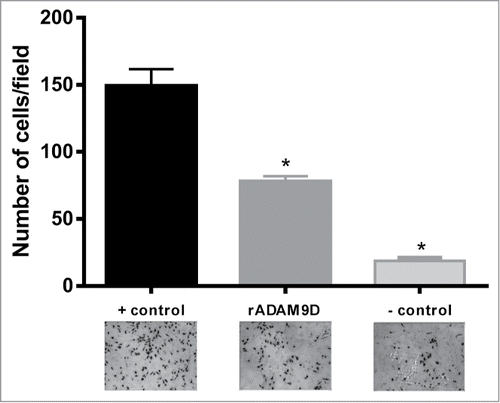
Figure 5. rADAM9D inhibits the migration of DU145 cells in a wound healing assay. (A) Cells (1 × 105 cells/ml) were plated in 24-wells plates and incubated properly until the culture reached 100% of confluence. Afterwards, a straight scratch was made with a pipette tip and cells were washed with culture medium to remove unbound cells and debris. Cells were incubated with ADAM9D (100, 500, 1000 and 2000 nM) for 24 h and 48 h. Only pictures from rADAMD9 500 and 2000 nM are represented. Central field was photographed at 0h, 24 h and 48 h (when cells close the scratch completely). (B) Closure area of migrating cells was measure using ImageJ software, and it was calculated the percentage of wound closure, comparing time zero and 24 hour. Results are expressed as percent of wound closure relative to control (untreated) cells. The results were obtained from 3 independent experiments in triplicate. The error bars show the SE of three samples with less deviation from the mean. The means that are significantly different from the control using ANOVA followed by post hoc Dunnett's test: *(P ≤ 0.05), **(P ≤ 0.01) and ***(P ≤ 0.001).
Discussion
The progression of malignant tumors results from invasion of the primary tumor to a secondary site, causing metastasis in a multi-step process. These steps can be summarized as follows: cell detachment from the primary tumor, migration and invasion into the ECM, intravasation into a blood or lymphatic vessel, survival within the vasculature, adherence of these tumor cells in the endothelium, extravasation, and formation of secondary tumors.Citation19-21 Therefore, metastasis necessitates disruption of cellular interactions with the tumor microenvironment, increased migratory and invasion capacity and the ability to overcome the pro-apoptotic signals provided by diminished intercellular and cell-ECM interactions mainly through integrin receptors.Citation22
Several members of the integrin family, including α1β1, α2β1, α3β1, α6β1, α7β1 and α6β4 heterodimers serve as laminin receptors on a variety of normal and tumor cell typesCitation23 and DU145 prostate cell line was reported to have abundant content of integrin β1, along with α1, α2, α3, α5 and α6 integrin subunits.Citation24,25 In this work, we have demonstrated that DU145 tumor cell line contains high amounts of α2β1, moderate amounts of αvβ5 and α6 and low quantities of αvβ3 and α4 integrins. Additionally, we showed that the recombinant disintegrin domain of ADAM9 (rADAM9D) was able to inhibit the adhesion of DU145 prostate cancer cells to laminin, mainly by binding to α6β1 integrin receptor, therefore rADAM9D may act at the beginning of metastatic cascade, disrupting the binding of prostate cells to the ECM laminin component. Furthermore, rADAM9D was able to inhibit DU145 cell invasion and migration of this tumor cell line.
It was demonstrated in the literature that ADAM9 transcripts are alternatively spliced to express a transmembrane protein (ADAM9-L) and a secreted variant (ADAM9-S) with opposite functions in breast cancer cells and that the balance between these isoforms is an important determinant in manifestation of aggressive migratory phenotypes associated with breast cancer progression. According to the authors, ADAM9-S is an enhancer, whereas ADAM9-L is a suppressor of cell migration.Citation26 Mazzocca and colleaguesCitation8 have demonstrated the presence of these alternatively spliced variants of ADAM9. In hepatic stellate cells, it was demonstrated that ADAM9-S induced a highly invasive phenotype and promotes tumor cell invasion. Moreover, they demonstrated that ADAM9-S binds directly to α6β4 and α2β1 integrins on the surface of colon carcinoma cells through the disintegrin domain.Citation8 Our results are not in accordance with these data since we have demonstrated that rADAM9D is an inhibitor of invasion and migration in prostate tumor cells. ADAM9-S is a construct composed by a signal sequence, a prodomain, a metalloproteinase domain and by disintegrin and cysteine-rich domains. In our work, however, only the disintegrin domain of ADAM9 was recombinantly produced in a bacterial system and this difference in protein structure may explain the differential biological activities of both molecules. Additionally, it is important to point out that the adhesion properties of the isolated rADAM9D domain are not sufficient to explain the molecular interactions and functional significance of the entire protein in a cellular and in vivo system. Moreover, the native disintegrin domain of ADAM9 has a N-glycosylation site,Citation27 which undoubtedly influences its adhesion properties. In this work, however, rADAM9D does not have the presence of this N-glycosylation, since bacterial systems to produce recombinant proteins are not able to perform such types of post-translational modifications. Furthermore, Takeda and co-workers,Citation11 resolving the crystal structure of VAP-1, a homolog of mammalian ADAMs, demonstrated that the binding area of the disintegrin domain in a C-type conformation is not accessible for protein binding and a Hyper-Variable Region (HVR) of the cysteine-rich domain may instead be involved in substrate interaction. Therefore, alone the disintegrin domain may only contribute, when in native conformation, but not mediate cell adhesion in physiological conditions.
On the other hand, Zigrino and coworkersCitation10 prepared a construct only with disintegrin and cysteine-rich domains of ADAM9 and reported that this protein functions as a cell adhesion molecule. These authors also demonstrated that the recombinant ADAM9 disintegrin-cysteine-rich domains specifically interact with the β1 integrin subunit on keratinocytes. However, engagement of integrin receptors by the ADAM9 disintegrin-cysteine-rich domains resulted in ERK phosphorylation and increased MMP9 synthesis. Additionally, keratinocytes adhering to the immobilized disintegrin and cysteine-rich domains showed increased motility, which was partially due to the induction of MMP9 secretion in this cell line.Citation10 In our work, instead, rADAM9D inhibited DU145 cell migration and invasion, again demonstrating the influence of different protein constructs on its functional activity.
This is the first time that the effects of a recombinant domain of an human ADAM are demonstrated on adhesion, migration and invasion of prostate tumor cells. Taken together, our results demonstrate that recombinant ADAM9D has specific migratory properties when compared with different constructs having additional domains, specially those of metalloproteinase and cysteine-rich domains. Furthermore, we showed that rADAM9D was able to inhibit cell adhesion, migration and invasion mainly through interacting with α6β1 in DU145 tumor cell line.
Conclusion
The potential of the rADAM9D as an anti-adhesive molecule can be explored as tool to combat metastasis and cancer progression and for the design of selective inhibitors against the adhesion and extravasation of cancer cells. Other studies, using animal models, should be conducted to confirm this hypothesis.
Material and Methods
Recombinant ADAM9 disintegrin domain (rADAM9D)
Expression and purification of the recombinant disintegrin domain of ADAM9 (rADAM9D) was performed as previously described.Citation9 Briefly, total RNA from a VMM12 human melanoma cell line was reverse transcribed and resulting cDNA was used for amplification of the disintegrin domain of human ADAM9 (ADAM9D) (GenBank accession no. NM003816). For ADAM9D expression it was used the pGEX-4T-1 vector which is classically used to produce GST fusion proteins. After the transformation of Escherichia coli DH5-α cells, the ampicillin-resistant recombinant plasmids were selected for restriction analysis. The confirmed recombinant plasmids (pADAM9D) were used to transform the E. coli AD494(DE3) expression strain. Cultures of E. coli AD494(DE3)pADAM9D were induced for expression by addition of 0.5 mM isopropyl thio-β-D-galactopyranoside (IPTG). Four hours after induction, cells were harvested by centrifugation (7000 rpm, 15 min, 4°C) and cell pellet was suspended in PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3) and lysed by sonication (5 times, 4°C, 1 min interval). ADAM9D was released from the fusion protein (GST) by thrombin cleavage. Thrombin was eliminated from samples containing ADAM9D by purification in a Benzamidine-Sepharose 4B column (GE Healthcare). Fractions were analyzed by SDS-PAGE with Coomassie brilliant blue staining and rADAM9D concentration was determined by BCA protein assay (Pierce).
Antibodies
For flow cytometry and competition assays the monoclonal blockers antibodies used against human α2 (MAB 1233), α6 (MAB13501), β1 (MAB17781) and αvβ5 (MAB2528) integrins, were obtained from R&D Systems (R&D Systems Inc.., Minneapolis, MN, USA). Antibodies against αvβ3 integrin (MAB1976) were from Chemicon, and antibodies to the α4 integrin subunit (I6528) were from Sigma-Aldrich. Control IgG was from Dako. Secondary antibody goat anti-mouse IgG-FITC (Fluorescein isothiocyanate, sc-2010), used for flow cytometry, were purchased from Santa Cruz Biotechnology.
Cell line and culture
DU145 human prostate tumor cell line was obtained from ATCC and maintained at 37°C in 5% CO2 in RPMI culture medium (Cultilab) containing 10% FBS, penicillin (100 UI/ml), streptomycin (100 µg/ml) and L-glutamine (2mM). Cell cultures and experiments were conducted in a humidified environment with 5% CO2 at 37°C.
Cell adhesion and antibody competition
Cell adhesion and inhibition of cell adhesion were performed as described before.Citation9 Briefly, collagen type I, collagen type IV, laminin, fibronectin (10 µg), rADAM9D (5, 10 and 50 μg) or 1% BSA were immobilized overnight at 4°C in 96 well plate. Wells were blocked with 1% BSA in adhesion buffer (HEPES 20mM, pH 7,35; containing NaCl 150 mM, KCl 5 mM, MgSO4 1 mM and MnCl2 1 mM) for 2 h. DU145 cells were labeled with 12.5 µM CMFDA (37°C for 30 min) and then 1 × 105 cells/well were plated and maintained for 30min at 37°C. Wells were washed and the remaining cells were lysed using 0.5% Triton X-100. Fluorescence was read in a fluorimeter Spectra-Max Gemini XS (Molecular Devices, Sunnyvale, CA, USA) with 485nm excitation and 530nm emission filters. For inhibition of adhesion assay, cells were labeled and measured as described above, except that the cells were pre-incubated with rADAM9D in different concentrations (100, 500, 1000 nM) and then plated on 10 µg of collagen type I (Sigma-Aldrich) in acetic acid (0.1%) or on 10 µg of laminin (Sigma-Aldrich) in adhesion buffer. For antibody competition assays CMFDA-labeled cells were incubated with different anti-integrin antibodies (αvβ3, αvβ5, β1, α2, α4, α6 at 10 µg/ml) and IgG control (10 µg/ml) before being plated on rADAM9D-coated wells.
Flow cytometry analysis
DU145 cells were submitted to flow cytometry analysis to verify their integrin cell profile. Cells (1 × 105cells/ml) were incubated for 40 min at 4°C with specific antibodies against αvβ3, αvβ5, β1, α2, α4, α6 and control IgG. Cells were washed and incubated with secondary antibody labeled with FITC, at same conditions described before, washed and fixed with FACs buffer contaning 1% phormaldehyde overnight at 4°C. To verify the interaction with integrins, rADAM9D (1 µM) was previously incubated (30 min at room temperature) with DU145 cells, before the addition of antibodies. Cells were analyzed in FACSCanto (BD Biosciences).
Invasion
Invasion assays were performed with DU145 prostate cancer cells incubated with rADAM9D using the BioCoat Matrigel Invasion Chambers (BD Biosciences). Briefly, DU145 cells (1.25 × 105 cells/ml) were seeded on the inserts (12 well-plate) of the invasion chamber in the presence or absence of rADAM9D (1 µM). Complete medium was used as a chemoattractant at the lower chamber. Plates were incubated for 22 h at 37°C and 5% CO2. Non-invading cells were removed with a cotton swab from the upper surface of the membrane and invading cells were fixed using 100% methanol and stained with 1% toluidine blue in 1% borax. Ten random fields from microscope slides were photographed and cells were counted using Image J software. Positive control was made in the presence of chemoattractant (10% FBS) at the lower chamber and negative control was FBS free medium at the lower chamber.
Wound healing
Cells (1 × 105 cells/ml) were plated in 24-wells plates and incubated properly until the culture reached 100% of confluence. Afterwards, a straight scratch was made with a pipette tip and cells were washed with culture medium to remove unbound cells and debris. Cells were incubated with rADAM9D (100, 500, 1000 and 2000 nM) for 24 h and 48 h. Central field was photographed at 0 h, 24 h and 48 h (when cells close the scratch completely). Closure area of migrating cells was measure using ImageJ software, and it was calculated the percentage of wound closure, comparing time zero and 24 hours later, using a formula from Yue et al.,Citation28 showing the difference between 0 h and 24 h:
Statistical analysis
All experiments were repeated 3 times in triplicate and the mean and standard deviation were calculated. Comparisons were made using one-way analysis (ANOVA) and when samples presented significance, Dunnet's post hoc analysis was applied to verify difference between groups.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The authors are grateful for the financial support of FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo, #2004/06986-6). Martin ACBM had a scholarship sponsored by CNPq (National Council of Research, #130539/2009-0).
References
- Maximum androgen blockade in advanced prostate cancer: an overview of the randomised trials. Prostate Cancer Trialists' Collaborative Group. Lancet 2000; 355:1491-8; PMID:10801170; http://dx.doi.org/10.1016/S0140-6736(00)02163-2
- Stone AL, Kroeger M, Sang QX. Structure-function analysis of the ADAM family of disintegrin-like and metalloproteinase-containing proteins (review). J Protein Chem 1999; 18:447-65; PMID:10449042; http://dx.doi.org/10.1023/A:1020692710029
- Klein T, Bischoff R. Active metalloproteases of the A Disintegrin and Metalloprotease (ADAM) family: biological function and structure. J Proteome Res 2011; 10:17-33; PMID:20849079; http://dx.doi.org/10.1021/pr100556z
- Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol 2005; 6:32-43; PMID:15688065; http://dx.doi.org/10.1038/nrm1548
- Nath D, Slocombe PM, Webster A, Stephens PE, Docherty AJ, Murphy G. Meltrin gamma(ADAM-9) mediates cellular adhesion through alpha(6)beta(1)integrin, leading to a marked induction of fibroblast cell motility. J Cell Sci 2000; 113 ( Pt 12):2319-28; PMID:10825303
- Zhou M, Graham R, Russell G, Croucher PI. MDC-9 (ADAM-9/Meltrin gamma) functions as an adhesion molecule by binding the alpha(v)beta(5) integrin. Biochem Biophys Res Commun 2001; 280:574-80; PMID:11162558; http://dx.doi.org/10.1006/bbrc.2000.4155
- Karadag A, Zhou M, Croucher PI. ADAM-9 (MDC-9/meltrin-gamma), a member of the a disintegrin and metalloproteinase family, regulates myeloma-cell-induced interleukin-6 production in osteoblasts by direct interaction with the alpha(v)beta5 integrin. Blood 2006; 107:3271-8; PMID:16373656; http://dx.doi.org/10.1182/blood-2005-09-3830
- Mazzocca A, Coppari R, De Franco R, Cho JY, Libermann TA, Pinzani M, et al. A secreted form of ADAM9 promotes carcinoma invasion through tumor-stromal interactions. Cancer Res 2005; 65:4728-38; PMID:15930291; http://dx.doi.org/10.1158/0008-5472.CAN-04-4449
- Cominetti MR, Martin AC, Ribeiro JU, Djaafri I, Fauvel-Lafeve F, Crepin M, et al. Inhibition of platelets and tumor cell adhesion by the disintegrin domain of human ADAM9 to collagen I under dynamic flow conditions. Biochimie 2009; 91:1045-52; PMID:19505527; http://dx.doi.org/10.1016/j.biochi.2009.05.012
- Zigrino P, Steiger J, Fox JW, Loffek S, Schild A, Nischt R, et al. Role of ADAM-9 disintegrin-cysteine-rich domains in human keratinocyte migration. JBiol Chem 2007; 282:30785-93; PMID:17704059; http://dx.doi.org/10.1074/jbc.M701658200
- Takeda S, Igarashi T, Mori H, Araki S. Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold. EMBO J 2006; 25:2388-96; PMID:16688218; http://dx.doi.org/10.1038/sj.emboj.7601131
- Fritzsche FR, Jung M, Tolle A, Wild P, Hartmann A, Wassermann K, et al. ADAM9 expression is a significant and independent prognostic marker of PSA relapse in prostate cancer. Euro Urol 2008; 54:1097-106; PMID:18061337; http://dx.doi.org/10.1016/j.eururo.2007.11.034
- Carl-McGrath S, Lendeckel U, Ebert M, Roessner A, Rocken C. The disintegrin-metalloproteinases ADAM9, ADAM12, and ADAM15 are upregulated in gastric cancer. Int JOncol 2005; 26:17-24; PMID:15586220
- Peduto L, Reuter VE, Shaffer DR, Scher HI, Blobel CP. Critical function for ADAM9 in mouse prostate cancer. Cancer research 2005; 65:9312-9; PMID:16230393; http://dx.doi.org/10.1158/0008-5472.CAN-05-1063
- Li J, Ji Z, Qiao C, Qi Y, Shi W. Overexpression of ADAM9 Promotes Colon Cancer Cells Invasion. J Invest Surg 2013; 26; PMID:23514059; http://dx.doi.org/10.3109/08941939.2012.728682
- Josson S, Anderson CS, Sung SY, Johnstone PA, Kubo H, Hsieh CL, et al. Inhibition of ADAM9 expression induces epithelial phenotypic alterations and sensitizes human prostate cancer cells to radiation and chemotherapy. Prostate 2011; 71:232-40; PMID:20672324; http://dx.doi.org/10.1002/pros.21237
- Peduto L. ADAM9 as a potential target molecule in cancer. Curr Pharm Des 2009; 15:2282-7; PMID:19601830; http://dx.doi.org/10.2174/138161209788682415
- Zhang J, Chen N, Qi J, Zhou B, Qiu X. HDGF and ADAM9 are novel molecular staging biomarkers, prognostic biomarkers and predictive biomarkers for adjuvant chemotherapy in surgically resected stage I non-small cell lung cancer. JCancer ResClin Oncol 2014; 140:1441-9; PMID:24770635; http://dx.doi.org/10.1007/s00432-014-1687-2
- Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer 2002; 2:91-100; PMID:12635172; http://dx.doi.org/10.1038/nrc727
- Pontier SM, Muller WJ. Integrins in breast cancer dormancy. APMIS 2008; 116:677-84; PMID: 18834411; http://dx.doi.org/10.1111/j.1600-0463.2008.01026.x
- Van den Eynden GG, Majeed AW, Illemann M, Vermeulen PB, Bird NC, Hoyer-Hansen G, et al. The multifaceted role of the microenvironment in liver metastasis: biology and clinical implications. Cancer Res 2013; 73:2031-43; PMID:23536564; http://dx.doi.org/10.1158/0008-5472.CAN-12-3931
- Sakamoto S, Kyprianou N. Targeting anoikis resistance in prostate cancer metastasis. Mol Aspects Med 2010; 31:205-14; PMID:20153362; http://dx.doi.org/10.1016/j.mam.2010.02.001
- Belkin AM, Stepp MA. Integrins as receptors for laminins. Microsc Res Techniq 2000; 51:280-301; PMID:NOT_FOUND; http://dx.doi.org/10.1002/1097-0029(20001101)51:3%3c280::AID-JEMT7%3e3.0.CO;2-O
- Skogseth H, Holt RU, Larsson E, Halgunset J. Tyrosine kinase inhibitors alter adhesivity of prostatic cancer cells to extracellular matrix components. APMIS 2006; 114:225-33; PMID:16643189; http://dx.doi.org/10.1111/j.1600-0463.2006.apm_365.x
- Witkowski CM, Rabinovitz I, Nagle RB, Affinito KS, Cress AE. Characterization of integrin subunits, cellular adhesion and tumorgenicity of four human prostate cell lines. J Cancer Res Clin Oncol 1993; 119:637-44; PMID:7688749; http://dx.doi.org/10.1007/BF01215981
- Fry JL, Toker A. Secreted and membrane-bound isoforms of protease ADAM9 have opposing effects on breast cancer cell migration. Cancer Res 2010; 70:8187-98; PMID:20736367; http://dx.doi.org/10.1158/0008-5472.CAN-09-4231
- Roghani M, Becherer JD, Moss ML, Atherton RE, Erdjument-Bromage H, Arribas J, et al. Metalloprotease-disintegrin MDC9: intracellular maturation and catalytic activity. J Biol Chem 1999; 274:3531-40; PMID:9920899; http://dx.doi.org/10.1074/jbc.274.6.3531
- Yue PY, Leung EP, Mak NK, Wong RN. A simplified method for quantifying cell migration/wound healing in 96-well plates. J Biomol Screen 2010; 15:427-33; PMID:20208035; http://dx.doi.org/10.1177/1087057110361772
