Abstract
A characteristic feature of the opportunistic foodborne pathogen Cronobacter sakazakii is its ability to survive in extremely arid environments, such as powdered infant formula, making it a dangerous opportunistic pathogen of individuals of all age groups, especially infants and neonates. Herein, we provide a brief overview of the pathogen; clinical manifestations, environmental reservoirs and our current understanding of stress response mechanisms and virulence factors which allow it to cause disease.
The Genus Cronobacter
Cronobacter spp, previously known as Enterobacter sakazakiiCitation1 are Gram-negative, rod-shaped, motile, facultative anaerobic bacteria.Citation2 The Cronobacter genus is genetically closely related to the Citrobacter (97.8%) and Enterobacter genera (97.0%)Citation3 and until recently consisted of 10 speciesCitation2,4,5: Cronobacter sakazakii, Cronobacter malonaticus, Cronobacter turicensis, Cronobacter muytjensii, Cronobacter dublinesis, Cronobacter condiment, Cronobacter universalis, Cronobacter pulveris, Cronobacter helveticus, Cronobacter zurichensis. Three of which; Cronobacter sakazakii, Cronobacter malonaticus and Cronobacter turicensis, have been linked to neonatal infections.Citation6,7 However, recent studies proposed that Cronobacter species C. helveticus, C. pulveris and C zurichensis form their own sub-clades, genetically distinct from Cronobacter.Citation8 The prototypical member of the Cronobacter family, C. sakazakii, is an opportunistic foodborne pathogen that has been linked with life-threatening conditions in neonates, immunocompromised infants, children (from 3 months to 4 y old) and elderly adults.Citation9-11
Clinical manifestations of C. sakazakii
While Cronobacter sakazakii is primarily a foodborne pathogen, its association with severe neonatal infections has significantly increased awareness and triggered a surge of interest in recent years.Citation12 C. sakazakii can cause bacteraemia and sepsis, cerebro-spinal and peritoneal fluid accumulation, brain abscesses, cyst formation, necrotizing enterocolitis (NEC), meningitis and intracerebral infarctions.Citation13,14 NEC follows colonisation of the intestine by C. sakazakii, with the incidence increasing in low birth weight or premature neonates.Citation15 NEC is characterized by inflammation and death of intestinal tissue and is one of the most common gastrointestinal conditions that can arise in neonates.Citation16 Indeed, this gastrointestinal pathogen is responsible for up to 80% of infant deaths associated with infection.Citation11 Infants that survive C. sakazakii infection often suffer delayed neurological symptoms, e.g., delayed brain development, brain abscesses or hydrocephalus.Citation17,18 Consequently, The International Commission on Microbiological Specification for FoodsCitation19 has classified C. sakazakii as a ‘severe hazard for restricted populations, life-threatening or with substantial chronic sequelae over long duration’. Furthermore, the World Health Organization and Food and Agriculture Organization in 2008 issued a joint call to the scientific community for more data on this organism (WHO/FAO).
Morbidity and mortality as a result of C. sakazakii infection are largely dependent on the immune status of the host. Neonates and infants, up to the age of 12 months, have an immunodeficiency as a result of an immature immune system that can be worsened by premature or traumatic delivery, maternal disease or certain drugs.Citation20 Furthermore, a innate immunity in the form of their competing intestinal microflora is not yet fully established. One of the most severe cases of C. sakazakii infections recorded to date took place in France in 1994 in a Neonatal Intensive Care Unit (NICU). 17 new-born children were infected, of which 7 resulted in necrotizing enterocolitis, 1 case of septicemia and 1 case of meningitis. No clinical symptoms were identified in 8 infants that were infected and colonized, yet 3 neonatal deaths occurred as a result of the infection. The cause of the outbreak was traced to the feeding practices employed in the NICU.Citation21 This study found the same Cronobacter strains in the intestinal tracts of healthy infants, as those presenting with symptoms.Citation21 Therefore, suggesting that the host immune response is a major factor in contributing to C. sakazakii associated disease. A further study which involved collecting faecal samples from both healthy neonates and adult carriers of C. sakazakii, found 3 C. sakazakii isolates in neonate faecal samples, whereas the adults’ faecal samples contained only one C. sakazakii strain and one C. malonaticus strain.Citation22 This suggests that C. sakazakii might be more likely to colonize neonatal intestinal tracts, most likely as a consequence of their comparatively naive immune systems. C. malonaticus on the other hand, was only found in adults suggesting that this species is associated more with adult than neonatal infection.Citation22
Antibiotic Resistance and Alternative Means of Control
Originally C. sakazakii has been reported to be susceptible to a wide range of antibiotics including β-lactams,Citation23 however, several new strains have been described that were found to be resistant to tetracycline,Citation24 neomycin and trimethoprimCitation25 and cephalotin.Citation26 Furthermore, recent studies have described a strain carrying an unusual ampC gene, conferring resistance to cephalosporineCitation27 and found environmental isolates of C. sakazakii from domestic kitchens exhibiting resistance to penicillin, tetracycline, ciprofloxacin and nalidixic acid.Citation28 A comprehensive analysis by next generation whole genome sequencing and annotation of the C. sakazakii strain SP291 indicates that C. sakazakii possesses a substantial number of genes associated with antibiotic resistance, including ampC (cephalosporine), fosA (fosfomycine), gyrA, gyrB, parC and parB conferring resistance to fluoroquinolones and encoding a multitude of multi-drug resistance mechanisms, mainly drug efflux pumps.Citation29 Increasing drug resistance in bacteria is a general problem and consequently the need to identify alternative means to control bacterial infections is pressing.Citation30 The need for new therapeutics and disinfectants has re-kindled interest in bacteriophages as a method of controlling bacterial growth.Citation31,32 In this context C. sakazakii infecting bacteriophages have been isolated, including the second largest known bacteriophage; phage GAP32.Citation33 The applicability of bacteriophages to control C. sakazakii infections in various systems has been the subject of several studies. Two studies in particular have characterized the ability of bacteriophages to reduce and control growth of C. sakazakii in PIF.Citation34,35 However, the application of bacteriophages is not limited to environmental control but can also be used in vivo, as has been demonstrated in insect and mouse infection models, where bacteriophages were shown to protect insect larvaeCitation36 or reduce bacterial load in urinary tract infections in mice.Citation37 Bacteriophages have furthermore been successfully used as immuno-stimulants to increase expression of pro-inflammatory host factors.Citation38
Environmental reservoirs
C. sakazakii is a ubiquitous bacterium found in a wide variety of reservoirs in the environment; from fliesCitation39 to plants such as wheat, rice, herbs and spicesCitation40 or meat and other common household foods.Citation41 The wide variety of environments in which C. sakazakii is present, indicates that it has developed various traits that increase survival in difficult conditions, e.g., resistance to UV irradiation,Citation42 the ability to adhere to various surfaces due to fimbriae formation,Citation42 biofilm formationCitation43 and the ability to survive desiccation.Citation7,9,44 The ability of C. sakazakii to survive in low water environments is a unique stress survival strategy which is linked with survival and persistence of the pathogen in powdered infant formula (PIF), subject to an aw of ∼0.2 (inhospitable to most micro-organisms). A 2004 study found that 8 out of 9 PIF factories investigated were contaminated with C. sakazakii.Citation45 The same study also identified C. sakazakii in households, where 5 isolates were found in 16 family homes. It is likely that contamination of PIF with C. sakazakii occurs during the manufacturing process. While C. sakazakii may not survive pasteurization it is likely that the PIF becomes contaminated after pasteurisation during the processing and addition of non-sterile ingredients to the PIF.Citation9,40,46 Furthermore, several reports have shown that C. sakazakii contamination can occur during the reconstitution of formula, both at home and in hospitals, and is therefore not limited to the production stages.Citation47 In an effort to prevent contamination at this stage the WHO/FAO issued guidelines to hospitals for the reconstitution and storage of PIF (WHO/FAO).
C. sakazakii osmotolerance
An improved understanding of C. sakazakii osmotolerance will provide valuable insights into the mechanisms which aid the survival of this neonatal pathogen as it transits the gastrointestinal tract, where it is subject to varying osmolality,Citation48 in addition to its survival in a dry environment such as PIF. Desiccation is an extreme form of osmotic stressCitation49 and the upregulation of osmotolerance genes is the first line of defense against drying for a bacterial cell.Citation44 It is also worth noting that while microorganisms acquire a state of tolerance to one type of stress, this state of acquired tolerance will also add resistance to other types of stress.Citation50-52 Trollmo et al demonstrated that Saccharomyces cerevisiae cells conditioned to osmotic dehydration were equally as thermotolerant as heat conditioned cells, however heat conditioned cells were not osmotolerant.Citation51 More recently Hoffman et al demonstrated that a single point mutation in the Listeria monocytogenes betL gene leads to improved osmotolerance and chill tolerance in this foodborne pathogen.Citation52,53 Indeed, this link between osmotolerance and other environmental stress resistance mechanisms highlights the multiple benefits of an improved understanding of C. sakazakii osmotolerance in addition to providing an insight into the mechanisms which facilitate its survival in PIF.
While few studies have focused on C. sakazakii osmotolerance, the ability of this organism to survive in such an extreme hyperosmotic environment is a unique adaptation which is of fundamental importance to its pathogenic potential.Citation54 Most microorganisms require an aw significantly higher than that of PIF for growth and survival, suggesting that C. sakazakii is armed with a vast array of osmotic stress survival mechanisms.Citation54 However, the growth phase of the organism has also been demonstrated to play a role in C. sakazakii osmotolerance, with stationary phase cells demonstrating a higher resistance to osmotic and dry stress in comparison to other known osmotolerant organisms such as E. coli and Salmonella.Citation44 Further elucidation of C. sakazakii osmotolerance is aided by the wealth of knowledge available on how other bacterial cells overcome hyperosmotic stress and it is believed that C. sakazakii responds in a similar way.Citation55-57
Bacterial cells in general need to maintain an intracellular osmotic pressure greater than that of the surrounding media in order to generate cell turgor and prevent plasmolysis and ultimately cell death.Citation58 Most bacteria survive osmotic stress through a biphasic response, which first involves the accumulation of potassium and its counter ion glutamate; representing the primary response.Citation55 However, given that high levels of potassium are detrimental to the normal functioning of the cell, a secondary response is triggered which stimulates the synthesis and/or uptake of osmoprotective compounds called compatible solutes,Citation55 so called because they are compatible with cellular physiology at high internal concentrations.Citation59-61 Compatible solutes are generally soluble molecules with no net charge at physiological pH and do not interact with proteins and other macromolecules, thus they do not disrupt vital cellular processes such as DNA repair, DNA-protein interactions or the cellular metabolic machinery.Citation55
There are a large variety of compatible solutes available in the environment which can be acquired by microorganisms under high osmotic stress. Most microorganisms harbour multiple osmoregulatory transporters with overlapping substrate specificity which allow them to overcome changes in the osmolality of the surrounding media.Citation62 One of the most extensively studied Gram-positive foodborne pathogens in this respect is L. monocytogenes.Citation57 This pathogen uses the compatible solutes glycine betaine,Citation63 prolineCitation64 and carnitineCitation65 to counteract the cytotoxic effects of elevated osmolality. Indeed, Gram-negative bacteria share similar preferences and mechanisms of compatible solute accumulation as those seen in Gram-positives. The compatible solute trehalose was previously demonstrated to play a role in the desiccation survival of C. sakazakii, illustrated by a more than five-fold increase in the trehalose concentration in dried stationary phase cells.Citation66 The mechanism of trehalose mediated osmoprotection is believed to play a significant role in the survival of microorganisms during dry stress as a result of the substitution of the water layer around the biomolecules by trehalose. This enables maintenance of the 3 dimensional structure of essential biological macromolecules; a mechanism referred to as the water replacement theory ().Citation67 However, experimental data have demonstrated that this substitution has an effect on the mobility of the macromolecule and further research is needed to determine the mechanism of stabilization by trehalose.Citation68 The existence of a trehalose biosynthesis pathway in C. sakazakii is demonstrated by the presence of a putative otsBA operonCitation54; suggesting that the mechanism of trehalose biosynthesis in C. sakazakii is similar to that of E. coli. Furthermore, in addition to trehalose, other potentially important C. sakazakii compatible solutes include proline, betaine and ectoine, accumulated via secondary transporters homologous to the ProP porter in E. coli. Indeed, while E. coli possesses only a single ProP homolog, multiple ProP homologues exist in the C. sakazakii genome; a feature which goes some way to explaining the increased osmotolerance of the pathogen in PIF compared to E. coli.Citation69
Figure 1. Structure stabilization of enzymes at elevated osmolarity. Preferential exclusion of compatible solutes (blue circles) from the protein surface helps to maintain enzyme structure at elevated osmolarity, while also helping to increase cell volume (adapted from Sleator and Hill Citation55).
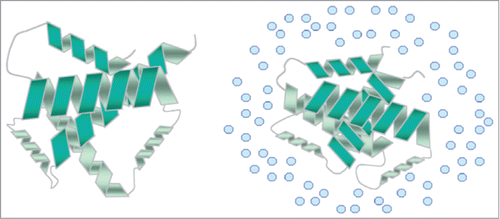
While relatively few studies have focused on the osmotolerance capacity of C. sakazakii at the physiological level, our knowledge of C. sakazakii osmotolerance has been significantly expanded by a recent in silico analysis of the C. sakazakii BAA-894 genome; revealing 53 putative osmotolerance loci, including both hypo- and hyper-osmotic stress response systems.Citation54 While homologues of all the principal E. coli osmotolerance loci were identified in the BAA-894 genome; multiple copies of certain osmotolerance loci; including 7 copies of the E. coli proP homolog were found. The ProP protein encoded by the gene locus tag ESA_02131 encodes a protein with 90% identity to E. coli ProP.Citation54 While the remaining 6 homologues encode proteins exhibiting features of classic secondary transporters, they were all 60–70 amino acids shorter than the E. coli ProP; lacking the extended carboxyl tail.Citation54 This structural inconsistency is unique and likely contributes to the increased osmotolerance of this pathogen compared to E. coli.
Gene expression analysis of each proP homolog demonstrated an increase in expression levels in C. sakazakii during an osmotic upshift, indicating that each homolog responds to osmotic stress conditions, at least at the level of transcription.Citation69 However, expression levels varied significantly between each of the proP homologues analyzed, with the ProP encoded by gene locus tag ESA_04214, a sequence homologous to the inner membrane transporter YhjE in E.coli, demonstrating the highest level of up-regulation when subject to hyperosmotic stress. Furthermore, functional analysis revealed that each of the 6 ProP homologues analyzed conferred increased osmotolerance when heterologously expressed against an osmotically sensitive E. coli host (E. coli MKH13), albeit to different degrees; suggesting that each of the 7 proteins might be tailored to specific osmotic conditions.
C. sakazakii virulence
While the unique osmotolerance phenotype associated of C. sakazakii likely aids the pathogenicity of the bacterium by providing access to a highly susceptible host via PIF, other factors influence the virulence of C. sakazakii. To cause systemic infection and some of the more severe clinical manifestations of Cronobacter infections, like NEC, sepsis or meningitis, C. sakazakii must either directly infect cells of the inner gut lining or cross this barrier to reach the bloodstream. Indeed, C. sakazakii has been shown to infect mucous membranes, gastric and intestinal epithelial, and endothelial tissues.Citation22,70,71 As such, the genes encoding C. sakazakii's ability to adhere to and invade cells of the inner gut lining represent major virulence factors.
Crossing the gut barrier–adhesion
The first step in the invasion of the gastrointestinal tract is adherence of bacteria to the surface of these tissues. Studying the invasion of internal epithelial barriers in situ is not trivial and several cell lines e.g., Caco-2 isolated from gut tissue have been established as laboratory models.Citation72-75 The first study to investigate the adherence of C. sakazakii to Caco-2 human epithelial cell lines involved 50 C. sakazakii strains from different environmental niches. Twenty-eight of these isolates were found to adhere to the surface of Caco-2 cells. Furthermore, this study outlined 3 basic adhesion patterns for C. sakazakii: diffuse adhesion, formation of localized clusters and a mixed phenotype.Citation71 While the ability of C. sakazakii to adhere to the tested cell models was independent of bacterial fimbrae formation,Citation71 Hartmann et al. investigated the adherence efficiency of selected mutants of C. sakazakii strain ES5 to Caco-2 cells and found that the absence of flagella reduced the adherence ability of the bacterium,Citation43 demonstrating that flagella are important for this process. Others investigated adherence of clinical isolates from an outbreak in France.Citation13 This was the first in vitro study that used clinical samples with associated patient information and therefore could link genotypes to symptoms and severity of the infection. The clinical manifestations associated with these genotypes were necrotising enterocolitis (NEC), bacteriaemia, sepsis and meningitis and the study found a link between the virulence of the strains and their ability to adhere to and invade Caco-2 cells.Citation13
To better understand the interactions on the cell surface that lead to invasion of C. sakazakii, significant efforts have been made to identify the host receptors for this pathogen-host interaction. Fibronectin, a glycoprotein which forms part of the extracellular matrix of eukaryotic tissue,Citation76 plays a role in host cell adhesion, movement, growth and differentiation, and is one of the primary targets in the adhesion process of several pathogenic organisms.Citation77 Studies identified binding of fibronectin as an important step in the adherence of C. sakazakii to the cells of the gastrointestinal tract.Citation71 Reduction of fibronectin levels has been shown to impair C. sakazakii attachment to INT-407 cells - an in vitro model of embryonic intestinal cells.Citation76 It is likely that this is due to interaction between host receptors and adhesins on the surface of the bacterial cells. The C. sakazakii outer membrane protein A (OmpA) was identified as a major fibronectin-binding protein which plays a significant role in the adherence of this gastrointestinal pathogen to neonatal and immunocompromised hosts.Citation76,78 However, due to the high diversity of the genus, not all Cronobacter spp. possess this ability.
Crossing the gut barrier–invasion
In many pathogenic bacteria, adherence to host tissues is followed by invasion of the tissue, allowing the pathogen to breach this first line of defense. While gastrointestinal invasion is common with other pathogenic bacteria such as Salmonella typhimurium (relative invasion rate of 60% in 60 minutes), only moderate invasion rates have been observed in C. sakazakii (0.2% in 60 min). This is similar to the moderately invasive gastrointestinal pathogen Campylobacter jejuni (0.1%–0.4%).Citation79
The mechanism of invasion in C. sakazakii is not yet fully understood, however, several host factors and bacterial membrane proteins seem to be involved in this process.
On the host side, the intercellular tight-junctions of the gut epithelia play a role in preventing molecules and bacteria from bypassing the epithelial cells of healthy individuals. It is therefore not surprising that invasion by C. sakazakii strain ATCC 29544 is greatly improved by the disruption of tight junctions,Citation70 which can be caused by lipopolysaccharides (LPS) from Gram-negative bacteria.Citation80 Furthermore, epithelium cell tight-junctions and gut microbial flora of neonates are not yet fully developed and therefore would not be as effective in preventing invasion of pathogens such as C. sakazakii from entering the blood stream.Citation81 Taken together with high levels of LPS found in PIF, and the contamination of PIF with C. sakazakii, this could provide an underlying mechanism for the observed tenfold higher incidence rate of NEC observed in infants fed with PIF compared to breast fed infants.Citation82
Outer membrane protein A (OmpA) of C. sakazakii has been shown to be necessary for invasion of Caco-2 and INT407 cells,Citation83-85 since the invasiveness of a ΔompA mutant decreased by 80% in Caco-2 and 87% in INT407 cells.Citation85 However, Kim and colleagues further demonstrated that another outer membrane protein, OmpX, also plays a major role in invasion of Caco-2 epithelial cellsCitation85 and has been shown to be necessary for the invasion of Hep-2 cells by Yersinia pestis and C. sakazakii.Citation86,87 As a consequence, it was observed that ompA/X deletion mutants were significantly less efficient in the dissemination of the bacterium to other organs such as the liver and spleen in vivo.Citation85
C. sakazakii is a member of a unique subset of microorganisms that can penetrate the blood-brain barrier. This characteristic is facilitated by the organism's ability to invade human brain microvascular endothelial cells (HBMEC), an essential feature in meningitis causing organisms. Indeed, Townsend et al. demonstrated that invasion of HBMEC by C. sakazakii strains was equivalent or greater than that of the neonatal E. coli meningitis strain K1.Citation13 A more recent study observed significant levels of transcytosis across a tight monolayer of HBMEC and included electron-microscopic analysis demonstrating the presence of Cronobacter within host vacuoles of HBMEC, in addition to being trancytosed at the basolateral surface.Citation88 While significant strain variation was noted, this study further contributes to our understanding of how this gastrointestinal pathogen penetrates the blood brain barrier (BBB) and ultimately causes meningitis. The previously mentioned OmpA protein, which plays a role in attachment of C. sakazakii to the gastrointestinal tract, has also been implicated in the invasion of HBMEC. Nair et al. demonstrated that a lack of this protein resulted in a reduction of HBMEC invasion by 83%.Citation78 While OmpA and OmpX likely play a role in the organism, penetrating the blood-brain barrier, the mechanism which leads to the death of the brain cells is as yet unknown and it is possible that the host's immune response may also play a role.Citation6 Furthermore, C. sakazakii possess zpx, a gene which encodes a zinc containing metalloprotease which is responsible for the lysis of collagen and may allow the pathogen to cross the blood brain barrier.Citation89 It is important to note however that not all C. sakazakii strains have been shown to cause meningitis. An extensive multilocus sequence typing (MLST) method identified C. sakazakii sequence type 4 as the predominant sequence type found in cerebro-spinal fluid from cases of neonatal meningitis.Citation6
Virulence factors beyond invasion – toxins, immune-evasion and immune-toxicity
The first putative virulence factors identified in Cronobacter were enterotoxins produced by 4 of the 18 isolates studied by Pagotto et al.Citation90 Purification and characterization identified a 66kDa protein which was most stable at pH 6. The ability of the toxin to withstand high temperatures (90°C for 30mins), coupled with its potent cell toxicity (LD50=56pg), represented a significant virulence factor for this foodbourne pathogen.Citation91 However, to date there remains a dearth of information available on enterotoxin production by C. sakazakii and its role in pathogenesis as other factors have been studied in more detail.
The ability of some C. sakazakii strains to evade the innate immune response by surviving and even replicating in macrophages,Citation17 represents another important virulence factor. While the survival of this organism in macrophages varies significantly between strains, the presence of putative sod genes, encoding a superoxide dismutase, is believed to aid survival in phagosomes, where the bacteria are subjected to acidic conditions and macrophage oxidase activity.Citation17 Furthermore, C. sakazakii has been found to persist in macrophages for up to 48 hours, suggesting that this organism is well equipped to survive within this extreme environmentCitation13; a stress survival strategy possibly aided by compatible solute accumulation.Citation55
It has been demonstrated more recently that the flagella of C. sakazakii ST1 and ST4 strains stimulate the activation of pro- and anti-inflammatory cytokines in human monocytes, such as TNF, IL8 and IL10, and this response is dependent on TLR5 recognition.Citation92 The role of these cytokines in the pathogenicity of C. sakazakii is not clear and as such further research is needed.
While the C. sakazakii genome encodes several virulence factors, the host also contributes to its pathogenicity through immune-related cell toxicity. Characterization of OmpA showed that disruption of the epithelium triggers release of various pro and anti-inflammatory chemokines, cytokines such as transforming growth factor β (TGF-β) and Nitric Oxide (NO). TGF-β is essential for proper immune functions, such as growth, repair and development, and is produced in high amounts by dendritic cells leading to tight junction disruption and cell death in the presence of the pathogen. Nitric Oxide has been associated with NEC as a result of intestinal barrier failure, caused by high localized concentrations of NO that trigger cell apoptosis or necrosis.Citation83
Conclusion and Future Prospects
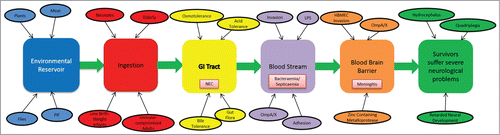
The severity of disease coupled with a high mortality rate (up to 80%) in C. sakazakii infected infants and neonates, highlights the need for further research into the virulence and environmental stress survival mechanisms of this gastrointestinal pathogen. Since the reclassification of Enterobacter sakazakii in 2007 there has been a surge in research aimed at elucidating the virulence factors of C. sakazakii, however our understanding of the mechanisms of its pathogenesis is far from complete. In particular the identification of surface proteins on C. sakazakii which facilitate adherence and invasion of the neonatal host tissue has made a significant contribution to the field. OmpA, a fibronectin binding protein, has been identified as playing a major role in the invasion of host cells of the gastrointestinal tract and is also implicated in facilitating the invasion. However, further elucidation of the structure and mechanism of OmpA and other proteins involved in attachment to host cells would significantly contribute to our understanding of the virulence of this organism and may indeed aid the development of adequate control measures. gives an overview of the discussed mechanisms, virulence factors and clinical manifestations of C. sakazakii infections.
Figure 2. Schematic representation of the different stages and organs/tissues involved in a C. sakazakii infection. The different stages are represented by colored boxes, with important factors and consequences associated with these stages indicated. Main clinical manifestations (e.g., NEC) linked to C. sakazakii infection in different organs/tissues are shown.
The progress made in C. sakazakii virulence and osmotic stress survival research was largely aided by genome sequence analysis. To date 16 C. sakazakii draft genomes are available, however only 4 of these are gap-less chromosomal sequences. It is certainly important to note that increasing the number of available whole genome sequences would significantly benefit C. sakazakii research in general, and in particular would facilitate the comparative analysis of closely related strains. Previous taxonomic revision of Cronobacter was based on DNA hybridization, 16s rDNA sequence analysis and biotyping. However, with the increasing number of available C. sakazakii genome sequences, MLST is the most favorable typing method to date due to its higher resolution and promising clinical significance.Citation93 MLST data has demonstrated the C. sakazakii sequence type (ST) 4 is the sequence type implicated most often in cases of neonatal meningitis.Citation94 Indeed, 75% of ST4 isolates correlated with cases of meningitis over a 50 y period in a study spanning 6 countries.Citation12,94 Improved availability of genomic sequence information would therefore be beneficial to the future of C. sakazakii research and will undoubtedly help to direct future studies toward the most pathogenic sequence types.
While sequence analysis studies are improving our knowledge of C. sakazakii virulence and osmotolerance, “wet lab” information available on C. sakazakii osmotolerance is still very limited. The osmotic stress survival mechanisms of C. sakazakii represent a fundamental survival strategy; enabeling the pathogen to survive in environments of low aw such as PIF, thus an improved knowledge of these mechanisms would have a significant impact on the manufacture of PIF and other dried food products. Furthermore, identification of the compatible solute uptake and synthesis systems utilized by pathogenic C. sakazakii strains may identify traits unique to pathogenic isolates, e.g., ST4, so that targeted inhibition analysis can be carried out. An improved understanding of the structures of the compatible solute transporters could provide valuable insights for the development of control measures using either small molecule research (known as smuggling technology), or protein-protein interactions which mimic transporter-compatible solute interactions but do not facilitate the uptake of compatible solutes and therefore do not have an osmoprotective effect. Such control measures may significantly reduce or prevent survival of C. sakazakii in low aw environments and would therefore significantly improve the safety of dry foods such as PIF.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
RDS is Coordinator of the EU FP7 ClouDx-i project which funds KK. AF is funded by an IRCSET EMBARK Postgraduate Scholarship RS/2010/2300. RO’C was funded by the Biological Sciences Department at CIT.
References
- Farmer JJ, III, Asbury MA, Hickman FW, Brenner DJ. The Enterobacteriaceae Study Group. Enterobacter sakazakii: a new species of "Enterobacteriaceae" isolated from clinical specimens. Int J Syst Bacteriol 1980; 30:569-84; http://dx.doi.org/10.1099/00207713-30-3-569
- Iversen C, Lehner A, Mullane N, Bidlas E, Cleenwerck I, Marugg J, Fanning S, Stephan R, Joosten H. The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov. Cronobacter sakazakii subsp. sakazakii, comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies 1. BMC Evol Biol 2007; 7:64; PMID:17439656; http://dx.doi.org/10.1186/1471-2148-7-64
- Iversen C, Waddington M, On SLW, Forsythe S. Identification and Phylogeny of Enterobacter sakazakii relative to Enterobacter and Citrobacter Species. J Clin Microbiol 2004; 42:5368-70; PMID:15528745; http://dx.doi.org/10.1128/JCM.42.11.5368-5370.2004
- Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol 2013; 36:309-19; PMID:23632228; http://dx.doi.org/10.1016/j.syapm.2013.03.005
- Joseph S, Desai P, Ji Y, Cummings CA, Shih R, Degoricija L, Rico A, Brzoska P, Hamby SE, Masood N, et al. Comparative analysis of genome sequences covering the seven Cronobacter species. PLoS One 2012; 7:e49455; PMID:23166675; http://dx.doi.org/10.1371/journal.pone.0049455
- Joseph S, Forsythe S. Insights into the emergent bacterial pathogen Cronobacter spp, generated by multilocus sequence typing and analysis. Front Microbiol 2012; 3:3; PMID:22279445; http://dx.doi.org/10.3389/fmicb.2012.00003
- Fakruddin M, Rahaman MM, Ahmed MM, Hoque MM. Cronobacter sakazakii (Enterobacter sakazakii): an emerging foodborne pathogen. IJBAR 2013; 4:349-59; http://dx.doi.org/10.7439%2Fijbar.v4i6.35610. 7439/ijbar.v4i6.356
- Stephan R, Grim CJ, Gopinath GR, Mammel MK, Sathyamoorthy V, Trach LH, Chase HR, Fanning S, Tall BD. Re-examination of the taxonomic status of Enterobacter helveticus., Enterobacter pulveris, and Enterobacter turicensis as members of Cronobacter and description of Siccibacter turicensis com. nov., Franconibacter helveticus comb. nov., and Franconibacter pulveris com. nov. Int J Syst Evol Microbiol 2014; 64(Pt 10):3402-10; PMID:25028159; http://dx.doi.org/10.1099/ijs.0.059832-0
- Barron JC, Forsythe SJ. Dry stress and survival time of Enterobacter sakazakii and other Enterobacteriaceae in dehydrated powdered infant formula. J Food Prot 2007; 70:2111-7; PMID:17900090
- Stoll BJ, Hansen N, Fanaroff AA, Lemons JA, for the National Institute of Child H, Human Development Neonatal Research N. Enterobacter sakazakii is a rare cause of neonatal septicemia or meningitis in vlbw infants. J. Pediat 2004; 144:821-3; PMID:15192634
- Lai KK. Enterobacter sakazakii infections among neonates, infants, children, and adults: case reports and a review of the literature. Medicine 2001; 80:113-22; PMID:11307587; http://dx.doi.org/10.1097/00005792-200103000-00004
- Joseph S, Forsythe SJ. Predominance of Cronobacter sakazakii sequence type 4 in neonatal infections. Emerg Infect Dis 2011; 17:1713-5; PMID:21888801; http://dx.doi.org/10.3201/eid1709.110260
- Townsend S, Hurrell E, Forsythe S. Virulence studies of Enterobacter sakazakii isolates associated with a neonatal intensive care unit outbreak. BMC Microbiol 2008; 8:64; PMID:18423002; http://dx.doi.org/10.1186/1471-2180-8-64
- Bowen AB BC. Invasive Enterobacter sakazakii disease in infants. Emerg Infect Dis 2006; 12(8):1185-9; PMID:16965695
- van Acker J, de Smet F, Muyldermans G, Bougatef A, Naessens A, Lauwers S. Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J Clin Microbiol 2001; 39:293-7; PMID:11136786; http://dx.doi.org/10.1128/JCM.39.1.293-297.2001
- Lin PW, Stoll BJ. Necrotising enterocolitis. Lancet 2006; 368:1271-83; PMID:17027734; http://dx.doi.org/10.1016/S0140-6736(06)69525-1
- Townsend SM, Hurrell E, Gonzalez-Gomez I, Lowe J, Frye JG, Forsythe S, Badger JL. Enterobacter sakazakii invades brain capillary endothelial cells, persists in human macrophages influencing cytokine secretion and induces severe brain pathology in the neonatal rat. Microbiol 2007; 153:3538-47; PMID:17906151; http://dx.doi.org/10.1099/mic.0.2007/009316-0
- Burdette JH SC. Enterobacter sakazakii brain abscess in the neonate: the importance of neuroradiologic imaging. Pediatr Radiol 2000; 30(1):30-2; PMID:10663505
- International Commission on Microbiological Specifications for Foods (ICMSF). Microorganisms in foods 7. Microbiological testing in food safety management. Kluwer academic/Plenum publishers 2002. ISBN 978-0-306-47262-6
- Joseph S, Forsythe SJ. Predominance of Cronobacter sakazakii Sequence Type 4 in Neonatal Infections. Emerging Infectious Diseases 2011;17(9):1713-1715. doi:10.3201/eid1709.110260.
- Caubilla-Barron J, Hurrell E, Townsend S, Cheetham P, Loc-Carrillo C, Fayet O, Prère M-F, Forsythe SJ. Genotypic and phenotypic analysis of enterobacter sakazakii strains from an outbreak resulting in fatalities in a neonatal intensive care unit in France. J Clin Microbiol 2007; 45:3979-85; PMID:17928419; http://dx.doi.org/10.1128/JCM.01075-07
- Liu H, Cui JH, Cui ZG, Hu GC, Yang YL, Li J, Shi YW. Cronobacter carriage in neonate and adult intestinal tracts. Biomed Environ Sci: BES 2013; 26:861-4; PMID:24215882; http://dx.doi.org/10.3967/bes2013.011
- Stock I, Wiedemann B. Natural antibiotic susceptibility of Enterobacter amnigenus, Enterobacter cancerogenus, Enterobacter gergoviae and Enterobacter sakazakii strains. Clin Microbiol Infec 2002; 8:564-78; PMID:12427217; http://dx.doi.org/10.1046/j.1469-0691.2002.00413.x
- Miranda CD, Kehrenberg C, Ulep C, Schwarz S, Roberts MC. Diversity of tetracycline resistance genes in bacteria from chilean salmon farms. Antimicrob Agents Ch 2003; 47:883-8; PMID:12604516; http://dx.doi.org/10.1128/AAC.47.3.883-888.2003
- El-Sharoud W, O'Brien S, Negredo C, Iversen C, Fanning S, Healy B. Characterization of Cronobacter recovered from dried milk and related products. BMC Microbiol 2009; 9:24; PMID:19187534; http://dx.doi.org/10.1186/1471-2180-9-24
- Chon J-W, Song K-Y, Kim S-Y, Hyeon J-Y, Seo K-H. Isolation and characterization of Cronobacter from desiccated foods in Korea. J Food Sci 2012; 77:M354-M8; PMID:22671692; http://dx.doi.org/10.1111/j.1750-3841.2012.02750.x
- Müller A, Hächler H, Stephan R, Lehner A. Presence of AmpC beta-lactamases, CSA-1, CSA-2, CMA-1, and CMA-2 conferring an unusual resistance phenotype in cronobacter sakazakii and Cronobacter malonaticus. Microb Drug Resist 2014; 20(4):275-80; PMID:24568164; http://dx.doi.org/10.1089/mdr.2013.0188
- Kilonzo-Nthenge A, Rotich E, Godwin S, Nahashon S, Chen F. Prevalence and antimicrobial resistance of Cronobacter sakazakii isolated from domestic kitchens in middle tennessee, United States. J Food Protect 2012; 75:1512-7; PMID:22856579; http://dx.doi.org/10.4315/0362-028X.JFP-11-442
- Yan Q, Power KA, Cooney S, Fox E, Gopinath GR, Grim CJ, Tall BD, Mccusker MP, Fanning S. Complete genome sequence and phenotype microarray analysis of Cronobacter sakazakii SP291: a persistent isolate cultured from a powdered infant formula production facility. Front Microbiol 2013; 4:4; PMID:23386843; http://dx.doi.org/10.3389/fmicb.2013.00004
- Van de Velde S, Carryn S, Van Bambeke F, Hill C, Tulkens PM, Sleator RD. Penicillin-binding proteins (PBP) and Lmo0441 (a PBP-like protein) play a role in Beta-lactam sensitivity of Listeria monocytogenes. Gut Pathog 2009; 1:23; PMID:20003484; http://dx.doi.org/10.1186/1757-4749-1-23
- Lu TK, Koeris MS. The next generation of bacteriophage therapy. Curr Opin Microbiol 2011; 14:524-31; PMID:21868281; http://dx.doi.org/10.1016/j.mib.2011.07.028
- O'Mahony J, Fenton M, Henry M, Sleator RD, Coffey A. Lysins to kill - a tale of viral weapons of mass destruction. Bioeng Bugs 2011; 2:306-8; PMID:22008941; http://dx.doi.org/10.4161/bbug.2.6.16804
- Abbasifar R, Griffiths MW, Sabour PM, Ackermann H-W, Vandersteegen K, Lavigne R, Noben J-P, Alanis Villa A, Abbasifar A, Nash JHE, et al. Supersize me: Cronobacter sakazakii phage GAP32. Virology 2014; 460–461:138-46; PMID:25010279; http://dx.doi.org/10.1016/j.virol.2014.05.003
- Zuber S, Boissin-Delaporte C, Michot L, Iversen C, Diep B, Brüssow H, Breeuwer P. Decreasing Enterobacter sakazakii (Cronobacter spp.) food contamination level with bacteriophages: prospects and problems. Microb Biotechnol 2008; 1:532-43; PMID:21261874; http://dx.doi.org/10.1111/j.1751-7915.2008.00058.x
- Kim K-P, Klumpp J, Loessner MJ. Enterobacter sakazakii bacteriophages can prevent bacterial growth in reconstituted infant formula. Int J Food Microbiol 2007; 115:195-203; PMID:17196280; http://dx.doi.org/10.1016/j.ijfoodmicro.2006.10.029
- Abbasifar R, Kropinski A, Sabour P, Chambers J, MacKinnon J, Malig T, Griffiths M. Efficiency of bacteriophage therapy against Cronobacter sakazakii in Galleria mellonella (greater wax moth) larvae. Arch Virol 2014; 159:2253-61; PMID:24705602; http://dx.doi.org/10.1007/s00705-014-2055-x
- Tothova L, Celec P, Babickova J, Gajdosova J, Al-Alami H, Kamodyova N, Drahovska H, Liptakova A, Turna J, Hodosy J. Phage therapy of Cronobacter-induced urinary tract infection in mice. Med Sci Monit 2011; 17:BR173-BR8; PMID:21709627; http://dx.doi.org/10.12659/MSM.881844
- An T-W, Kim S-J, Lee Y-D, Park J-H, Chang H-I. The immune-enhancing effect of the Cronobacter sakazakii ES2 phage results in the activation of nuclear factor-κB and dendritic cell maturation via the activation of IL-12p40 in the mouse bone marrow. Immunol Lett 2014; 157:1-8; PMID:24184907; http://dx.doi.org/10.1016/j.imlet.2013.10.007
- Mramba F, Broce A, Zurek L. Isolation of Enterobacter sakazakii from stable flies, Stomoxys calcitrans L.(Diptera: Muscidae). J Food Prot 2006; 69:671-3; PMID:16541702
- Osaili T, Forsythe S. Desiccation resistance and persistence of Cronobacter species in infant formula. Int J Food Microbiol 2009; 136:214-20; PMID:19720413; http://dx.doi.org/10.1016/j.ijfoodmicro.2009.08.006
- Friedemann M. Enterobacter sakazakii in food and beverages (other than infant formula and milk powder). Int J Food Microbiol 2007; 116:1-10; PMID:17331606; http://dx.doi.org/10.1016/j.ijfoodmicro.2006.12.018
- Jo S-H, Baek S-B, Ha J-H, Ha S-D. Maturation and survival of Cronobacter biofilms on silicone, polycarbonate, and stainless steel after UV light and ethanol immersion treatments. J Food Prot 2010; 73:952-6; PMID:20501047
- Hartmann I, Carranza P, Lehner A, Stephan R, Eberl L, Riedel K. Genes involved in cronobacter sakazakii biofilm formation. Appl Environ Microbiol 2010; 76:2251-61; PMID:20118366; http://dx.doi.org/10.1128/AEM.00930-09
- Breeuwer P, Lardeau A, Peterz M, Joosten HM. Desiccation and heat tolerance of Enterobacter sakazakii. J Appl Microbiol 2003; 95:967-73; PMID:14633024; http://dx.doi.org/10.1046/j.1365-2672.2003.02067.x
- Kandhai MC, Reij MW, Gorris LG, Guillaume-Gentil O, van Schothorst M. Occurrence of Enterobacter sakazakii in food production environments and households. Lancet 2004; 363:39-40; PMID:14723994; http://dx.doi.org/10.1016/S0140-6736(03)15169-0
- Breeuwer P, Lardeau A, Peterz M, Joosten H. Desiccation and heat tolerance of Enterobacter sakazakii. J Appl Microbiol 2003; 95:967-73; PMID:14633024; http://dx.doi.org/10.1046/j.1365-2672.2003.02067.x
- Friedemann M. Epidemiology of invasive neonatal Cronobacter (Enterobacter sakazakii) infections. Eur J Clin Microbiol Infect Dis 2009; 28:1297-304; PMID:19662446; http://dx.doi.org/10.1007/s10096-009-0779-4
- Sleator RD, Clifford T, Hill C. Gut osmolarity: a key environmental cue initiating the gastrointestinal phase of Listeria monocytogenes infection? Med Hypotheses 2007; 69:1090-2; PMID:17433559; http://dx.doi.org/10.1016/j.mehy.2007.02.028
- Dreux N, Albagnac C, Sleator RD, Hill C, Carlin F, Morris CE, Nguyen-the C. Glycine betaine improves Listeria monocytogenes tolerance to desiccation on parsley leaves independent of the osmolyte transporters BetL, Gbu and OpuC. J Appl Microbiol 2008; 104:1221-7; PMID:17976173; http://dx.doi.org/10.1111/j.1365-2672.2007.03623.x
- Sharma SC. A possible role of trehalose in osmotolerance and ethanol tolerance in Saccharomyces cerevisiae. FEMS Microbiol Lett 1997; 152:11-5; PMID:9228764; http://dx.doi.org/10.1111/j.1574-6968.1997.tb10402.x
- Trollmo C, Andre L, Blomberg A, Adler L. Physiological overlap between osmotolerance and thermotolerance in Saccharomyces cerevisiae. FEMS Microbiol Lett 1988; 56:321-5; http://dx.doi.org/10.1111/j.1574-6968.1988.tb03200.x
- Hoffmann RF, McLernon S, Feeney A, Hill C, Sleator RD. A single point mutation in the listerial betL σA-dependent promoter leads to improved osmo-and chill-tolerance and a morphological shift at elevated osmolarity. Bioengineered 2013; 4:401; PMID:23478432; http://dx.doi.org/10.4161/bioe.24094
- Sleator RD, Wood JM, Hill C. Transcriptional regulation and posttranslational activity of the betaine transporter BetL in Listeria monocytogenes are controlled by environmental salinity. J Bacteriol 2003; 185:7140-4; PMID:14645273; http://dx.doi.org/10.1128/JB.185.24.7140-7144.2003
- Feeney A, Sleator RD. An in silico analysis of osmotolerance in the emerging gastrointestinal pathogen Cronobacter sakazakii. Bioeng Bugs 2011; 2:260-70; PMID:21918371; http://dx.doi.org/10.4161/bbug.2.5.17238
- Sleator RD, Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev 2002; 26:49-71; PMID:12007642; http://dx.doi.org/10.1111/j.1574-6976.2002.tb00598.x
- Wemekamp-Kamphuis HH, Sleator RD, Wouters JA, Hill C, Abee T. Molecular and physiological analysis of the role of osmolyte transporters BetL, Gbu, and OpuC in growth of Listeria monocytogenes at low temperatures. Appl Environ Microbiol 2004; 70:2912-8; PMID:15128551; http://dx.doi.org/10.1128/AEM.70.5.2912-2918.2004
- Sleator RD, Gahan CG, Hill C. A postgenomic appraisal of osmotolerance in Listeria monocytogenes. Appl Environ Microbiol 2003; 69:1-9; PMID:12513970; http://dx.doi.org/10.1128/AEM.69.1.1-9.2003
- Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev 1989; 53:121-47; PMID:2651863
- Brown AD. Microbial water stress. Bacteriol Rev 1976; 40:803-46; PMID:1008746
- Sleator RD, Hill C. Compatible solutes: the key to Listeria's success as a versatile gastrointestinal pathogen? Gut Pathog 2010; 2:20; PMID:21143981; http://dx.doi.org/10.1186/1757-4749-2-20
- Sleator RD, Hill C. Compatible solutes: a listerial passe-partout? Gut Microbes 2010; 1:77-9; PMID:21326913; http://dx.doi.org/10.4161/gmic.1.2.10968
- Wood JM, Bremer E, Csonka LN, Kraemer R, Poolman B, van der Heide T, Smith LT. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp Biochem Physiol A Mol Integr Physiol 2001; 130:437-60; PMID:11913457; http://dx.doi.org/10.1016/S1095-6433(01)00442-1
- Sleator RD, Gahan CGM, Abee T, Hill C. Identification and disruption of BetL, a secondary glycine betaine transport system linked to the salt tolerance of listeria monocytogenes LO28. Appl Environ Microbiol 1999; 65:2078-83; PMID:10224004
- Sleator RD, Gahan CGM, Hill C. Identification and disruption of theproBA locus in listeria monocytogenes: role of proline biosynthesis in salt tolerance and murine infection. Appl Environ Microbiol 2001; 67:2571-7; PMID:11375165; http://dx.doi.org/10.1128/AEM.67.6.2571-2577.2001
- Sleator RD, Wouters J, Gahan CG, Abee T, Hill C. Analysis of the role of OpuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes. Appl Environ Microbiol 2001; 67:2692-8; PMID:11375182; http://dx.doi.org/10.1128/AEM.67.6.2692-2698.2001
- Breeuwer P, Lardeau A, Peterz M, Joosten HM. Desiccation and heat tolerance of Enterobacter sakazakii. J Appl Microbiol 2003; 95:967-73; PMID:14633024; http://dx.doi.org/10.1046/j.1365-2672.2003.02067.x
- Jain NK, Roy I. Effect of trehalose on protein structure. Protein Sci 2009; 18:24-36; PMID:19177348; http://dx.doi.org/10.1002/pro.3
- Roy I, Sharma A, Gupta MN. Obtaining higher transesterification rates with subtilisin Carlsberg in nonaqueous media. Bioorg Med Chem Lett 2004; 14:887-9; PMID:15012987; http://dx.doi.org/10.1016/j.bmcl.2003.12.021
- Feeney A, Johnston CD, Govender R, O’Mahony J, Coffey A, Sleator RD. Analysis of the role of the Cronobacter sakazakii ProP homologues in osmotolerance. Gut Pathog 2014; 6:15; PMID:24910715; http://dx.doi.org/10.1186/1757-4749-6-15
- Kim K-P, Loessner MJ. Enterobacter sakazakii invasion in human intestinal Caco-2 cells requires the host cell cytoskeleton and is enhanced by disruption of tight junction. Infect Immun 2008; 76:562-70; PMID:18070906; http://dx.doi.org/10.1128/IAI.00937-07
- Mange J-P, Stephan R, Borel N, Wild P, Kim KS, Pospischil A, Lehner A. Adhesive properties of Enterobacter sakazakii to human epithelial and brain microvascular endothelial cells. BMC Microbiol 2006; 6:58; PMID:16800879; http://dx.doi.org/10.1186/1471-2180-6-58
- Jumarie C, Malo C. Caco-2 cells cultured in serum-free medium as a model for the study of enterocytic differentiation in vitro. J Cell Physiol 1991; 149:24-33; PMID:1939345; http://dx.doi.org/10.1002/jcp.1041490105
- Gahring LC, Heffron F, Finlay B, Falkow S. Invasion and replication of Salmonella typhimurium in animal cells. Infect Immun 1990; 58:443-8; PMID:2404872
- Harmsen HJM, Wildeboer–Veloo ACM, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr 2000; 30:61-7; PMID:10630441; http://dx.doi.org/10.1097/00005176-200001000-00019
- Sambuy Y, De Angelis I, Ranaldi G, Scarino M, Stammati A, Zucco F. The Caco-2 cell line as a model of the intestinal barrier: influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol Toxicol 2005; 21:1-26; PMID:15868485; http://dx.doi.org/10.1007/s10565-005-0085-6
- Nair MKM, Venkitanarayanan K. Role of bacterial OmpA and host cytoskeleton in the invasion of human intestinal epithelial cells by Enterobacter sakazakii. Pediatric Res 2007; 62:664-9; PMID:17957161; http://dx.doi.org/10.1203/PDR.0b013e3181587864
- Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci 2002; 115:3861-3; PMID:12244123; http://dx.doi.org/10.1242/jcs.00059
- Mohan Nair MK, Venkitanarayanan K, Silbart LK, Kim KS. Outer membrane protein A (OmpA) of Cronobacter sakazakii binds fibronectin and contributes to invasion of human brain microvascular endothelial cells. Foodborne Pathog Dis 2009; 6:495-501; PMID:19415974; http://dx.doi.org/10.1089/fpd.2008.0228
- Hu L, Kopecko DJ. Campylobacter jejuni 81-176 associates with microtubules and dynein during invasion of human intestinal cells. Infect Immun 1999; 67:4171-82; PMID:10417189
- Moriez R, Salvador-Cartier C, Theodorou V, Fioramonti J, Eutamene H, Bueno L. Myosin light chain kinase is involved in lipopolysaccharide-induced disruption of colonic epithelial barrier and bacterial translocation in rats. Am J Pathol 2005; 167:1071-9; PMID:16192642; http://dx.doi.org/10.1016/S0002-9440(10)61196-0
- Anderson R, Dalziel J, Gopal P, Bassett S, Ellis A, Roy N. The Role of Intestinal Barrier Function in Early Life in the Development of Colitis, Colitis, Dr Fukata (Ed.), ISBN: 978-953-307-799-4, InTech, DOI: 10.5772/25753.
- Iversen C, Forsythe S. Risk profile of Enterobacter sakazakii, an emergent pathogen associated with infant milk formula. Trends Food Sci Technol 2003; 14:443-54; http://dx.doi.org/10.1016/S0924-2244(03)00155-9
- Emami CN, Mittal R, Wang L, Ford HR, Prasadarao NV. Recruitment of dendritic cells is responsible for intestinal epithelial damage in the pathogenesis of necrotizing enterocolitis by Cronobacter sakazakii. J Immunol 2011; 186:7067-79; PMID:21551359; http://dx.doi.org/10.4049/jimmunol.1100108
- Verhoeff-Bakkenes L, Hazeleger W, Zwietering M, De Jonge R. Lack of response of INT-407 cells to the presence of non-culturable Campylobacter jejuni. Epidemiol Infect 2008; 136:1401-6; PMID:18081950; http://dx.doi.org/10.1017/S0950268807000040
- Kim K, Kim K-P, Choi J, Lim J-A, Lee J, Hwang S, Ryu S. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl Environ Microbiol 2010; 76:5188-98; PMID:20543055; http://dx.doi.org/10.1128/AEM.02498-09
- Kolodziejek AM, Sinclair DJ, Seo KS, Schnider DR, Deobald CF, Rohde HN, Viall AK, Minnich SS, Hovde CJ, Minnich SA. Phenotypic characterization of OmpX, an Ail homologue of Yersinia pestis KIM. Microbiology 2007; 153:2941-51; PMID:17768237; http://dx.doi.org/10.1099/mic.0.2006/005694-0
- De Kort G, Bolton A, Martin G, Stephen J, Van De Klundert J. Invasion of rabbit ileal tissue by Enterobacter cloacae varies with the concentration of OmpX in the outer membrane. Infect Immun 1994; 62:4722-6; PMID:7927747
- Giri CP, Shima K, Tall BD, Curtis S, Sathyamoorthy V, Hanisch B, Kim KS, Kopecko DJ. Cronobacter spp.(previously Enterobacter sakazakii) invade and translocate across both cultured human intestinal epithelial cells and human brain microvascular endothelial cells. Microb Pathog 2012; 52:140-7; PMID:22023990; http://dx.doi.org/10.1016/j.micpath.2011.10.003
- Kothary MH, McCardell BA, Frazar CD, Deer D, Tall BD. Characterization of the zinc-containing metalloprotease encoded by zpx and development of a species-specific detection method for Enterobacter sakazakii. Appl Environ Microb 2007; 73:4142-51; PMID:17483271; http://dx.doi.org/10.1128/AEM.02729-06
- Pagotto FJ, Nazarowec-White M, Bidawid S, Farber JM. Enterobacter sakazakii: infectivity and enterotoxin production in vitro and in vivo. J Food Prot 2003; 66:370-5; PMID:12636287
- Raghav M, Aggarwal PK. Purification and characterization of Enterobacter sakazakii enterotoxin. Can J Microbiol 2007; 53:750-5; PMID:17668035; http://dx.doi.org/10.1139/W07-037
- Cruz-Córdova A, Rocha-Ramírez LM, Ochoa SA, Gónzalez-Pedrajo B, Espinosa N, Eslava C, Hernández-Chiñas U, Mendoza-Hernández G, Rodríguez-Leviz A, Valencia-Mayoral P. Flagella from five Cronobacter species induce pro-inflammatory cytokines in macrophage derivatives from human monocytes. PloS One 2012; 7:e52091; PMID:23284883; http://dx.doi.org/10.1371/journal.pone.0052091
- Baldwin A, Loughlin M, Caubilla-Barron J, Kucerova E, Manning G, Dowson C, Forsythe S. Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance which do not correlate with biotypes. BMC Microbiol 2009; 9:223; PMID:19852808; http://dx.doi.org/10.1186/1471-2180-9-223
- Joseph S, Sonbol H, Hariri S, Desai P, McClelland M, Forsythe SJ. Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J Clin Microbiol 2012; 50:3031-9; PMID:22785185; http://dx.doi.org/10.1128/JCM.00905-12
