Abstract
Cells of infected organisms transport disease defense-related molecules along actin filaments to deliver them to their sites of action to combat the pathogen. To accommodate higher demand for intracellular traffic, plant F-actin density increases transiently during infection or treatment of Arabidopsis with pathogen-associated molecules. Many animal and plant pathogens interfere with actin polymerization and depolymerization to avoid immune responses. Pseudomonas syringae, a plant extracellular pathogen, injects HopW1 effector into host cells to disrupt the actin cytoskeleton and reduce vesicle movement in order to elude defense responses. In some Arabidopsis accessions, however, HopW1 is recognized and causes resistance via an actin-independent mechanism. HopW1 targets isoform 7 of vegetative actin (ACT7) that is regulated by phytohormones and environmental factors. We hypothesize that dynamic changes of ACT7 filaments are involved in plant immunity.
Actin is Targeted by Pathogen Effectors to Disrupt Intracellular Traffic and Immunity
Many pathogens target actin to remodel the host cytoskeleton. Most often actin targeting is observed in intracellular pathogens to enable their entry to the cell. Animal/human pathogens such as Salmonella or plant fungal and oomycete pathogens induce actin filament rearrangement leading to microbial uptake or penetration of a host cell by pathogen hyphaeCitation1-4. Similarly, enteropathogenic E. coli, although extracellular, rearranges the host cytoskeleton to build pedestals to which it attaches and multiplies.Citation5 However, the cytoskeleton is also a network used to deliver antimicrobial agents to sites of infection e.g. secrete antimicrobial compounds, reinforce plant cell wall by callose deposition or deliver pathogen-associated molecular pattern (PAMP) receptors and other elements of the immune response machinery to the plasma membrane.Citation6 Some effectors from pathogenic bacteria interfere with these defense processes: for example VipA of Legionella or YopE of Yersinia target actin in animal cells to disrupt intracellular trafficking to avoid the immune response and phagocytosis.Citation7-11 Inhibition of endosomal trafficking and phagocytosis is achieved by different ways of interference with actin dynamics: VipACitation7 is an actin nucleator, whereas YopECitation9,10 has Rho GAP activity that disrupts actin filaments.
HopW1 of Pseudomonas Syringae Disrupts Actin in Plants
HopW1 of P. syringae PmaES2643 is the first bacterial effector shown to target actin in plants.Citation12 P. syringae is an extracellular pathogen that injects effectors into plant cells via a type III secretion system in order to suppress immune responses and create an environment supporting bacterial growth. HopW1 binds and solubilizes F-actin and disrupts actin-dependent trafficking, such as endocytosis and vacuole targeting in infected plants. This activity of HopW1 leads to increased virulence of P. syringae in the Col accession of Arabidopsis. We showed that the C-terminal domain of HopW1 disrupts cytosolic actin filaments in vitro and the full-length protein expressed in plants cells disorganizes the cytoskeleton to such a degree that actin filaments visualized by Lifeact-GFP (a protein that binds actin fused to GFP to allow fluorescence imaging in living cells) cannot be detected.Citation12
HopW1 has no similarity to any proteins of known functions but its N-terminal domain is also found in the N-terminal region of HopAE from P. syringae pv. syrinagae and its C-terminal domain is present in a predicted effector (EspW) from pathogenic E. coli.Citation13 It is possible that EspW is another E. coli effector that targets actin and may have filament-disrupting activity similar to HopW1.
Dynamic Changes of the Actin Cytoskeleton are Necessary for Plant Defense
Cytoskeleton changes have been implicated in immune responses in plants. Actin depolymerization factor 4 (ADF4) is necessary for transcriptional activation of the resistance gene RPS5 and MAP kinase signaling in response to the P. syringae effector AvrPphB.Citation14,15 During infection with virulent P. syringae DC3000 or treatment with PAMPs, transient increase of F-actin density was observed, probably to satisfy the requirement for increased intracellular traffic.Citation16,17 Responses to one of PAMPs, a fragment of bacterial elongation factor (efl26), were linked to inhibition of ADF4.Citation17 A recent publication reported that pathogen infection also increases the numbers of multivesicular bodies and exocyst vesicles related to relocation of defense compounds and that endosomal trafficking is essential for basal defense against P. syrinage.Citation18
Actin depolymerization by 10 μM latrunculin B (LatB) induces plant susceptibility to bacterial infectionCitation12,16, however treatment with 200 nM LatB was shown to induce salicylic acid (SA)-dependent gene transcription (pathogen growth in plants treated with low concentrations of LatB was not testedCitation19). SA is a major defense regulator in plants and induces transcription of genes correlated with resistance. High concentrations of SA (1 mM) caused F-actin disruption.Citation19
Effectors and Toxins that Target Actin may also Induce Immune Responses
Consistent with induced susceptibility, HopW1 that disrupts F-actin in plant cells is a virulence factor during infection of susceptible Arabidopsis (Col accession). However in the Ws accession of Arabidopsis, HopW1 is recognized and induces defense responses via WIN2 and WIN3 proteins that interact with HopW1. Defense activation in Ws and WIN protein binding were attributed to the C-terminal domain of HopW1,Citation20 the same domain that binds and disrupts F-actin.Citation12
Several effectors or toxins of animal pathogens that target actin also induce immune responses (reviewed inCitation21) and actin depolymerization was linked to NFκB activation.Citation22
Recognition of HopW1 in a Resistant Arabidopsis Accession is not Related to Actin Disruption
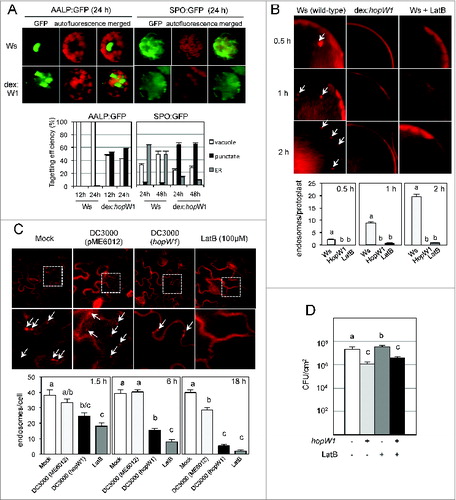
The defense-inducing activity (avirulence) of many plant effectors results from detection of perturbation of their targets, therefore it was possible that disruption of F-actin could be monitored (guarded) in Ws and induce resistance. We tested if HopW1 interfered with actin-dependent intracellular trafficking in Ws, similarly to what we found in the susceptible Col accession. Effects of expression of HopW1 in Ws protoplasts () were the same as in Col or as treatment with the actin-depolymerizing agent LatBCitation12: HopW1 reduced proper targeting of marker proteins to the ER and vacuole () and inhibited endocytosis (). During infection with PtoDC3000 expressing HopW1, endocytosis was also reduced in Ws in comparison to infection with the Pto strain carrying empty vector () or mock-treated control. The effect of HopW1 on actin-dependent processes in Ws was similar to the effect of LatB treatment. However F-actin depolymerization by LatB slightly increased the growth of virulent strain PtoDC3000 in Ws (), similarly as in Col, while HopW1 expression reduced Pto growth in Ws. Therefore recognition and the avirulent effect of HopW1 is not related to F-actin disruption, and actin depolymerization seems to increase susceptibility to P. syringae in different Arabidopsis accessions.
Figure 1 (See previous page). Effects of HopW1 on actin dependent processes and bacterial growth in Arabidospsis Ws. (A) HopW1 inhibits vacuolar and ER trafficking. Wild-type and dex:hopW1 protoplasts were transfected with AALP:GFP or SPO:GFP, incubated in 0.2 μM of dexamethasone for indicated times and imaged by confocal microscopy. AALP:GFP was targeted to the central vacuole and SPO:GFP localized to the ER and vacuole in wild-type protoplasts (upper row), as previously documented for Col.Citation12 In the presence of HopW1, many protoplasts transfected with the AALP:GFP and SPO:GFP showed notable punctate patterns. Localization patterns were quantified from at least 100 images, see Citation12 for comparison with Col protoplasts and the effect of LatB. Bars indicate SEM. χ2 tests indicated that the distributions were significantly different between the wild-type and dex:hopW1 at each time point (P < 0.0001, n ≥ 100 per genotype/fusion construct). (B) HopW1 inhibits endocytosis in protoplastas. Representative microscopic images show effects of HopW1 and LatB on endocytic vesicle formation. Wild-type and dex:hopW1 protoplasts were treated with 0.2 μM dexamethasone, stained with FM4–64 and visualized by confocal microscopy. Ten μM LatB was used to disrupt the actin cytoskeleton. After over-night incubation with dexamethasone and/or LatB, protoplasts were labeled with FM4–64 and viewed after 0.5, 1 and 2 h. Arrows point to some of the FM4–64-stained endosomes. Endosomes were quantified in at least 20 protoplasts per treatment, per time-point in 3 independent experiments. Bars indicate SEM, and letters indicate significantly different number of endosomes (P < 0.0001, ANOVA/Neuman-Keuls test). (C) HopW1 inhibits endocytosis during infection. Cotyledons of Arabidopsis Ws seedlings grown on MS plates were infected with PtoDC3000 carrying either empty vector (pME6012) or vector with the HopW1 gene at OD600 = 0.01. 100 μM LatB was used as an actin cytoskeleton-disrupting control. After infections and treatments for the indicated times, cotyledons were labeled for 1 h with FM4–64 and viewed. Examples of microscopic images of tissue 6h after infection are shown. Lower row is magnification of fragments from pictures in upper row. Arrows indicate some of the FM4–64-labeled endosomes. Enodosomes per cell were manually counted in at least 8 images per treatment, per timepoint, from 2 biological repeats. Bars indicate SEM and letters indicate significantly different numbers of endosomes for given treatments (P < 0.05, ANOVA/Neuman-Keuls test). (D) Disruption of actin cytoskeleton does not induce resistance in Arabidopsis Ws. Growth of PtoDC3000/vector or PtoDC3000/HopW1 (OD600 = 0.0001) 3 d post-inoculation of Ws accessions was monitored in the presence or absence of 10 μM LatB. Different letters indicate significantly different growth (ANOVA/Neuman-Keuls test, P < 0.05). LatB did not trigger resistance in Ws, whereas HopW1 did, suggesting that the mechanism of resistance does not involve disruption of the actin cytoskeleton. Average of all results with SEM from 3 independent experiments (n=9) is shown. All experiments were repeated 3 times with similar results. Detailed methods are described in ref.Citation12
Plants have Multiple Actin Isotypes
In our previous study,Citation12 we found that the C-terminal domain of HopW1 disrupts animal cytoplasmic (non-muscle) F-actin, but has no activity toward muscle actin. Plants have multiple actin isoforms, e.g., Arabidopsis has at least 8 functional actin genes (plus 2 pseudogenes) divided into 2 major functional groups: vegetative (expressed in all vegetative tissues: ACT 2, 8 and 7) and reproductive (expressed in reproductive organs and meristems: ACT 1, 3, 4, 12 and 11) (reviewed in Citation23). Animal cytoplasmic and muscle actins or plant vegetative and reproductive actins are not interchangeable i.e. animal cytoplasmic actin mutant is not rescued by muscle actin and plant vegetative actin mutant is not fully rescued by reproductive actin. Plant actins are not more phylogenetically related to animal cytoplasmic than muscle actins however, vegetative actins seem to be functionally related to animal cytosolic actins and ancestral actins of unicellular organisms. Vegetative plant actin (act7/8 mutant in Arabidopsis) can be complemented by cytoplasmic (non-muscle) human/animal actins and protist actins, but not by muscle actins.Citation24
HopW1 Targets Vegetative Actin 7 that Responds to Hormones and Environment
Most of the actin peptides found in a complex with HopW1 in N. benthamianaCitation12 match multiple actin isotypes, however 2 peptides are specific for a vegetative actin 7 (ACT7) and ACT7 is the only isotype that all other peptides also match (). Both HopW1 activity on animal cytosolic F-actin and proteomics analysis of HopW1 complexes in planta suggest that HopW1 has specificity toward vegetative actin(s) in host cells. Although the identity of actin isotypes involved in plant response to pathogens was not determined, these are very likely vegetative actins involved in vesicle trafficking and transport in the cells of infected tissue. ACT7 targeted by HopW1 seems to be the only plant actin found responsive to external stimuli and phytohormones. ACT7 expression is induced by auxin, light and wounding and suppressed by abscisic acid.Citation25,26 It would be intriguing to know if ACT7 dynamics are specifically affected by infection and if this actin isotype is responsive to salicylic acid or other hormones involved in defenses.
Table 1. Actin peptides identified in HopW1 complex immunoprecipitated from N. benthamiana by LC-MS/MS and search of whole Genbank protein sequences. Identity to Arabidopsis actin isoforms is indicated (actin isotypes are annotated in Arabidopsis and some other species but not in N. benthamiana). *This peptide has 100% identity to Glycyrrhiza and Mimosa sp. actin.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by National Science Foundation Grant 2010: IOS0822393, and in part by Korea Research Foundation Grant KRF-2009-F00006.
References
- Carabeo R. Bacterial subversion of host actin dynamics at the plasma membrane. Cell microbiol 2011; 13:1460-9; PMID:21790944; http://dx.doi.org/10.1111/j.1462-5822.2011.01651.x
- Dunn JD, Valdivia RH. Uncivil engineers: Chlamydia, Salmonella and Shigella alter cytoskeleton architecture to invade epithelial cells. Future Microbiol 2010; 5:1210-32; PMID:20722600; http://dx.doi.org/10.2217/fmb.10.77
- Shimada C, Lipka V, O'Connell R, Okuno T, Schulze-Lefert P, Takano Y. Nonhost resistance in Arabidopsis-Colletotrichum interactions acts at the cell periphery and requires actin filament function. Mol Plant-Microbe Interact 2006; 19:270-9; PMID:16570657; http://dx.doi.org/10.1094/MPMI-19-0270
- Miklis M, Consonni C, Bhat RA, Lipka V, Schulze-Lefert P, Panstruga R. Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol 2007; 144:1132-43; PMID:17449647; http://dx.doi.org/10.1104/pp.107.098897
- Campellone KG. Cytoskeleton-modulating effectors of enteropathogenic and enterohaemorrhagic Escherichia coli: Tir, EspFU and actin pedestal assembly. The FEBS J 2010; 277:2390-402; PMID:20477869; http://dx.doi.org/10.1111/j.1742-4658.2010.07653.x
- Day B, Henty JL, Porter KJ, Staiger CJ. The pathogen-actin connection: a platform for defense signaling in plants. Annu Rev Phytopathol 2011; 49:483-506; PMID:21495845; http://dx.doi.org/10.1146/annurev-phyto-072910-095426
- Franco IS, Shohdy N, Shuman HA. The Legionella pneumophila effector VipA is an actin nucleator that alters host cell organelle trafficking. PLoS Pathog 2012; 8:e1002546; PMID:22383880; http://dx.doi.org/10.1371/journal.ppat.1002546
- Franco IS, Shuman HA. A pathogen's journey in the host cell: Bridges between actin and traffic. BioArchitecture 2012; 2:38-42; PMID:22754628; http://dx.doi.org/10.4161/bioa.20422
- Rosqvist R, Forsberg A, Wolf-Watz H. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect immun 1991; 59:4562-9; PMID:1937815
- Black DS, Bliska JB. The RhoGAP activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence. Mol microbiol 2000; 37:515-27; PMID:10931345; http://dx.doi.org/10.1046/j.1365-2958.2000.02021.x
- Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature 2007; 449:827-34; PMID:17943119; http://dx.doi.org/10.1038/nature06247
- Kang YS, Jelenska J, Cecchini NM, Li YJ, Lee MW, Kovar DR, Greenberg JT. HopW1 from Pseudomonas syringae Disrupts the Actin Cytoskeleton to Promote Virulence in Arabidopsis. PLoS Pathog 2014; 10:e1004232; PMID:24968323; http://dx.doi.org/10.1371/journal.ppat.1004232
- Greenberg JT, Vinatzer BA. Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr Opin Microbiol 2003; 6:20-8; PMID:12615215; http://dx.doi.org/10.1016/S1369-5274(02)00004-8
- Tian M, Chaudhry F, Ruzicka DR, Meagher RB, Staiger CJ, Day B. Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiol 2009; 150:815-24; PMID:19346440; http://dx.doi.org/10.1104/pp.109.137604
- Porter K, Shimono M, Tian M, Day B. Arabidopsis Actin-Depolymerizing Factor-4 links pathogen perception, defense activation and transcription to cytoskeletal dynamics. PLoS Pathog 2012; 8:e1003006; PMID:23144618; http://dx.doi.org/10.1371/journal.ppat.1003006
- Henty-Ridilla JL, Shimono M, Li J, Chang JH, Day B, Staiger CJ. The plant actin cytoskeleton responds to signals from microbe-associated molecular patterns. PLoS Pathog 2013; 9:e1003290; PMID:23593000; http://dx.doi.org/10.1371/journal.ppat.1003290
- Henty-Ridilla JL, Li J, Day B, Staiger CJ. ACTIN DEPOLYMERIZING FACTOR4 regulates actin dynamics during innate immune signaling in Arabidopsis. Plant Cell 2014; 26:340-52; PMID:24464292; http://dx.doi.org/10.1105/tpc.113.122499
- Wang F, Shang Y, Fan B, Yu JQ, Chen Z. Arabidopsis LIP5, a positive regulator of multivesicular body biogenesis, is a critical target of pathogen-responsive MAPK cascade in plant basal defense. PLoS Pathog 2014; 10:e1004243; PMID:25010425; http://dx.doi.org/10.1371/journal.ppat.1004243
- Matouskova J, Janda M, Fiser R, Sasek V, Kocourkova D, Burketova L, Duskova J, Martinec J, Valentova O. Changes in actin dynamics are involved in salicylic acid signaling pathway. Plant Sci 2014; 223:36-44; PMID:24767113; http://dx.doi.org/10.1016/j.plantsci.2014.03.002
- Lee MW, Jelenska J, Greenberg JT. Arabidopsis proteins important for modulating defense responses to Pseudomonas syringae that secrete HopW1-1. Plant J 2008; 54:452-65; PMID:18266921; http://dx.doi.org/10.1111/j.1365-313X.2008.03439.x
- Rajamuthiah R, Mylonakis E. Effector triggered immunity: Activation of innate immunity in metazoans by bacterial effectors. Virulence 2014; 5:1–6; PMID:24802615; http://dx.doi.org/10.4161/viru.29091
- Nemeth ZH, Deitch EA, Davidson MT, Szabo C, Vizi ES, Hasko G. Disruption of the actin cytoskeleton results in nuclear factor-kappaB activation and inflammatory mediator production in cultured human intestinal epithelial cells. J Cell Physiol 2004; 200:71-81; PMID:15137059; http://dx.doi.org/10.1002/jcp.10477
- Slajcherova K, Fiserova J, Fischer L, Schwarzerova K. Multiple actin isotypes in plants: diverse genes for diverse roles? Front plant sci 2012; 3:226; PMID:23091476; http://dx.doi.org/10.3389/fpls.2012.00226
- Kandasamy MK, McKinney EC, Roy E, Meagher RB. Plant vegetative and animal cytoplasmic actins share functional competence for spatial development with protists. Plant Cell 2012; 24:2041-57; PMID:22589468; http://dx.doi.org/10.1105/tpc.111.095281
- Kandasamy MK, Gilliland LU, McKinney EC, Meagher RB. One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 2001; 13:1541-54; PMID:11449050; http://dx.doi.org/10.1105/tpc.13.7.1541
- McDowell JM, An YQ, Huang S, McKinney EC, Meagher RB. The arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol 1996; 111:699-711; PMID:8754679; http://dx.doi.org/10.1104/pp.111.3.699
